Abstract
Numerically addressing scientific questions such as simulating drug diffusion through the human stratum corneum is a challenging task requiring complex codes and plenty of computational resources. The UG4 framework is used for such simulations, and though empirical tests have shown good scalability so far, its sheer size precludes analytical modeling of the entire code. We have developed a process which combines the power of our automated performance modeling method and the workflow manager JUBE to create insightful models for entire UG4 simulations. Examining three typical use cases, we identified and resolved a previously unknown latent scalability bottleneck. In collaboration with the code developers, we validated the performance expectations in each of the use cases, creating over 10,000 models in less than a minute, a feat previously impossible without our automation techniques.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
A broad variety of research questions in natural sciences is formulated in terms of partial differential equations. The range of applications reaches from classical continuum field descriptions - such as fluid dynamics, electromagnetism, or structure mechanics - over biological settings - e.g., drug diffusion through the human skin or computational neuroscience - to non-physical settings such as computational finance. Numerical simulations can be used to predict or compare with measured physical behavior and help to gain insight into the underlying physical processes. A software framework focusing on the grid-based solutions of these problems is UG4 [28].
Such simulation codes demand increased computational resources to perform larger and more refined simulations. Therefore, they must scale to the largest computing clusters to benefit from available computing power. However, code developers face two challenges: First, the source code is large, making manual analysis and optimization of the code time consuming and error prone. This creates a strong need for an automated workflow supporting scaling analysis. Second, code developers have to consume lots of computing resources for testing and can only run tests up to their currently available process counts. This requires a workflow that allows performance modeling using data from smaller process counts and hence offers the possibility to resolve performance bottlenecks at an early stage of code development. As a byproduct, the models for the resource consumption provide users with an estimate for the requirements of production runs.
We expanded the automated performance modeling approach by Calotoiu et al. [7] to meet the mentioned requirements. This approach creates performance models from a small number of test measurements with a small numbers of processes. The models are used to detect potential performance bottlenecks and to predict the resource consumption at larger core counts. We have combined this approach with the workflow manager JUBE [30] to facilitate the submission and collection of numerous test simulations that serve as inputs for the performance modeling approach. In this paper we focus on the applicability of our approach in realistic code development scenarios and show how scalability issues are detected.
We demonstrate the power of our automated performance modeling process by applying it to the software framework UG4. Given its approximately half a million lines of C++ code, manually modeling the performance of UG4 is practically impossible, which is why it provides a good example for the benefits of our approach. The major contributions of our work are:
-
An automated modeling approach in combination with an automated workflow manager for a fast and streamlined detection of scalability issues.
-
Demonstration of the tool chain by applying it to the large simulation framework UG4 focusing on human skin permeation simulations.
-
Discussion of two performance issues detected by our approach.
-
Validation of the UG4 scaling behavior.
The remainder of this paper is organized as follows. In Sect. 2, the UG4 simulation environment is presented, Sect. 3 outlines the modeling approach and Sect. 4 gives an overview on the benchmark environment JUBE. Then, in Sect. 5 we present three test cases where the tools are used in order to analyze the UG4 simulation code. Sections 6 and 7 are dedicated to related work and concluding remarks.
2 The UG4 Simulation Framework
The UG4 simulation framework (unstructured grids 4.0) [28] addresses the numerical solution of partial differential equations and is implemented as a C++ library. It uses grid-based discretization methods such as the finite element method or the vertex-centered finite volume method [6]. Complex physical geometries are resolved by hybrid, unstructured, adaptive, hierarchical grids in up to three space dimensions. In addition, a strong focus of the software framework is on efficient and highly scalable solvers, using algebraic and geometric multigrid methods. The framework is parallelized using MPI. To simplify the usage, a separate library called PCL (parallel communication layer) has been developed, which encapsulates the MPI calls and which provides lightweight structures for graph-based parallelization. A key feature of PCL is that parallel copies of objects are not identified by global IDs. Instead, containers, called interfaces, are used to store the parallel copies on each process in a well-defined order such that identification can be done by these interfaces in an efficient way [20, 21, 28].
A typical simulation run consists of several phases, each with its own character, especially with respect to parallelization. At first, a computing grid is required. In this specific work, we proceed as follows: A coarse grid describing the domain is loaded onto one process. The grid is refined, creating new levels of the multigrid hierarchy and after some refinements the finest grid level is distributed to empty processes, proceeding with the refinement in parallel. This process can be iterated, successively creating a tree structure of processes holding parts of the hierarchical grid. The grid refinement is mainly performed process-wise and communication is only needed at redistribution stages [20]. An illustration of the resulting hierarchy for a 1d distribution is shown in Fig. 1.
On the grid, the partial differential equations are discretized by assembling large sparse matrices and corresponding vectors based on the grid element contributions. Using only lower-dimensional parallel overlap (i.e., each full-dimensional element is present on exactly one process, but the lower-dimensional boundary has parallel copies on several processes), the assembly process can be performed by traversing the full-dimensional elements only and therefore it is an inherent parallel process. Given optimal load balancing, i.e., an equal distribution of the elements across the processes, perfect scalability is expected for the assembly.
The most difficult part, from a parallelization perspective, is the subsequent solution of the matrix equation. Since the algebraic structures are distributed, solvers naturally involve parallel communication. Multigrid methods are of optimal complexity (linear in the degrees of freedom) and thus a good candidate for weak scaling. They compute corrections iteratively to approximate the solution. On every level, simple iterative schemes, called smoothers, are applied and the problem is transferred to coarser grids in order to compute coarse corrections [6, 13]. Our multigrid solver is based on the above-mentioned hierarchically distributed multigrid. Especially on coarser grid levels, where less computational work has to be done, fewer processes are involved in the solution algorithm. In addition, Krylov methods such as CG and BiCGStab are implemented [14]. Their parallelization is mainly based on the parallelization of the matrix-vector and vector-vector products that appear in their formulation.
3 Automated Performance Modeling
We developed an automatic performance-model generator [7, 8] for the purpose of simplifying the identification of scalability bottlenecks in complex codes. Our targets are scalability bugs defined as parts of a code that scale worse than expected. To this end, we create performance models for each part of the code at the level of function calls to better identify potential problems. Our focus is to create simple, easy to read, insightful models quickly, as opposed to detailed, precise models. In our studies, not only execution time is considered as a performance metric, but also requirements such as the number of bytes injected into the network or the number of floating-point operations are taken into account. This helps developers not only to uncover the existence of potential scalability bottlenecks, but also to explain their causes. For brevity, we will only present a short overview of the method.
When conducting a scalability study, our tool takes measurements of metrics (e.g., time, flops, bytes sent, ...) at different processor counts \(\{p_1, \ldots , p_{max}\}\) for each individual program region (e.g., function call) as input. This is accomplished by instrumenting the application and generating parallel profiles at runtime, which are then analyzed post-mortem. Models describing the growth are generated for each region, called kernel, and can be analyzed either in an interactive GUI, which displays a call tree of the application annotated with performance-model information, or in text form as a ranked list, ordered by either predicted execution time at a larger scale \(p_t > p_{max}\), or asymptotic by behavior.
3.1 Model Generation
Our model generator rests on the observation that the models describing the behavior of parallel programs as a function of the number of processes are usually finite combinations of terms composed of polynomials and logarithms. For practical purposes, models with two or three terms are often sufficient. The performance model normal form (PMNF) below describes our representation, which covers the practical cases encountered so far by virtue of the way that computer algorithms are designed.
Moreover, the sets \(I, J \subset \mathbb {Q}\) from which the exponents \(i_k\) and \(j_k\) are chosen from can be quite small and still allow a large number of different behaviors to be modeled. After creating the sets I and J and choosing n, all possible model assignments, called model hypotheses, can be tried and the best candidate is then selected via cross-validation [19].
3.2 Recursive Multigrid Extension
One of the core assumptions of our method is that a code will generate the same call tree for each of the different processor counts \(\{p_1, \ldots , p_{max}\}\). This allows us to traverse the call tree and compare each individual function call and its behavior. However, within a weak scaling study, the number of grid levels increases with the process count. Since the multigrid algorithm is based on recursive calls for each grid level, the involved code kernels are visited recursively more often. This leads to a different call tree for different processor counts, which required us to develop a special method to be able to analyze multigrid applications. To handle this issue, we developed an extension to our method that compares the different performance measurements and creates a call tree containing only such kernels which are present in all measurements. The information of kernels which have to be removed is not lost, but rather added to the parent kernel of the one pruned from the call tree.
4 Automated Benchmarking Environment
The automated modeling of numerical software codes demands numerous experiments with varying execution parameters – such as process counts, used solvers, or physical parameters – and multiple repetitions, in order to ensure statistical significance. Configuring, compiling, running, verifying its correctness, and collecting results means a lot of administrative work and produces a large amount of data to be processed. Without a benchmarking environment, all these steps have to be performed manually. To facilitate all these tasks, Forschungszentrum Jülich provided and improved JUBE (Juelich Benchmarking Environment) [30], a script-based framework created to easily perform benchmark runs for different sets of parameters, execution sizes, compilation options, computer systems, and to evaluate the results thereafter.
JUBE workflow ([30])
Figure 2 shows the steps that are performed by JUBE in sequence: preparation, compilation, and execution, where each step might exist multiple times. Each of these steps can be adjusted to a given code or application by modifying XML-based setup scripts. The created runs can be verified and parsed by automatic pre- and post-processing scripts that filter out the desired information and store it in a more compact form for manual interpretation. With JUBE, it is easy to create combinatorial runs of multiple parameters. For example, in a scaling experiment, one can simply specify multiple numbers of processes, different solver setups, and physical parameters. JUBE will create one experiment for each possible combination, submit all of them to the resource manager, collect all results, and display them together.
5 Results
Using the tools from Sects. 3 and 4, we analyze the UG4 code in three substudies: In the first two tests, we focus on modeling drug diffusion through the human skin. First, we analyze the code behavior under weak scaling, then we vary the diffusivity of the skin cells over several ranges of magnitude. In the third study, we compare two different types of solvers, again under weak scaling: the geometric multigrid solver and the unpreconditioned conjugate gradient (CG) method.
Drug Diffusion though the Human Skin. The outermost part of the epidermis (stratum corneum) consists of flattened, dead cells (corneocytes), that are surrounded by an inter-cellular lipid. The stratum corneum is the natural barrier to protect underlying tissue, but still allows for the throughput of certain concentrations (e.g., drugs, medicine). The latter process can be modeled by a diffusion process, in which the diffusion coefficient within the corneocytes differs from the one in the lipid. Different geometric representations of the stratum corneum have been used to compute the diffusional throughput [17].
In the following two studies, we use a brick-and-mortar model (Fig. 3). Assuming diffusion driven transport in the two subdomains \(s \in \{cor, lip\}\) (corneocyte, lipid), the governing equation is given by
The diffusion coefficient \(\mathbf {D}_s\) is assumed to be constant within each subdomain \(s \in \{cor, lip\}\), but may differ between subdomains. For the scalability analysis, we compute the steady state of the concentration distribution.
As solver, we employ a geometric multigrid method, accelerated by an outer conjugate gradient method. The multigrid uses a damped Jacobi smoother, two (resp. three) smoothing steps in 2d (resp. 3d), a V-cycle, and an LU base solver. The iterations are completed once an absolute residuum reduction of \(10^{-10}\) is achieved. The main difficulty of this problem is the bad aspect ratio of the computational domain (\(0.1\,\mu m\) vs. \(30\,\mu m\) for the lipid channels). This is resolved by three (resp. five) steps of anisotropic refinement to enhance those ratios. Base solvers are applied at a level where ratios are satisfactory.
Weak-scaling Analysis of the 3d Skin Model. Using the 3d skin model described above, we fix the diffusion parameter to \(\mathbf {D}_{cor} = 10^{-3}\). Table 1 shows models for a scalability issue we detected. In these kernels, we create MPI communicator groups for each level of the multigrid hierarchy, excluding processes from the group that do not own a grid part on the level. In order to inform every process on these memberships, we employ an MPI_Allreduce for an array of length p, resulting in a \(p \cdot \mathcal {O}\)(MPI_Allreduce) dependency, that will lead to scalability issues for large process counts. In these kernels, we substituted MPI_Comm_split for MPI_Allreduce, also eliminating the linearly growing input. First tests do not show a significant improvement in runtime, however now the dependency is \(\mathcal {O}\)(MPI_Comm_split), whose scaling properties have been analyzed for exascale purposes [22]. Enhanced algorithms for MPI_Comm_split are known to scale with \(\mathcal {O}(\log ^2 p)\) [24].
Left: Measured wallclock times (marks) and models (lines) for the assembly, the multigrid solver initialization, and the solution of the skin 3d problem. Right, top: Number of grid refinements (L), degrees of freedom (DoF) and number of iterations of the solver (\(n_{\text {gmg}}\)). Right, bottom: Performance models for the kernels
Besides the above-mentioned issue, no further scalability bugs were detected, i.e., no kernel scales worse than logarithmically (see Table 1 for examples). The accumulated wallclock times for coarse-grain kernels (Fig. 4) show good scaling behavior, and bounded iteration counts are observed. Our empirical approach even reveals a rather small but apparent \(\mathcal {O}(\log _2^2 p)\) dependent kernel during solver initialization where the matrix diagonal is communicated.
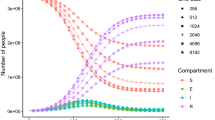
Varying the Diffusion Parameter. Our second substudy highlights the demand for a workflow manager. Biological case studies can require a variation of input parameters over 10 orders of magnitude. Combining this with 5–10 different process counts in scaling studies, several solver setups and repetitions for jitter reduction, easily hundreds of measurement runs have to be performed. We use the JUBE manager for this task. This allows us to easily schedule, collect, and analyze these runs. As an illustration, we present a study resembling results by Nägel et al. [17]: Fixing the lipid diffusion coefficient to \(\mathbf {D}_{lip} = 1\), we vary the diffusion in the corneocytes in the range of \(\mathbf {D}_{cor} = 10^{2}, 10^{1}, \dots , 10^{-7}, 10^{-8}\). Figure 5 illustrates the solution at an early time step and the stationary case. The biologically interesting fluxes at the bottom of the domain, \(\mathbf {F}_{\text {bot}} := \int _{\partial \varOmega _{\text {bot}}} \nabla c\; dS\), and the iteration count for the multigrid solver are collected using JUBE (Table 2). The relatively constant iteration count over the whole range of physical parameters shows the robustness of the solver. The performance validation of the solver could have never been so thorough without the use of our automated process, allowing us to handle, analyze, and refine hundreds of experimental runs and to provide insights to developers as quickly as possible.
5.1 Analysis of Algebraic Solvers
This section demonstrates the usability of the presented approach to validate performance expectations. We analyze two solvers with known weak scaling properties: the nicely scaling multigrid method and the unpreconditioned conjugate gradient method with known weak-scaling issues. Our tests will confirm the theoretical expectations.
Left: Measured times (marks) and models (lines) for the assembling and solver execution for the conjugate gradient (CG) and multigrid (GMG) methods. Right, top: Number of grid refinements (L), degrees of freedom (DoF) and number of solver iterations (\(n_{\text {cg}}\), \(n_{\text {gmg}}\)). Right, bottom: Performance models for the kernels
Weak Scaling Comparison of Multigrid and Conjugate Gradient. To allow a theoretical analysis, we choose a well known test problem: For the model equation \(- \varDelta c(\mathbf {x}) = f(\mathbf {x}), \mathbf {x} \in [0,1]^2\), discretized on a regular grid with mesh size h, it is known that the extreme eigenvalues of the resulting matrix are given by \(\lambda _{min} = 8 h^{-2} \sin ^2(\pi h / 2)\) and \(\lambda _{max} = 8 h^{-2} \cos ^2(\pi h / 2)\) and therefore, the condition number is given by \(\kappa := \lambda _{max} / \lambda _{min} = \tan ^{-2}(\pi h / 2)\) [14]. For the CG method, it is known that the error reduction factor in each iteration step can be estimated by \(\frac{\sqrt{\kappa } - 1}{\sqrt{\kappa } + 1}\) [14] and the number of iterations needed to achieve a prescribed reduction of the initial error by a factor of \(\delta \) can be estimated by \(n_{iter}(\delta ) \le \frac{1}{2} \sqrt{\kappa } \ln (\frac{2}{\delta }) + 1\). For the model problem under consideration and a fixed reduction factor \(\delta \), one can use the known condition number, the Taylor-series approximation of tan, and the fact that \(\frac{1}{h}\) is proportional to \(2^{n_{\text {ref}}}\), where \(n_{\text {ref}}\) is the number of refinements of the unit square, to estimate that the number of iterations \(n_{iter} \sim \sqrt{\kappa } = \tan ^{-1}(\pi h / 2) \approx \frac{2}{\pi h} \sim \frac{1}{h} \sim 2^{n_{\text {ref}}}\) is related to the grid refinement and will increase roughly by a factor of two with each refinement. In contrast, for the multigrid method it is known that the reduction rate is independent of the mesh size and, thus, a constant number of iterations can be expected [13].
The multigrid results are equivalent to the skin tests. However, for the unpreconditioned conjugate gradient method, our empirical performance models reveal an \(\mathcal {O}(\sqrt{p} )\) dependency, expected via the explanation above. We increase the process count and work load by a factor of four under weak scaling. Ideally, a constant time is expected, but due to the increase by a factor of two for the iteration count, models as shown in Table 3 are observed. We emphasize that one invocation of matrix-vector or vector-vector products does scale and the increase is due to the iteration count increase. A remedy of this issue can not be achieved by implementation alone, but must be achieved by a change of the mathematical method, e.g., using multigrid. Figure 6 shows a wall-clock time comparison.
6 Related Work
Performance modeling has a long history. Manual models proved to be very effective in describing many qualities and characteristics of applications, systems, and even entire tool chains [5, 18]. Recent approaches advocate source-code annotations [27] or specialized languages [25] to support developers in the creation of analytical performance models.
Various automated modeling methods exist. Many of these focus on learning the performance characteristics automatically using various machine-learning approaches [15]. Zhai et al. extrapolate single-node performance to complex parallel machines using a trace-driven network simulator [32], and Wu and Müller extrapolate traces to predict communications at larger scale [31]. Similar to our method, Carrington et al. extrapolate trace-based performance measurements using a set of canonical functions [9].
Numerous codes for the solution of partial differential equations exist, and several employ multigrid methods. There are a number of highly scalable geometric multigrid methods [4, 12, 23, 26, 29] and highly scalable algebraic multigrid [1–3]. Gahvari and Gropp model the performance of geometric [11] and algebraic multigrid methods [10]. Nägel et al. present an overview of how to treat skin permeation numerically [16].
7 Conclusion
UG4 is a framework with around half a million lines of code employed to solve problems such as drug diffusion through the human skin. With UG4, we have demonstrated the power of our performance modeling process as a fast and streamlined way to detect scalability bugs and validate performance expectations of simulation codes. The JUBE workflow manager vastly simplifies and accelerates the acquisition of performance measurements and our performance modeling method automates model creation. After removing a previously unknown performance bottleneck and validating the scalability of entire simulations, we can confidently claim that UG4 is ready for exascale.
References
Baker, A., Falgout, R., Kolev, T., Yang, U.: Multigrid smoothers for ultra-parallel computing. SIAM J. Sci. Comput 33, 2864–2887 (2011)
Baker, A.H., Falgout, R.D., Gamblin, T., Kolev, T.V., Schulz, M., Yang, U.M.: Scaling algebraic multigrid solvers: on the road to exascale. In: Competence in High Performance Computing 2010, pp. 215–226. Springer (2012)
Bastian, P., Blatt, M., Scheichl, R.: Algebraic multigrid for discontinuous galerkin discretizations of heterogeneous elliptic problems. Numer. Linear Algebra Appl. 19(2), 367–388 (2012)
Bergen, B., Gradl, T., Rude, U., Hulsemann, F.: A massively parallel multigrid method for finite elements. Comput. Sci. Eng. 8(6), 56–62 (2006)
Boyd, E.L., Azeem, W., Lee, H.H., Shih, T.P., Hung, S.H., Davidson, E.S.: A hierarchical approach to modeling and improving the performance of scientific applications on the KSR1. In: Proceedings of the International Conference on Parallel Processing (ICPP), pp. 188–192 (1994)
Braess, D.: Finite elements: Theory, Fast Solvers, and Applications in Solid Mechanics. Cambridge University Press, Cambridge (2001)
Calotoiu, A., Hoefler, T., Poke, M., Wolf, F.: Using automated performance modeling to find scalability bugs in complex codes. In: Proceedings of the ACM/IEEE Conference on Supercomputing (SC13). ACM, Denver, CO, USA, November 2013
Calotoiu, A., Hoefler, T., Wolf, F.: Mass-producing insightful performance models. In: Workshop on Modeling and Simulation of Systems and Applications. University of Washington, Seattle, Washington, August 2014
Carrington, L., Laurenzano, M., Tiwari, A.: Characterizing large-scale HPC applications through trace extrapolation. Parallel Process. Lett. 23(4), 1340008 (2013)
Gahvari, H., Baker, A.H., Schulz, M., Yang, U.M., Jordan, K.E., Gropp, W.: Modeling the performance of an algebraic multigrid cycle on HPC platforms. In: Proceedings of the International Conference on Supercomputing, pp. 172–181. ACM (2011)
Gahvari, H., Gropp, W.: An introductory exascale feasibility study for FFTs and multigrid. In: International Symposium on Parallel and Distributed Processing (IPDPS), pp. 1–9. IEEE (2010)
Gmeiner, B., Köstler, H., Stürmer, M., Rüde, U.: Parallel multigrid on hierarchical hybrid grids: a performance study on current high performance computing clusters. Concurrency Comput. Pract. Experience 26(1), 217–240 (2014)
Hackbusch, W.: Multi-grid Methods and Applications, vol. 4. Springer, Heidelberg (1985)
Hackbusch, W.: Iterative Solution of Large Sparse Systems of Equations. Springer, New York (1994)
Lee, B.C., Brooks, D.M., de Supinski, B.R., Schulz, M., Singh, K., McKee, S.A.: Methods of inference and learning for performance modeling of parallel applications. In: Proceedings of the 12th ACM SIGPLAN Symposium on Principles and Practice of Parallel Programming (PPoPP 2007), pp. 249–258 (2007)
Nägel, A., Heisig, M., Wittum, G.: Detailed modeling of skin penetration—an overview. Adv. Drug Deliv. Rev. 65(2), 191–207 (2013)
Nägel, A., Heisig, M., Wittum, G.: A comparison of two- and three-dimensional models for the simulation of the permeability of human stratum corneum. Eur. J. Pharm. Biopharm. 72(2), 332–338 (2009)
Petrini, F., Kerbyson, D.J., Pakin, S.: The case of the missing supercomputer performance: achieving optimal performance on the 8,192 processors of ASCI Q. In: Proceedings of the ACM/IEEE Conference on Supercomputing (SC 2003), p. 55 (2003)
Picard, R.R., Cook, R.D.: Cross-validation of regression models. J. Am. Statist. Assoc. 79(387), 575–583 (1984)
Reiter, S.: Efficient algorithms and data structures for the realization of adaptive, hierarchical grids on massively parallel systems. Ph.D. thesis, University of Frankfurt, Germany (2014)
Reiter, S., Vogel, A., Heppner, I., Rupp, M., Wittum, G.: A massively parallel geometric multigrid solver on hierarchically distributed grids. Comp. Vis. Sci. 16(4), 151–164 (2013)
Sack, P., Gropp, W.: A scalable MPI\_Comm\_split algorithm for exascale computing. In: Keller, R., Gabriel, E., Resch, M., Dongarra, J. (eds.) EuroMPI 2010. LNCS, vol. 6305, pp. 1–10. Springer, Heidelberg (2010)
Sampath, R., Biros, G.: A parallel geometric multigrid method for finite elements on octree meshes. SIAM J. Sci. Comput. 32, 1361–1392 (2010)
Siebert, C., Wolf, F.: Parallel sorting with minimal data. In: Cotronis, Y., Danalis, A., Nikolopoulos, D.S., Dongarra, J. (eds.) EuroMPI 2011. LNCS, vol. 6960, pp. 170–177. Springer, Heidelberg (2011)
Spafford, K.L., Vetter, J.S.: Aspen: a domain specific language for performance modeling. In: Proceedings of the International Conference on High Performance Computing, Networking, Storage and Analysis, SC 2012, pp. 84:1–84:11. IEEE Computer Society Press, Los Alamitos (2012)
Sundar, H., Biros, G., Burstedde, C., Rudi, J., Ghattas, O., Stadler, G.: Parallel geometric-algebraic multigrid on unstructured forests of octrees. In: Proceedings of the International Conference on High Performance Computing, Networking, Storage and Analysis. p. 43. IEEE Computer Society Press (2012)
Tallent, N.R., Hoisie, A.: Palm: easing the burden of analytical performance modeling. In: Proceedings of the International Conference on Supercomputing (ICS), pp. 221–230 (2014)
Vogel, A., Reiter, S., Rupp, M., Nägel, A., Wittum, G.: UG 4: a novel flexible software system for simulating PDE based models on high performance computers. Comp. Vis. Sci. 16(4), 165–179 (2013)
Williams, S., Lijewski, M., Almgren, A., Straalen, B.V., Carson, E., Knight, N., Demmel, J.: s-step Krylov subspace methods as bottom solvers for geometric multigrid. In: 28th International Parallel and Distributed Processing Symposium, pp. 1149–1158. IEEE (2014)
Wolf, F., Bischof, C., Hoefler, T., Mohr, B., Wittum, G., Calotoiu, A., Iwainsky, C., Strube, A., Vogel, A.: Catwalk: a quick development path for performance models. In: Lopes, L., et al. (eds.) Euro-Par 2014, Part II. LNCS, vol. 8806, pp. 589–600. Springer, Heidelberg (2014)
Wu, X., Mueller, F.: ScalaExtrap: trace-based communication extrapolation for SPMD programs. In: Proceedings of the 16th ACM Symposium on Principles and Practice of Parallel Programming (PPoPP 2011), pp. 113–122 (2011)
Zhai, J., Chen, W., Zheng, W.: Phantom: predicting performance of parallel applications on large-scale parallel machines using a single node. SIGPLAN Not. 45(5), 305–314 (2010)
Acknowledgment
Financial support from the DFG Priority Program 1648 Software for Exascale Computing (SPPEXA) is gratefully acknowledged. The authors also thank the Gauss Centre for Supercomputing (GCS) for providing computing time on the GCS share of the supercomputer JUQUEEN at Jülich Supercomputing Centre (JSC).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Vogel, A. et al. (2015). 10,000 Performance Models per Minute – Scalability of the UG4 Simulation Framework. In: Träff, J., Hunold, S., Versaci, F. (eds) Euro-Par 2015: Parallel Processing. Euro-Par 2015. Lecture Notes in Computer Science(), vol 9233. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-662-48096-0_40
Download citation
DOI: https://doi.org/10.1007/978-3-662-48096-0_40
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-662-48095-3
Online ISBN: 978-3-662-48096-0
eBook Packages: Computer ScienceComputer Science (R0)