Abstract
We describe a probabilistic technique for diagnostic prediction of first-episode schizophrenia patients based on their brain diffusion MRI data. The method begins by transforming each voxel from a high-dimensional diffusion weighted signal to a low-dimensional diffusion tensor representation. Three orthogonal diffusion measures (fractional anisotropy, norm, mode) that capture different aspects of the local tissue properties are derived from this diffusion tensor representation. Next, we compute a one-dimensional probability density function of each of the diffusion measures with values obtained from the entire brain. This representation is affine invariant, thus obviating the need for registration of the images. We then use a Parzen window classifier to estimate the likelihood of a new patient belonging to either group. To demonstrate the technique, we apply it to the analysis of 22 first-episode schizophrenic patients and 20 normal control subjects. With leave-one-out cross validation, we find a detection rate of 90.91 % (10 % false positives). We also provide several error bounds on the performance of the classifier.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- Fractional Anisotropy
- Diffusion Tensor Image
- Diffusion Tensor
- Probability Density Function
- White Matter Region
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
A recent World Health Organization (WHO) report estimates that nearly 11 % of the population world-wide is affected by some form of brain disorder. These illnesses can often be psychologically and financially devastating to patients, their families and the larger community. Nearly 1 % of the population in the US is affected by schizophrenia. A growing body of evidence suggests that the early stages of schizophrenia (and many other brain disorders) are critical in forming and predicting the course and outcome of the disorder. The classification tools proposed in this work can serve as a first step towards early detection of schizophrenia, which may result in a better prognosis and functional outcome.
Both, post-mortem and neuroimaging studies have contributed significantly to what we know about the brain. Moreover, MRI studies of volumetric reduction in several brain regions in schizophrenia have been particularly informative with respect to confirming early speculations that the brain is disordered in schizophrenia. This work is largely the result of advances in neuroimaging that allowed for more careful measurement of regions of interest within the brain (see review in [15, 22]). In particular, a shape based framework was recently developed by the authors in [8], which utilized volumetric differences as discriminatory features for distinguishing a population of schizophrenia from normal controls (NC). Another recent work in this direction was proposed in [20], where the authors used the affine parameters obtained during registration of each subject to a given atlas as a discriminant feature. However, both works, used structural MRI data, as opposed to diffusion MRI being used in this study.
The advent of diffusion magnetic resonance imaging (dMRI) has provided the opportunity for non-invasive investigation of neural architecture of the brain. Using this imaging technique, neuroscientists want to ask how neurons originating from one region connect to other regions, or how well-defined those connections may be. One of the models that is widely used to analyze dMRI images is the diffusion tensor model. Diffusion tensors represent the diffusion of water molecules in three orthogonal directions with the principal direction aligned with the fiber orientation. Several scalar measures derived from this model are used to assess the strength of connectivity in neural fiber bundles.
Experimental evidence suggests that the tissue component responsible for the observed orientational anisotropy in white matter is principally the spatial organization of cellular membranes, which is modulated by the degree of myelinization of the individual axons and the density of cellular packing [6]. As a result, fractional anisotropy (FA) is the most popular measure used to study abnormalities in white matter [15].
While there has been an explosion in the number of studies reporting statistical differences in various regions of the brain (see [15] and the references therein), very few works have addressed the problem of classifying schizophrenic patients from healthy subjects. Apart from structural MRI data being used by Davatzikos et al. [8] and Pohl and Sabuncu [20], there has been work done by Caan et al. [4], where the authors use dimensionality reduction followed by linear discriminant analysis for classification of patients with schizophrenia (chronic). They use the fractional anisotropy and linear anisotropy images derived from single tensor estimation as discriminant features.
In all of the approaches listed above [4, 8], the authors first perform a registration of all the subjects to a common atlas space and subsequent analysis is done on this dataset. However, such an analysis is computationally expensive and requires a very good registration algorithm to spatially normalize all the subjects in one common co-ordinate system. In the proposed work, we compute the probability distribution of several diffusion measures over the entire brain white matter, which makes it un-necessary to register the diffusion images, thus reducing computational complexity. Note that, the probability distribution (by its definition) is invariant to affine-transformations (see Sect. 2.2 for more details).
2 Methods
The overall outline of the proposed algorithm for computing the probabilistic representation of each subject is shown in Fig. 1. From the dMRI scan of a subject, diffusion tensors are first estimated. Three orthogonal diffusion measures (fractional anisotropy (FA), norm (N), mode (M d )) [14] that form the discriminatory features of our classifier are then computed at each voxel in the white matter region. A nonparametric density estimator is then used to convert each of these measures of each subject into a probabilistic representation, which is affine invariant. Note that, we compute a 1D probability distribution function (pdf) of each of the diffusion measures (FA, N, M d ) from values obtained throughout the white matter. These three one-dimensional pdf’s form the features for each subject. This representation is subsequently used by a Parzen window classifier to compute the probability of a previously unseen subject being FE or NC in a cross-validation scheme. Details on each of these steps are given in the subsequent sections.
2.1 Preliminaries
In diffusion weighted imaging, image contrast is related to the strength of water diffusion. At each image voxel, diffusion is measured along a set of distinct gradients, \(\mathbf{u}_{1},\ldots,\mathbf{u}_{n} \in \mathbb{S}^{2}\) (on the unit sphere), producing the corresponding signal, \(\mathbf{s} = [\,s_{1},\ldots,s_{n}\,]^{T} \in \mathbb{R}^{n}\). The diffusion tensor is related to the signal using the following relation [3, 16]:
where s 0 is a baseline signal intensity, b is an acquisition-specific constant, and D is a tensor describing the diffusion pattern. D can be estimated using a weighted least-squares approach [1].
Several scalar measures derived from the single tensor model have been proposed in the literature [14, 18, 24]. In particular, we use a set of three orthogonal invariants studied in [14], namely the norm N, fractional anisotropy FA and mode M d . These measures capture different (orthogonal) aspects of the shape of the tensor. Given, a diffusion tensor D, these measures can be computed as follows:
where, | . | denotes the determinant, tr(. ) is the trace and ∥ . ∥ denotes the frobenius norm of a matrix. Thus, FA measures how the shape of the tensor deviates from that of a sphere (diameter of the sphere is given by the average length of the axes of the ellipsoid (tensor)). M d indicates the mode of the tensor, i.e. \(M_{d} = -1\) indicates planar anisotropy, M d = 0 indicates an orthotropic tensor and M d = 1 indicates linear anisotropic tensor. Norm N measures the “size” of the diffusion tensor. Of these measures, only FA has been widely used to study white matter abnormalities in schizophrenia (see references in [15]). From the above discussion, at voxel r, we compute the following 3-dimensional vector
2.2 Probabilistic Representations
Probability density functions (PDF) are invariant to translation, rotation, scale and shear of an image, i.e. PDF’s are invariant under linear transformation of the coordinates of an image. A nonparametric estimate of the PDF can be computed using the following expression [19]:
where I(x) is a scalar image at spatial location x, M is the number of data points, G is a Gaussian kernel and h denotes the bandwidth of the kernel. An affinely transformed image \(\tilde{I}\) is related to the original image using the relation \(\tilde{I}(Ax) = I(x)\), where A is an affine transformation. Notice that only the co-ordinates of the image I change without changing the image intensities (scalar values). By applying a change of variable in Eq. (3), one can easily see that the PDF p(z) is invariant under affine (linear) transformations.
The proposed set of diffusion measures f(r) lives in a 3-dimensional space. Computing the joint PDF of the 3-dimensional space is computationally intensive. Further, the measures N, FA, M d are mutually orthogonal. As such, we compute a 1D PDF for each measure separately using (3). Note that, each of these measures captures different aspects of the variation in “shape” of the diffusion tensor and thus are independent of the orientation.
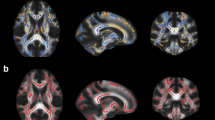
Several schizophrenia studies [15] have shown abnormalities in the white matter region of the brain. We thus choose this entire region (white matter) to compute the PDF. Specifically, a diffusion tensor is estimated at each voxel and FA is computed in the entire image volume. Regions of the brain that have FA ≥ 0. 4 are selected for further analysis (see Fig. 2). This roughly corresponds to the single fiber white matter region in the brain. Note that, we chose a threshold of 0.4 in order to exclude regions that have crossing fiber bundles (which would result in lower FA). Such crossing regions cannot be correctly represented using a single diffusion tensor. All the other features (such as, M d , N) are computed in this region (with FA > 0. 4).
We should note that since FA is a discriminatory feature between the two populations (first-episode schizophrenics and healthy controls), thresholding the image in itself amounts to a feature selection step. For example, if one group in general has lower FA than another, this would lead to a difference in the estimated pdf which would be useful during classification.
Using (3), we compute the PDF for each of the three discriminatory measures and combine them into a matrix representation denoted by \(\mathbf{p} = [p_{n}\;p_{fa}\;p_{md}]\). Thus, each patient scan i can now been transformed into a probabilistic representation (matrix) p i of dimension n b × 3, where n b is the number of bins used in the pdf computation. In our subsequent discussions, we will use this representation in our classifier.
Figure 3a–c show the PDF’s for 22 first-episode (FE) schizophrenic patients (red) along with 20 age-matched normal controls (NC) (blue). A visual inspection shows differences between the two groups (blue and red) for each of these measures. Figure 4 shows a cross-section of the two groups for a certain value of FA. This figure confirms the existence of two distinct clusters in the data (albeit with overlap).
2.3 Parzen Window Classifier
The Parzen window classifier was first introduced by Jain and Ramaswami [11]. In this method, a Parzen window based density estimate [9] is used to compute the likelihood that a new data point belongs to one of the groups in the training data set.
Let \(\{\mathbf{p}_{fe}^{i}\}_{i=1}^{N_{fe}}\) and \(\{\mathbf{p}_{nc}^{i}\}_{i=1}^{N_{nc}}\) be the set of N fe FE and N nc NC subjects in the training data set. Given a test data point \(\mathbf{\hat{p}}\), the likelihood (probability) that it belongs to either group can be computed using the Parzen window density estimator as follows:
where K(. , . ) is a Gaussian kernel given by
with m = [ N, FA, M d ]T as described earlier, and i, j represent the indices for ith and jth subject. Note that, we assume that the PDF’s of each of the diffusion measures for a subject are independent, due to the fact that these measures themselves are orthogonal.
2.3.1 Design Choices
For each of the two groups, we choose σ m using the following relation:
where N is N fe for the group of FE patients and N = N nc for NC subjects. Thus a different set of \(\{\sigma _{m}\}_{m=1}^{3}\) is computed separately for each group in the training data set. The constant c m is a scalar that is computed so that the training error is minimized. Typical values for c m that give a good generalization of the sampled data while reducing the risk of over fitting lie in the range \(c_{m} \in [1.5,\;2]\), as has been noted in [7]. In numerical experiments, we discretize c m in the range [1. 5, 2] at an interval of 0.1. The value of c m that minimizes the training error is chosen for a given training data set. We should note that this is the only parameter one needs to choose in our entire classification system.
This data driven approach of choosing σ m is quite common in the literature and has been used in other works as well [7]. This choice of σ m is guided by the following considerations: (1) σ m varies appropriately with the scaling of each of the components of m, (2) It minimizes the training error of the classifier, (3) it respects the distribution of points within the clusters (whether the points are spread out or densely packed).
Thus, from the probabilities obtained in (4), we obtain the following simple classification rule:
3 Results
3.1 Data Acquisition Protocol
Our dataset consisted of 22 FE patients with average age 20. 89 ± 4. 8 years and 20 NC with average age 22. 3 ± 4. 2 years (p = 0. 21). All the subjects were scanned as part of Dr. Martha Shenton’s NIH grant (R01 MH 50740) on a 3-T GE system using an echo planar imaging (EPI) diffusion weighted image sequence. A double echo option was used to reduce eddy-current related distortions. To reduce impact of EPI spatial distortion, an eight channel coil was used to perform parallel imaging using Array Spatial Sensitivity Encoding Techniques (GE) with a SENSE-factor (speed-up) of 2. Acquisitions have 51 gradient directions with b-value = 900 and eight baseline scans with b = 0. The original GE sequence was modified to increase spatial resolution, and to further minimize image artifacts. The following scan parameters were used: TR 17,000 ms, TE 78 ms, FOV 24 cm, 144 × 144 encoding steps, 1.7 mm slice thickness. All scans had 85 axial slices parallel to the AC-PC line covering the whole brain.
The raw diffusion weighted images were preprocessed using the Rician noise removal algorithm of [2] followed by eddy current and head motion correction algorithm [13] (part of the FSL package – http://www.fmrib.ox.ac.uk/fsl/flirt/).
4 Classification Results
4.1 Leave-One-Out Cross-Validation
Leave-one-out (LOO) is an unbiased technique for cross-validation of classification results especially when the training data set is small [5, 23]. This is one of the techniques we use to test our classifier. In this method, one subject is removed from the dataset and the classifier is trained on the remaining samples. This procedure is repeated for all available samples and classification results are computed.
In our case, the data samples are the matrices p i of dimension (n b × 3), with each column representing a discretized pdf of the feature vectors. Here, n b is the number of bins, which we fix to 300 in all experiments. Given the matrices p i for all subjects, the probability of a previously unseen subject is computed using Eq. (4). This procedure is repeated by removing one datum each time and using the remaining samples as training data set. Thus, one sample is used as test while the remaining samples are used to train the classifier. The correct detection rate is then computed by counting the number of times the test sample was correctly identified (FE or NC) while testing all the subjects (in our case it is 42). The false positive rate is given by the number of subjects that were “predicted” by the classifier as FE, whereas they were NC. The overall classification error is given by the number incorrect classifications “predicted” by the classifier. In our experiments, the detection rate (true positives) obtained for LOO cross-validation is 90.91 %, while the false positive rate is 10.0 %. The overall classification error is 9.52 %.
As has been pointed out by the authors in [10], for small sample size, it is not enough to validate the results using LOO experiment. Instead, one should compute confidence intervals that give a lower and upper bound on the performance of the classifier. Several methods have been proposed in the literature to compute these bounds for small sample size, of which the Bayesian and Binomial bounds are most popular.
Table 1 gives the 95 % Bayesian and Binomial (Exact Wald and Adjusted Wald) [17, 21] confidence intervals (upper and lower limit) on the overall performance of the classifier. Intuitively, a 95 % confidence interval indicates that in 95 out of 100 experiments, the overall performance of the classifier will fall within the stated confidence interval. These confidence intervals are also a function of the number of samples in the data set. Thus, as the number of samples tested increases, the confidence interval becomes narrow and converges to the “true” estimate [10, 12]. The Exact method was designed to guarantee at least 95 % coverage, whereas the approximate methods (adjusted Wald) provide an average coverage of 95 % only when a large number of samples are available.
The above LOO experiment included all the three components of vector f as features. Table 2 shows classification results for LOO experiment, but with different number of features. As is clear, including all the three features does improve the performance of the classifier. Adding more features such as radial diffusivity, linear anisotropy, etc. did not improve the performance of the classifier.
5 Discussion
In this paper, we proposed a novel probabilistic classification method for separating first-episode schizophrenic patients from age-matched normal controls using anisotropic measures derived from diffusion tensor images. The output of the classifier is a probabilistic score of a previously unseen subject being FE or NC. We validate the proposed classifier using a leave-one-out experiment obtaining a sensitivity of 90.91 % and specificity of 90 %. In this work, we chose the entire white matter to perform classification. However, individual fiber tracts such as corpus callosum, fornix, cingulum bundle, etc. may be able to provide more information regarding the variation of these fiber bundles in either population. Our future work entails examining these fiber tracts to detect abnormalities and subsequently use them for probabilistic classification. We should note that the methodology presented here is quite general and can be applied for classification of many other types of brain disorders (bipolar disorder, schizotypal personality disorder, etc.).
This work is a first step towards early detection of schizophrenia, which can result in better patient care. Further, the probabilistic methodology proposed in this work could be used to study the effect of medication by analyzing changes in white matter anisotropy.
References
Aitken, A.: On least squares and linear combination of observations. In: Proceedings of the Royal Society of Edinburgh, vol. 55, pp. 42–48 (1934)
Aja-Fernandez, S., Niethammer, M., Kubicki, M., Shenton, M.E., Westin, C.F.: Restoration of DWI data using a rician LMMSE estimator. IEEE Trans. Med. Imaging 27, 1389–1403 (2008)
Basser, P., Mattiello, J., LeBihan, D.: MR diffusion tensor spectroscopy and imaging. Biophys. J. 66(1), 259–267 (1994)
Caan, M., Vermeer, K., van Vliet, L., Majoie, C., Peters, B., den Heeten, G., Vos, F.: Shaving diffusion tensor images in discriminant analysis: a study into schizophrenia. Med. Image Anal. 10(6), 841–849 (2006)
Cawley, G., Talbot, N.: Efficient leave-one-out cross-validation of kernel Fisher discriminant classifiers. Pattern Recognit. 36(11), 2585–2592 (2003)
Chenevert, T., Brunberg, J., Pipe, J.: Anisotropic diffusion in human white matter: demonstration with MR techniques in vivo. Radiology 177(2), 401–405 (1990)
Cremers, D., Kohlberger, T., Schnrr, C.: Nonlinear shape statistics in mumford-shah based segmentation. In: 7th ECCV ’02, Copenhagen, vol. 2351, pp. 93–108, (2002)
Davatzikos, C., Shen, D., Gur, R., Wu, X., Liu, D., Fan, Y., Hughett, P., Turetsky, B., Gur, R.: Whole-brain morphometric study of schizophrenia revealing a spatially complex set of focal abnormalities. Arch. Gen. Psychiatry 62(11), 1218–1227 (2005)
Girolami, M.: Orthogonal series density estimation and the kernel eigenvalue problem. Neural Comput. 14(3), 669–688 (2002)
Isaksson, A., Wallman, M., Goransson, H., Gustafsson, M.: Cross-validation and bootstrapping are unreliable in small sample classification. Pattern Recognit. Lett. 29(14), 1960–1965 (2008)
Jain, A., Ramaswami, M.: Classifier design with Parzen windows. In: Pattern Recognition and Artificial Intelligence, North Holland, pp. 211–228 (1988)
Jaynes, E.: Confidence intervals vs. Bayesian intervals. Found. Probab. Theory Stat. Inference Stat. Theor. Sci. 2, 175–257 (1976)
Jenkinson, M., Bannister, P., Brady, M., Smith, S.: Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage 17(2), 825–841 (2002)
Kindlmann, G., Ennis, D., Whitaker, R., Westin, C.: Diffusion tensor analysis with invariant gradients and rotation tangents. TMI 26(11), 1483–1499 (2007)
Kubicki, M., McCarley, R., Westin, C.-F., Park, H.-J., Maier, S., Kikinis, R., Jolesz, F., Shenton, M.: A review of diffusion tensor imaging studies in schizophrenia. J. Psychiatr. Res. 41, 15–30 (2007)
LeBihan, D., Mangin, J., Poupon, C., Clark, C., Pappata, S., Molko, N., Chabriat, H.: Diffusion tensor imaging: concepts and applications. J. Magn. Reson. Imaging 13, 534–546 (2001)
Lewis, J., Sauro, J.: When 100% really Isn’t 100%: improving the accuracy of small-sample estimates of completion rates. J. Usability Stud. 1(3), 136–150 (2006)
Ozarslan, E., Vemuri, B., Mareci, T.: Generalized scalar measures for diffusion MRI using trace, variance, and entropy. Magn. Reson. Med. 53(4), 866–876 (2005)
Parzen, E.: On estimation of a probability density function and mode. Ann. Math. Stat. 33(3), 1065–1076 (1962)
Pohl, K.M., Sabuncu, M.R.: A unified framework for MR based disease classification. In: Prince, J.L., Pham, D.L., Myers, K.J. (eds.) Information Processing in Medical Imaging. Lecture Notes in Computer Science, vol. 5636, pp. 300–313. Springer (2009). ISBN 978-3-642-02497-9
Sauro, J., Lewis, J.: Estimating completion rates from small samples using binomial confidence intervals: comparisons and recommendations. In: Human Factors & Ergonomics Society (ed.) Proceedings of the Human Factors and Ergonomics Society: 49th Annual Meeting, Orlando (2005)
Shenton, M., Dickey, C., Frumin, M., McCarley, R.: A review of MRI findings in schizophrenia. Schizophr. Res. 49(1–2), 1–52 (2001)
Vapnik, V.: The Nature of Statistical Learning Theory. Springer, New York (2000)
Westin, C.-F., Maier, S.E., Mamata, H., Nabavi, A., Jolesz, F.A., Kikinis, R.: Processing and visualization of diffusion tensor MRI. Med. Image Anal. 6(2), 93–108 (2002)
Acknowledgements
This work has been supported in part by a Department of Veteran Affairs Merit Award (Dr. Martha Shenton), the VA Schizophrenia Center Grant (MS), NIH grant R01MH50740 (MS), R01MH097979 (Rathi) and R01 MH074794(Westin).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Rathi, Y., Shenton, M.E., Westin, CF. (2014). Preliminary Findings in Diagnostic Prediction of Schizophrenia Using Diffusion Tensor Imaging. In: Westin, CF., Vilanova, A., Burgeth, B. (eds) Visualization and Processing of Tensors and Higher Order Descriptors for Multi-Valued Data. Mathematics and Visualization. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-54301-2_14
Download citation
DOI: https://doi.org/10.1007/978-3-642-54301-2_14
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-54300-5
Online ISBN: 978-3-642-54301-2
eBook Packages: Mathematics and StatisticsMathematics and Statistics (R0)