Abstract
Chromosomally encoded type II toxin–antitoxin (TA) systems generally consist of two adjacent genes in an operon encoding a stable toxin and a less stable, protease-sensitive cognate antitoxin. While the toxin and the antitoxin form a stable complex under normal growth conditions, the degradation of the antitoxin by stress-proteases under certain conditions leads to activation of the toxin and subsequent growth inhibition. Such stress-responsive TA systems have been associated with various cellular processes, including stabilization of genomic regions, protection against foreign DNA, biofilm formation, persistence, and control of the stress response. Yet, the contribution of chromosomal TA to bacterial virulence is presently unknown. Herein, we investigate the potential role of multiple chromosomally encoded TA systems in virulence, focusing on the tuberculosis agent Mycobacterium tuberculosis, which contains more than 70 TA loci in its genome. We describe what is currently known about the multiple TA families present in this bacterium, with emphasis on the recently discovered atypical stress-responsive toxin–antitoxin-chaperone (TAC) system, a TA system controlled by a SecB-like chaperone.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
17.1 Introduction
Toxin–antitoxin (TA) systems are two partner modules that were first discovered as plasmid-borne loci involved in plasmid maintenance by a mechanism called “postsegregational killing”. This phenomenon relies on a stable toxin that inhibits cell growth and a labile protease-sensitive antitoxin that antagonizes the toxin function, both encoded by the plasmid. When the plasmid is lost, the antitoxin is degraded resulting in toxin-mediated cell growth inhibition and eventual cell death (Yamaguchi et al. 2011). Since then, many TA loci have been found in numerous bacterial and archeal chromosomes, suggesting alternative functions, such as stabilization of chromosomal regions or growth modulation in response to environmental insults (Gerdes et al. 2005).
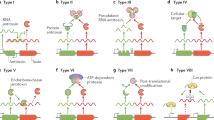
Three types of TA systems are defined based on the nature of the antitoxin and on the mechanism of neutralization of the toxin, which is always a protein. In type I TA systems, the antitoxin is a small antisense RNA that forms a duplex with a specific region of the toxin mRNA resulting in the inhibition of toxin production. Type II antitoxins are proteins that inhibit toxin function by a protein–protein interaction. The type III TA system is represented by only one characterized system to date, ToxIN from Petrobacterium atrosepticum, whose RNA antitoxin ToxI inactivates the ToxN toxin by direct binding (Yamaguchi et al. 2011). This chapter will focus on chromosomally encoded type II TA systems, with emphasis on the multiple TA systems present in the major human pathogen M. tuberculosis and their potential implication in virulence.
17.2 General Features of Type II TA Systems
Type II TA systems are encoded in an operon consisting of two small genes whose products, a toxin and its cognate antitoxin, form a complex in which the toxin is inactive under normal growth conditions. Type II antitoxins generally carry a DNA binding domain which binds directly on the promoter region of the operon, and acts as a repressor, often together with the toxin. Under certain conditions, the antitoxin is degraded by stress-proteases, resulting in toxin activation, growth inhibition, and eventual cell death (Gerdes et al. 2005). Free toxins from type II systems have been shown to target essential cellular processes such as replication, cell wall synthesis, cell division, or translation (Gerdes et al. 2005; Yamaguchi et al. 2011, Mutschler et al. 2011).
Chromosomal type II systems are widely distributed in prokaryotic genomes and often clustered in genomic islands (Makarova et al. 2009). This is in agreement with the fact that TA modules are often found on mobile genetic elements such as plasmid, phages, or transposable elements, thus mostly belonging to the prokaryotic mobilome (Makarova et al. 2009). For this reason, the content of type II TA systems is highly variable between species and also from one strain to another (http://bioinfo-mml.sjtu.edu.cn/TADB/). A study of 126 sequenced prokaryotic genomes revealed that free-living organisms have many TA loci, whereas obligate intracellular organisms seem to have fewer (Pandey and Gerdes 2005). However, some Rickettsia species, which are strict intracellular bacteria, contain multiple TA systems (Audoly et al. 2011). In addition, it has been proposed that human pathogens generally contain more TA systems than their non-pathogenic counterparts (Georgiades and Raoult 2011), but the differences are often subtle and multiple TA modules are found in non-pathogenic species as well (Makarova et al. 2009). Thus, to date there is no obvious correlation between TA abundance and organism lifestyle or virulence.
17.3 Role of Chromosomal Type II TA Systems and Possible Implication in Virulence
To date, the biological role of chromosomal TA systems is still unclear and under debate. Given their apparent dissemination by horizontal gene transfer, it has been proposed that TA systems could in some cases be junk DNA conserved by virtue of their intrinsic properties of addiction (Van Melderen 2010). Nevertheless, many chromosomal TA systems are induced under stress conditions and some seem to have important physiological functions, such as stabilization of genomic regions, anti-addiction against similar plasmid-borne toxins, protection against phage infection, biofilm formation, persistence, and control of the stress response (Van Melderen 2010; Yamaguchi and Inouye 2011). Therefore, the presence of multiple chromosomal TA systems in many bacteria suggests that these systems could function as intricate cellular networks of reactive elements that facilitate adaptive response to various environmental changes.
There is little or no evidence of a potential implication of TA systems in virulence. However, several important pathogens including Yersinia pestis, Streptococcus pneumoniae, Staphylococcus aureus, Pseudomonas aeruginosa, Vibrio cholerae, Rickettsia felis, and M. tuberculosis encode functional TA systems (Goulard et al. 2010; Mutschler et al. 2011; Yoshizumi et al. 2009; Hood et al. 2010; Wozniak and Waldor 2009; Audoly et al. 2011; Ramage et al. 2009). Interestingly, the pezAT locus of S. pneumoniae is located in a pathogenicity island and the PezT toxin was demonstrated to accelerate the progression of pneumococcal infections in mice, suggesting a role in virulence (Mutschler et al. 2011). In the case of V. cholerae, the mosAT TA locus was shown to promote the maintenance of SXT, an integrative and conjugative element that confers resistance to multiple antibiotics (Wozniak and Waldor 2009). In P. aeruginosa, the functional Tse2 toxin is addressed to other bacteria via the type VI secretion system, suggesting a potential role in biofilm formation, a key process in the chronic infection of cystic fibrosis patients by this bacterium (Hood et al. 2010). Remarkably, a recent study showed that VapC toxins from Rickettsia species could trigger apoptotic response in human cells, implying that TA toxins could be directly involved in virulence by poisoning host cells (Audoly et al. 2011).
Several studies suggest that TA systems could be involved in the formation of persisters, a phenomenon thought to be responsible for chronic infections (Lewis 2010). Persisters are specialized cells which constitute a subset of any bacterial population and are phenotypically drug-tolerant due to a slowed metabolism (Lewis 2010). Remarkably, the antitoxin gene mqsA is the most persistence-induced gene in Escherichia coli (Shah et al. 2006). The MqsA antitoxin from E. coli is known to antagonize the ribosome-independent RNase toxin MqsR and to regulate other stress genes than the mqsRA operon, including the major stress response regulator encoding gene rpoS (Wang et al. 2011). Consistent with the idea that TA modules contribute to persistence, an E. coli strain deleted for 10 TA loci was shown to produce reduced levels of persister cells (Maisonneuve et al. 2011). In M. tuberculosis, the causal agent of tuberculosis, a recent study of the drug-tolerant persisters transcriptome revealed that in addition to a massive shutdown of expression of the metabolic and biosynthetic pathways, a set of 10 TA systems were significantly upregulated together with other genes potentially involved in persisters formation (Keren et al. 2011). For this pathogen, persistence in hypoxic macrophages is a key step in the infectious cycle. This phenomenon is thought to be responsible for latent tuberculosis, an asymptomatic form of the disease concerning one third of the human population according to the WHO, which constitutes the major tuberculosis reservoir (www.who.int; Barry et al. 2009). The particularly high number of stress-responsive TA loci found in the M. tuberculosis chromosome and the ability of toxins to slow down bacterial growth raises the question of their implication in the setting up and/or the maintenance of the persistent state.
17.4 The Multiple TA Systems of Mycobacterium tuberculosis
The M. tuberculosis H37Rv chromosome potentially contains 75 TA systems (Fig. 17.1; Ramage et al. 2009; Gupta 2009; http://bioinfo-mml.sjtu.edu.cn/TADB/index.php). As it is generally the case, these systems are not randomly distributed across the M. tuberculosis chromosome but mostly clustered in hot spot regions (Makarova et al. 2009). Six well described TA pair families are represented: vapBC (50 systems), mazEF (10 systems), yefM/yoeB (1 system), relBE (2 systems), higBA (3 systems), and parDE (2 systems), while 7 systems could not be classified. A considerable subset of these TA systems (i.e., 63) has been experimentally tested, essentially in the bacterial models E. coli and M. smegmatis, for growth inhibition by the putative toxin and its neutralization by the putative antitoxin. The results are summarized in Fig. 17.1 (Gupta 2009; Ramage et al. 2009; Huang and He 2010; Singh et al. 2010; Zhu et al. 2010; Ahidjo et al. 2011). These studies identified a total of 37 TA systems that are functional in at least one of the tested conditions. In this part, we will describe each TA family present in M. tuberculosis, their mechanism of action and their potential implication in persistence.
Chromosomal map of M. tuberculosis H37Rv TA systems. Type II TA systems are annotated according to the tuberculist database (http://tuberculist.epfl.ch/index.html) except from VapBC49 (Rv2018-Rv2019), VapBC50 (Rv3181c-Rv3180c), VapBC51 (Rv3750c-Rv3749c), HigBA2 (Rv2022c-Rv2021c), HigBA3 (Rv3182-Rv3183), YefM/YoeB (Rv3357-Rv3358) and MazEF10 (Rv0298-Rv0299). For each system, the activity of the toxin is represented by colored circles. The first circle from the left represents experiments performed in E. coli, the second in M. smegmatis, and the third in M. tuberculosis; red color stands for “inhibition of growth”, green for “no inhibition of growth” and white for “not tested”. Systems that have never been tested experimentally display no circle. The 10 most induced TA systems in drug-tolerant persister cells are highlighted on dark gray background. The large genomic island containing TAC is depicted as a wide orange line and TAC is indicated by an asterisk
17.4.1 The vapBC Family
The most abundant TA loci in M. tuberculosis are from the VapBC (Virulence associated protein) family, characterized by a toxin with a PIN domain (homologous to PilT N-terminal domain). PIN domain proteins are found in all kingdoms of life and are generally involved in mRNA metabolism (Arcus et al. 2011). VapC toxins from diverse organisms, including M. tuberculosis VapC1, VapC2, VapC5, VapC11, and VapC29, were shown to have ribonuclease activity in vitro (Miallau et al. 2009; Ahidjo et al. 2011; Ramage et al. 2009), whereas M. tuberculosis VapC4 toxicity seems to be due to stable RNA binding (Sharp et al. 2012). Winther and Gerdes (2011) showed that two VapC from the entero-pathogenic bacteria Shigella flexneri and Salmonella enterica are specific tRNAses that cleave the initiator tRNA. Thus, VapC toxins appear to act by distinct mechanisms, all of them targeting RNA.
The structure of the M. tuberculosis VapBC5 complex was solved and revealed an asymmetric complex with a ratio of one antitoxin for one toxin (Miallau et al. 2009). Yet, the VapB5 antitoxin inhibits the toxin by a mechanism which seems to be conserved, where the antitoxin prevents efficient binding of Mg2+ at the active site of VapC (Miallau et al. 2009). The M. tuberculosis VapB antitoxins are generally related to families of transcriptional regulators or DNA binding domains already reported to be associated with VapC toxins (Makarova et al. 2009), i.e., 33 RHH, 8 Phd, 2 ArbR, and 1 MerR. Yet, 5 other VapB do not present any conserved domain.
In M. tuberculosis several VapBC were activated in response to stresses encountered during infection: hypoxia (VapBC15, VapBC7, and VapBC25) and in IFN-γ-stimulated murine bone marrow-derived macrophages (VapBC11, VapBC3, and VapBC47) (Ramage et al. 2009). Moreover, VapBC3, VapBC31, and VapBC50 were shown to be among the 10 TA systems the most upregulated in drug-tolerant persisters, suggesting a possible role in this physiological state (Keren et al. 2011). Remarkably, a recent study in the closely related bacterium M. smegmatis revealed that its unique VapC toxin is an RNase involved in coupling the rate of glycerol utilization to bacterial growth via post-transcriptional regulation of genes involved in sugar transport (McKenzie et al. 2012). Therefore, it is conceivable that the multiple stress-responsive VapBC present in the M. tuberculosis chromosome could act as global regulators of translation in response to very diverse environmental stresses.
17.4.2 The mazEF Family
In E. coli, overexpression of the sequence-specific endoribonuclease MazF leads to growth arrest via cleavage of almost all cellular mRNAs (Yamaguchi and Inouye 2011). The E. coli MazEF complex, in which the MazF toxin is inactive, is a linear hetero-hexamer composed of a dimer of the MazE antitoxin symmetrically bound to two MazF dimers (Blower et al. 2011). Many MazF specific cleavage sites have been reported, for example E. coli MazF cleaves at ACA sequences (Zhang et al. 2005) and Bacillus subtilis MazF (EndoA) cleaves UACAU sequences (Park et al. 2011). From the 10 MazF family members present in M. tuberculosis, 7 were shown to affect E. coli and/or M. smegmatis growth, 2 exhibited non-observable phenotype, and one was never tested (Fig. 17.1; Ramage et al. 2009; Gupta 2009; Zhu et al. 2006). It has been shown that MazF9 cleaves UAC sequences, MazF3 cleaves U rich regions (Zhu et al. 2006), MazF6 cleaves at UUCCU or CUCCU sequences, and MazF4 at UCGCU (Zhu et al. 2008). Such differences in the size of the cleavage sites imply a variation in the cleavage frequency. This suggests that recognition of 3 or 5 bases by MazF may not trigger the same type of response, the first leading more likely to a global translation inhibition, whereas the latter, having less frequent recognition sites, could regulate specific mRNAs in the cell. In this case, differential activation of TA systems depending on environmental conditions would lead to the appropriate adaptive response. In addition to mRNA recognition, the E. coli MazF was further shown to generate specialized ribosomes by cleavage of the 3′ region of the 16S rRNA, which contains the anti-Shine-Dalgarno sequence, and subsequently favor leaderless mRNAs translation (Vesper et al. 2011). Such a mechanism would lead to an alternative translation pathway, which could promote stress adaptation.
The expression of mazEF from E. coli was shown to be induced in various stress conditions, including DNA damage, heat shock and oxidative stress in a RelA dependent manner (Hazan et al. 2004). Similarly, M. tuberculosis MazF2 is downregulated after four hours of nutrient starvation, whereas, MazF3 is upregulated during amino acid starvation in a relA deleted strain (Betts et al. 2002; Dahl et al. 2003). These results suggest that mazEF modules from M. tuberculosis could be involved in the stringent response to amino acid starvation.
The M. tuberculosis MazF9 toxin was shown to be neutralized by the non-cognate antitoxins MazE6, VapB27, and VapB40 (Zhu et al. 2010). This is in agreement with the very low conservation between the different MazEs from M. tuberculosis (4–22 % of identity) and with the fact that VapB27 and VapB40 are closer to the E. coli MazE than any other MazE present in M. tuberculosis. In contrast, M. tuberculosis MazF toxins are well conserved with up to 46 % sequence identity. Together, these intriguing findings suggest that complex and fine-tuned cellular networks of toxin–antitoxin systems may exist in this pathogen. Finally, it has been shown that the mycobacterial MazF4 toxin and DNA topoisomerase I interact and mutually inhibit each other (Huang and He 2010). Since the DNA topoisomerase I gene is essential in M. tuberculosis (Sassetti et al. 2003), this suggests that MazF4 could act by two alternative mechanisms to inhibit mycobacterial growth.
17.4.3 The yefM/yoeB and relBE Families
The YefM/YoeB family, named after its E. coli member, was first described as homologous of the Axe-Txe addiction system of the multi-drug resistance plasmid pRUM of Enterococcus faecium (Grady and Hayes 2003). The YefM antitoxins are homologous to Phd, the antitoxin of the Phd-Doc module of phage P1, whereas the YoeB toxins belong to the RelE superfamily (Yamaguchi and Inouye 2011). In M. tuberculosis, the TA modules Rv1247c-Rv1246c, Rv2865-Rv2866, and Rv3357-Rv3358 are often referred to as RelBE1, RelBE2, and RelBE3, respectively, since the toxins belong to the RelE superfamily. Yet, the RelBE3 protein sequences are very close to those of the E. coli yefM/yoeB system (58 and 68 % sequence similarities on protein levels, respectively), with the mycobacterial RelE3 toxin containing the conserved C-terminal histidine and tyrosine residues of YoeB involved in RNase activity (Kamada and Hanaoka 2005). Therefore, in contrast with relBE1 and 2, the relBE3 system clearly belongs to the yefM/yoeB family and was thus renamed yefM/yoeB. In the cases of relBE1 and relBE2, even if the antitoxins present YefM domains (PF02604), the toxins do not contain the YoeB conserved residues and the original appellation was thus retained.
Both RelE and YoeB from E. coli are ribosome-dependent ribonucleases that cleave mRNA at the ribosomal A site (Yamaguchi et al. 2011). Nevertheless, despite sequence and structural homology, E. coli RelE and YoeB seem to act by distinct mechanisms: RelE binds to the 30S subunit of 70S ribosomes and inhibits translation elongation, whereas YoeB binds to the 50S subunit and inhibits translation initiation (Yamaguchi et al. 2011). Gel filtration analyses of the M. tuberculosis YefM-YoeB complex suggest a heterotrimeric complex, as it is the case for the E. coli homologues (Kumar et al. 2008). The structure of M. tuberculosis YefM was solved and revealed a well-structured protein with a flexible C-terminal domain which superimposes well with the E. coli YefM from the complex structure (Kumar et al. 2008). As for YoeB, inactivation of E. coli RelE upon binding to RelB seems to occur via a conformational shift in the catalytic site of the toxin (Blower et al. 2011; Kamada and Hanaoka 2005). Both RelE and YoeB exhibit a microbial RNase fold, called RelE-like fold, also shared by other RNase toxins such as MqsR (Brown et al. 2009).
yefM/yoeB and the two relBE of M. tuberculosis were shown to be functional bona fide TA systems in E. coli, M. smegmatis as well as in M. tuberculosis (Korch et al. 2009; Singh et al. 2010). Yang and colleagues (2010) showed that there is complex cross regulation between these three systems with different patterns of cross interaction and thus of promoter regulation. For example, they showed that YefM interacts only with its cognate toxin YoeB, whereas RelB1 and RelB2 are able to interact with any of the RelE1, RelE2, or YoeB toxins. Moreover, YefM can bind its promoter on its own, whereas RelB1 and RelB2 need to be part of the TA complex, but not necessarily with their cognate toxin (Yang et al. 2010).
Interestingly, relBE2 is among the 10 most induced TA systems in M. tuberculosis persister cells (Keren et al. 2011). Transcriptional analyzes also revealed that yefM/yoeB, relBE1, and relBE2 genes are all expressed during growth in broth medium, whereas only relE1, relB2, and yoeB transcripts were detected in human macrophages in late stages of infection (Korch et al. 2009). The relE1, relE2 and yoeB genes were also shown to be upregulated in response to antibiotic treatment and in lung tissues of infected mice (Singh et al. 2010). Remarkably, individual overexpression of the three toxin genes led to increased levels of persisters after antibiotic treatment, and deletion of relE2 or yoeB affected persisters formation by 4–9 fold, while relE1 deletion had no effect (Singh et al. 2010). However, single mutant strains of each toxin were not affected in in vivo growth or survival, indicating that they do not individually have direct roles in the infection (Singh et al. 2010).
17.4.4 The parDE Family
parDE was first identified on the broad-host-range plasmid RK2 as a stabilization system (Roberts and Helinski 1992). The ParE toxin acts by inhibiting DNA gyrase, thereby blocking DNA replication (Jiang et al. 2002). The crystal structure of the ParDE1 complex from Caulobacter crescentus revealed a heterotetrameric complex composed of two homodimers of toxin and antitoxin (Blower et al. 2011). As C. crescentus ParD1, M. tuberculosis ParD1 is predicted to have an RHH DNA binding domain, whereas ParD2 does not contain any conserved domains. With the exception of the unclassified toxins (Fig. 17.1) it is thus likely that ParE1 and ParE2 are the only toxins of M. tuberculosis that do not target RNA. Ectopic expression of ParE2 inhibited both E. coli and M. smegmatis growth whereas under the same conditions, ParE1 only affected E. coli growth (Ramage et al. 2009; Gupta 2009). So far, none of these systems were tested in M. tuberculosis.
17.4.5 Unclassified TA Modules
Among the 7 unclassified TA systems of M. tuberculosis, three were experimentally tested and two were functional in M. smegmatis (Ramage et al. 2009; Fig. 17.1). Four of these unclassified TA modules were identified as part of the 10 most induced TA systems in persister cells, but none of them have been tested for TA functions (Keren et al. 2011; Fig. 17.1). Except from the Rv2653c-Rv2654c system, all the unclassified modules contain conserved domains, and are paired together as follows (NCD is for No Conserved Domain): SRPBCC-NCD (Rv0910-Rv0909, Rv1546-Rv1545), GNAT-RHH (Rv0919-Rv0918), COG5654-Xre (Rv1989c-Rv1990, Rv3189-Rv3188), and COG3832-ArsR (Rv2035-Rv2034). The last three pairs of conserved domains have been predicted as putative TA families by Makarova and colleagues (2009).
17.4.6 The higBA Family
The first higBA locus (host inhibition of growth) was found on the Rts1 plasmid of Proteus vulgaris where it functions as an addiction module (Tian et al. 1996). The HigB toxin from Rts1 plasmid was shown to act as a ribosome-dependent ribonuclease with a mechanism distinct from RelE and YoeB, in which the toxin binds the ribosome 50S subunit and cleaves at AAA sequences on the processing mRNAs (Hurley and Woychik 2009). HigB1 and HigB2 from V. cholerae were shown to have different, less specific cleavage patterns (Christensen-Dalsgaard and Gerdes 2006). The HigA antitoxins present a C-terminal DNA binding domain from the HTH-Xre family, clearly visible in the crystal structure of E. coli HigA (Arbing et al. 2010).
Inspection of TA systems in M. tuberculosis revealed that two systems predicted as relBE (http://bioinfo-mml.sjtu.edu.cn/TADB/) are in fact homologous of the hitherto unique M. tuberculosis HigBA1 system, Rv1955-Rv1956, and were thus herein renamed HigBA2 (Rv2022c-Rv2021c) and HigBA3 (Rv3182-Rv3183). The three higBA loci of M. tuberculosis present homology with the gp49-gp48 locus from linear prophage N15 of E. coli which consists of a RelE-like toxin located upstream of an HTH Xre-domain containing antitoxin in the operon, an organization characteristic of HigBA modules (Gerdes et al. 2005; Makarova et al. 2009). While HigB2 has not been investigated yet, conditional expression of the HigB3 toxin in M. smegmatis displayed no apparent inhibition of growth (Fig. 17.1; Ramage et al. 2009). In contrast, HigB1 severely inhibited E. coli, M. smegmatis and M. tuberculosis growth (Gupta 2009; Ramage et al. 2009; Fivian-Hugues and Davis 2010). Remarkably, both higBA1 and higBA2 are among the 10 most upregulated TA systems in M. tuberculosis drug-tolerant persisters and are part of a large genomic island comprising other genes potentially involved in dormancy (Fig. 17.2a; Keren et al. 2011; Stinear et al. 2008). The higBA1 locus is atypical in that it is part of an operon comprising two other genes: the upstream Rv1954a gene of unknown function and the downstream Rv1957 gene encoding a SecB-like molecular chaperone (Fig. 17.2b; Smollet et al. 2009; Bordes et al. 2011). The proposed mechanism of action and activation as well as the evolutionary history of such novel tripartite toxin-antitoxin-chaperone, named TAC system, are presented below.
Genomic context of the M. tuberculosis TAC system. a To-scale representation of the genomic island containing the TAC system, extending from the gene Rv1942 to the gene Rv2028. TA loci are reported under the gene symbols colored in red for the toxin gene and blue for the antitoxin gene. The chaperone encoding gene Rv1957 is colored in green. Genes with potential implication in virulence are highlighted in light gray for the mce3 operon and in black for genes belonging to the dormancy regulon. Genes encoding putative transposases are also shown. b To-scale representation of the operon organization of the genes encoding the TAC system members: higB (378 bp), higA (450 bp) and Rv1957 (576 bp). The promoters P1 and P2 are depicted, as well as the Rv1954a gene of unknown function
17.5 The Atypical Toxin-Antitoxin-Chaperone System of Mycobacterium tuberculosis
The TAC system HigB1-HigA1-Rv1957 of M. tuberculosis is part of a genomic island of approximately 80,760 bp (Stinear et al. 2008), which contains nine other TA systems, including five vapBC, 2 mazEF, 1 unclassified, parDE1, and higBA2 (Fig. 17.2a). Among these, five are functional, and three are among the 10 most induced TA systems in persister cells (Fig. 17.1; Keren et al. 2011). Noticeably, this genomic island also contains potential pathogenicity determinants: one of the four mce operons of M. tuberculosis, mce3, which contains genes thought to be involved in lipid transport, as well as nine genes identified as being part of the dormancy regulon (Fig. 17.2a; de la Paz Santangelo et al. 2009; Voskuil et al. 2003).
17.5.1 Chaperone-Mediated Control of HigB1 Activation Cascade
It was recently shown that both in E. coli and in mycobacteria, the strong inhibition of growth induced by the mycobacterial toxin HigB1 from TAC is fully suppressed by the concerted action of the HigA1 antitoxin and the SecB-like chaperone Rv1957 (Bordes et al. 2011). In this case, the chaperone directly interacts with the HigA1 antitoxin and efficiently protects it from both aggregation and degradation by unknown protease(s) in vivo (Bordes et al. 2011). In agreement with such data, single deletion of the antitoxin gene higA1 in M. tuberculosis is lethal (Fivian-Hughes and Davis 2010), while disruption of Rv1957 alone exhibits a slow growth phenotype most likely due to a reduced antitoxin activity (Sassetti et al. 2003). These results indicate that the SecB-like chaperone Rv1957 indeed facilitates folding of the antitoxin and its subsequent interaction with the toxin (Fig. 17.3a). On the basis of these observations, it seems very likely that the HigB1 activation cascade could be triggered either by a decrease in expression or by direct hijacking of the chaperone at the post-translational level (see proposed model in Fig. 17.3b).
Proposed model for TAC function in M. tuberculosis. a Under permissive conditions, the chaperone (C, green) interacts early with the nascent antitoxin (A, blue), protecting it from both aggregation and degradation by unknown protease(s). The chaperone-bound antitoxin is then competent for toxin (T, red) binding and neutralization. Yet, it is not known whether the chaperone Rv1957 is part of the inactive TA complex or not. As shown at the bottom of the figure, the growth of an E. coli strain expressing the three TAC partners (TAC) in a spot test experiment (dilutions 10−3–10−7) was not affected. b Specific stress conditions could induce a decrease in chaperone level or availability, rapidly resulting in antitoxin degradation and subsequent toxin activation (see text for details). Note that previous results suggest that aggregation may not be physiologically relevant in the presence of active degradation, hence the dotted arrow. A spot test experiment showing the growth inhibition of E. coli expressing higB-higA without chaperone (TA) is shown at the bottom of the figure (adapted from Bordes et al. 2011)
As stated above, Rv1957 shares significant sequence similarities with the export chaperone SecB. In most Gram-negative bacteria, the homotetrameric chaperone SecB binds non-native precursor proteins and specifically transfers them to the SecA subunit of the Sec translocase, thus facilitating their export (Randall and Hardy 2002). Remarkably, the SecB-like chaperone Rv1957 from TAC functionally replaces the chaperone SecB during protein export in E. coli (Bordes et al. 2011). Furthermore, as observed for SecB, Rv1957 is a homotetrameric chaperone, thus reinforcing the fact that both proteins might be evolutionarily related (Bordes et al. 2011). The presence of a well-defined outer membrane and a remarkably large number of putative outer membrane proteins in M. tuberculosis suggests that this bacterium could make use of a functional export chaperone under certain circumstances (Zuber et al. 2008; Niederweis et al. 2010). An attractive hypothesis is that the SecB-like chaperone represents an intimate link between export and activation of stress-responsive toxin–antitoxin systems. In this case, the cytoplasmic accumulation of precursor proteins or the synthesis of stress-specific preproteins could result in recruitment of Rv1957 to assist their efficient targeting to the Sec translocon. Increasing presecretory clients could compete with HigA1 for binding to the chaperone, thus facilitating degradation of the free antitoxin and the subsequent activation of the toxin until normal conditions resume (see proposed model in Fig. 17.3b). Yet, how the mycobacterial SecB-like chaperone responds to stress and controls the HigB1 toxin activation cascade, and to what extent such activation is important for M. tuberculosis virulence and pathogenesis are so far unresolved questions.
17.5.2 Stress Regulation of the TAC Operon
A number of experimental evidences point toward a role for TAC in M. tuberculosis stress adaptive response. Indeed, it has been shown that transcription of the higB1-higA1-Rv1957 operon is induced by several relevant stress conditions including DNA damage (Smollett et al. 2009), heat shock (Stewart et al. 2002), nutrient starvation (Betts et al. 2002), hypoxia (Ramage et al. 2009), drug persistence (Keren et al. 2011), and in host phagocytes (Tailleux et al. 2008).
The TAC operon is under the control of two promoters identified by 5’-RACE (Smollet et al. 2009): higBP1 located 51 nucleotides upstream of the start codon of higB, thus controlling the expression of higB1-higA1-Rv1957, and the more distal promoter higBP2 located 29 nucleotides upstream of the ATG start codon of the upstream Rv1954a gene of unknown function, to control the whole operon (Fig. 17.2b). The DNA damage inducible P1 promoter possesses a RecA-NDp motif characteristic of LexA/RecA-independent genes in M. tuberculosis (Smollett et al. 2009). Interestingly, the Clp protease gene regulator (ClpR) Rv2745c of M. tuberculosis was shown to specifically bind the conserved RecA-NDp motif of several DNA repair genes and to activate transcription of clpC1 and clpP1P2 protease genes as well (Sherrid et al. 2010, Estorninho et al. 2010). These results suggest that ClpR could be part of the regulatory network that controls TAC activation via degradation of the antitoxin (see below). Noticeably, such a mechanism could be of particular interest since in many cases, stress conditions that induce ClpR expression in M. tuberculosis often induce higB (Rustad et al. 2008; Mehra et al. 2010).
As with most TA loci, TAC is autorepressed and this is likely to occur via the binding of HigA1 to a palindromic motif overlapping the −35 element of the higBP2 promoter (Fivian-Hugues and Davis 2010). Yet, it is not established whether HigB1 and/or Rv1957 also participate in this process. The activity of HigA as a global transcriptional regulator was also investigated. Microarray analysis of the ΔTAC mutant revealed that the fadB, Rv3173c, and Rv3662c genes might be directly or indirectly activated by HigA (Fivian-Hugues and Davis 2010). In addition, a one-hybrid reporter system showed that HigA was able to interact specifically with the promoter region of gene clusters containing stress response genes whiB7, rubA, rubB, and fatty acid metabolism genes echA20, fadE28, fadE29, ufaA2, and fad26 (Guo et al. 2009). Finally, ChIP-Seq analysis using FLAG-tagged HigA expressed in M. tuberculosis H37Rv revealed 30 potential HigA binding sites in intergenic regions, including the promoter region of higBP2 and another HigBA locus, higBA3 (http://genome.tbdb.org/annotation/genome/tbdb/RegulatoryNetwork.html). Taken together, these data suggest that as for MqsA in E. coli (Wang et al. 2011), HigA could specifically regulate targets other than its own operon and thus be part of a network of stress-associated regulons involved in M. tuberculosis pathogenesis.
17.5.3 Role of Proteases in TAC Activation
Classical type II toxins are activated following degradation of their cognate antitoxins by stress proteases Lon and/or Clp (Gerdes et al. 2005). However, in the case of TAC, the additional presence of a SecB-like chaperone that assists the antitoxin suggests a more competitive mode of regulation (Fig. 17.3). One of the main questions is thus how the HigB toxin is activated and to what extent stress proteases are involved. Previous work showed that in the absence of the chaperone, the HigA antitoxin was hardly detected in cell extracts, indicating that depletion of the chaperone triggers antitoxin degradation, even in the absence of activating stress conditions (Bordes et al. 2011).
The major stress protease families present in M. tuberculosis significantly differ from those of E. coli. The most striking difference is that M. tuberculosis possesses a 20S proteasome-like degradation machinery to which proteins are addressed by addition of an ubiquitine-like tag called Pup (Prokaryotic ubiquitin-like protein), giving the name of “Pupylation” to this pathway (Pearce et al. 2008). Yet, no evidence for a role of the proteasome in controlling TA activation has been described and none of the TAC components appeared to be pupylated (Poulsen et al. 2010), thus suggesting that the proteasome may not be involved in this case.
In contrast with E. coli, M. tuberculosis and closely related species do not have the AAA+ proteases Lon or ClpYQ. However, they do have two clpP genes, clpP1 and clpP2, encoding the proteolytic subunit, and three ClpP-associated regulatory subunits encoding genes, clpX, clpC1, and clpC2, involved in substrate binding and specificity (Ribeiro-Guimaraes and Pessolani 2007). Remarkably, as for higA1 and higB1, the clpC2 gene is strongly induced in drug-tolerant persisters from M. tuberculosis (Keren et al. 2011), suggesting that ClpC2ClpP AAA+ stress protease could participate in antitoxin degradation upon chaperone depletion or hijacking, as proposed (Fig. 17.2b). Yet, the understanding of the underlying mechanism will first require the identification of such protease(s) involved in the degradation of the HigA1 antitoxin.
17.5.4 Evolutionary Analysis of TAC Systems
In order to study the evolutionary history of the M. tuberculosis TAC system and to analyze the occurrence of such system in the taxonomy, we searched for similar systems using iterative BLASTp, on the basis of sequence similarity and conserved organization and size. Rapidly, we noticed that sequence similarities were often weak (e-value around 10−03). Nevertheless, when the organization and size criteria were filled and when at least two of the three partners showed even weak sequence similarities, the genes were then annotated as a putative TAC system. This approach allowed us to identify 50 putative TAC systems in both the complete and in progress genomes available in the NCBI database (http://www.ncbi.nlm.nih.gov/; Table 17.1). TAC systems are distributed in six phyla: Proteobacteria, Firmicutes, Actinobacteria, Thermotogae, Verrumicrobia, and Synergistetes. Strains containing TAC systems are always in the minority when analyzing phyla, except in Thermotogae. This wide and sparse pattern of distribution through the taxonomy cannot be easily explained by events of vertical transfer and loss, but rather suggests that the TAC systems were acquired by horizontal gene transfer. This is in agreement with TAC localization on a large genomic island in strains from the M. tuberculosis complex (Fig. 17.2a). Moreover, 4 TAC systems are plasmid-borne and thus potentially involved in plasmid stability (Table 17.1). Given these observations, the evolutionary history of TAC systems is likely to be highly complex and largely mediated by mobile genetic elements (manuscript in preparation).
We explored existing links between solitary SecB and chaperones from TAC and found that all TAC chaperones present significant homology links with the SecB chaperone family (Pfam02556; http://pfam.sanger.ac.uk). In addition, we noticed that in some δ-proteobacteria TAC chaperones were very similar to the known SecB consensus and were already annotated as SecB on these genomes (manuscript in preparation). Taken together, these observations further confirm the evolutionarily link between the SecB and TAC chaperones.
We also classified the TAC antitoxins according to their sequence similarity and found that three well-defined antitoxin families could be associated with SecB-like chaperones, namely HigA, MqsA, and HicB (manuscript in preparation). Interestingly, all the TA systems associated with a SecB-like chaperone identified to date present an unusual genetic organization with the toxin gene being upstream of the antitoxin gene in the operon. This organization is thought to be unfavorable to the formation of a “protective” TA complex since the stable toxin is synthesized before the unstable and protease-sensitive antitoxin. In the case of TAC, the dedicated chaperone could maintain a pool of native antitoxin protected from proteolysis as part of the antitoxin-chaperone complex and thus competent for the neutralization of the toxin as soon as it emerges from the ribosome.
17.6 Concluding Remarks
The remarkable prevalence of multiple chromosomally encoded type II TA systems in bacterial genomes raises questions about their role in cell physiology. The fact that many TA systems respond to stress conditions encountered during the infection process and that expression of toxins often leads to persistence phenomena alludes to possible roles in bacterial virulence. Remarkably, one of the main features of M. tuberculosis pathogenesis is its ability to persist long term in the host granulomas in a non-replicating and drug-tolerant state, and later awaken to cause disease. The presence of at least 75 TA systems in its chromosome, of which 37 appear to be functional in vivo (Fig. 17.1), suggests that such intricate networks of stress-responsive elements could indeed participate in M. tuberculosis virulence and thus represent potentially promising new drug targets. Further studies are clearly warranted to address such fundamental questions.
References
Ahidjo, B. A., Kuhnert, D., McKenzie, J. L., Machowski, E. E., Gordhan, B. G., Arcus, V., et al. (2011). VapC toxins from Mycobacterium tuberculosis are ribonucleases that differentially inhibit growth and are neutralized by cognate VapB antitoxins. PLoS ONE, 6, e21738.
Arbing, M. A., Handelman, S. K., Kuzin, A. P., Verdon, G., Wang, C., Su, M., et al. (2010). Crystal structures of Phd-Doc, HigA, and YeeU establish multiple evolutionary links between microbial growth-regulating toxin–antitoxin systems. Structure, 18, 996–1010.
Arcus, V. L., McKenzie, J. L., Robson, J., & Cook, G. M. (2011). The PIN-domain ribonucleases and the prokaryotic VapBC toxin–antitoxin array. Protein Engineering, Design & Selection, 24, 33–40.
Audoly, G., Vincentelli, R., Edouard, S., Georgiades, K., Mediannikov, O., Gimenez, G., et al. (2011). Effect of rickettsial toxin VapC on its eukaryotic host. PLoS ONE, 6, e26528.
Barry, C. E., 3rd, Boshoff, H. I., Dartois, V., Dick, T., Ehrt, S., Flynn, J., et al. (2009). The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nature Reviews Microbiology, 7, 845–855.
Betts, J. C., Lukey, P. T., Robb, L. C., McAdam, R. A., & Duncan, K. (2002). Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Molecular Microbiology, 43, 717–731.
Blower, T. R., Salmond, G. P., & Luisi, B. F. (2011). Balancing at survival’s edge: the structure and adaptive benefits of prokaryotic toxin–antitoxin partners. Current Opinion in Structural Biology, 21, 109–118.
Bordes, P., Cirinesi, A. M., Ummels, R., Sala, A., Sakr, S., Bitter, W., et al. (2011). SecB-like chaperone controls a toxin–antitoxin stress-responsive system in Mycobacterium tuberculosis. Proceedings of the National Academy of Sciences of the United States of America, 108, 8438–8443.
Brown, B. L., Grigoriu, S., Kim, Y., Arruda, J. M., Davenport, A., Wood, T. K., et al. (2009). Three dimensional structure of the MqsR:MqsA complex: a novel TA pair comprised of a toxin homologous to RelE and an antitoxin with unique properties. PLoS Pathogens, 5, e1000706.
Christensen-Dalsgaard, M., & Gerdes, K. (2006). Two higBA loci in the Vibrio cholerae superintegron encode mRNA cleaving enzymes and can stabilize plasmids. Molecular Microbiology, 62, 397–411.
Dahl, J. L., Kraus, C. N., Boshoff, H. I., Doan, B., Foley, K., Avarbock, D., et al. (2003). The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proceedings of the National Academy of Sciences of the United States of America, 100, 10026–10031.
de la Paz Santangelo, M., Klepp, L., Nunez-Garcia, J., Blanco, F. C., Soria, M., Garcia-Pelayo, M. C., et al. (2009). Mce3R, a TetR-type transcriptional repressor, controls the expression of a regulon involved in lipid metabolism in Mycobacterium tuberculosis. Microbiology, 155, 2245–2255.
Estorninho, M., Smith, H., Thole, J., Harders-Westerveen, J., Kierzek, A., Butler, R. E., et al. (2010). ClgR regulation of chaperone and protease systems is essential for Mycobacterium tuberculosis parasitism of the macrophage. Microbiology, 156, 3445–3455.
Fivian-Hughes, A. S., & Davis, E. O. (2010). Analyzing the regulatory role of the HigA antitoxin within Mycobacterium tuberculosis. Journal of Bacteriology, 192, 4348–4356.
Georgiades, K., & Raoult, D. (2011). Genomes of the most dangerous epidemic bacteria have a virulence repertoire characterized by fewer genes but more toxin–antitoxin modules. PLoS ONE, 6, e17962.
Gerdes, K., Christensen, S. K., & Lobner-Olesen, A. (2005). Prokaryotic toxin–antitoxin stress response loci. Nature Reviews Microbiology, 3, 371–382.
Goulard, C., Langrand, S., Carniel, E., & Chauvaux, S. (2010). The Yersinia pestis chromosome encodes active addiction toxins. Journal of Bacteriology, 192, 3669–3677.
Grady, R., & Hayes, F. (2003). Axe-Txe, a broad-spectrum proteic toxin–antitoxin system specified by a multidrug-resistant, clinical isolate of Enterococcus faecium. Molecular Microbiology, 47, 1419–1432.
Guo, M., Feng, H., Zhang, J., Wang, W., Wang, Y., Li, Y., et al. (2009). Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Research, 19, 1301–1308.
Gupta, A. (2009). Killing activity and rescue function of genome-wide toxin–antitoxin loci of Mycobacterium tuberculosis. FEMS Microbiology Letters, 290, 45–53.
Hazan, R., Sat, B., & Engelberg-Kulka, H. (2004). Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. Journal of Bacteriology, 186, 3663–3669.
Hood, R. D., Singh, P., Hsu, F., Guvener, T., Carl, M. A., Trinidad, R. R., et al. (2010). A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host & Microbe, 7, 25–37.
Huang, F., & He, Z. G. (2010). Characterization of an interplay between a Mycobacterium tuberculosis MazF homolog, Rv1495 and its sole DNA topoisomerase I. Nucleic Acids Research, 38, 8219–8230.
Hurley, J. M., & Woychik, N. A. (2009). Bacterial toxin HigB associates with ribosomes and mediates translation-dependent mRNA cleavage at A-rich sites. Journal of Biological Chemistry, 284, 18605–18613.
Jiang, Y., Pogliano, J., Helinski, D. R., & Konieczny, I. (2002). ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Molecular Microbiology, 44, 971–979.
Kamada, K., & Hanaoka, F. (2005). Conformational change in the catalytic site of the ribonuclease YoeB toxin by YefM antitoxin. Molecular Cell, 19, 497–509.
Keren, I., Minami, S., Rubin, E., & Lewis, K. (2011). Characterization and transcriptome analysis of Mycobacterium tuberculosis persisters. MBio, 2, 00100–00111.
Korch, S. B., Contreras, H., & Clark-Curtiss, J. E. (2009). Three Mycobacterium tuberculosis Rel toxin–antitoxin modules inhibit mycobacterial growth and are expressed in infected human macrophages. Journal of Bacteriology, 191, 1618–1630.
Kumar, P., Issac, B., Dodson, E. J., Turkenburg, J. P., & Mande, S. C. (2008). Crystal structure of Mycobacterium tuberculosis YefM antitoxin reveals that it is not an intrinsically unstructured protein. Journal of Molecular Biology, 383, 482–493.
Lewis, K. (2010). Persister cells. Annual Review of Microbiology, 64, 357–372.
Maisonneuve, E., Shakespeare, L. J., Jorgensen, M. G., & Gerdes, K. (2011). Bacterial persistence by RNA endonucleases. Proceedings of the National Academy of Sciences of the United States of America, 108, 13206–13211.
Makarova, K. S., Wolf, Y. I., & Koonin, E. V. (2009). Comprehensive comparative-genomic analysis of type 2 toxin–antitoxin systems and related mobile stress response systems in prokaryotes. Biology Direct, 4, 19.
McKenzie, J.L., Robson, J., Berney, M., Smith, T.C., Ruthe, A., Gardner, P.P., et al. (2012) A VapBC toxin-antitoxin module is a post-transcriptional regulator of metabolic flux in mycobacteria. Journal of Bacteriology, 194, 2189–21204.
Mehra, S., Pahar, B., Dutta, N. K., Conerly, C. N., Philippi-Falkenstein, K., Alvarez, X., et al. (2010). Transcriptional reprogramming in nonhuman primate (rhesus macaque) tuberculosis granulomas. PLoS ONE, 5, e12266.
Miallau, L., Faller, M., Chiang, J., Arbing, M., Guo, F., Cascio, D., et al. (2009). Structure and proposed activity of a member of the VapBC family of toxin–antitoxin systems. VapBC-5 from Mycobacterium tuberculosis. Journal of Biological Chemistry, 284, 276–283.
Mutschler, H., Gebhardt, M., Shoeman, R. L., & Meinhart, A. (2011). A novel mechanism of programmed cell death in bacteria by toxin–antitoxin systems corrupts peptidoglycan synthesis. PLoS Biology, 9, e1001033.
Niederweis, M., Danilchanka, O., Huff, J., Hoffmann, C., & Engelhardt, H. (2010). Mycobacterial outer membranes: in search of proteins. Trends in Microbiology, 18, 109–116.
Pandey, D. P., & Gerdes, K. (2005). Toxin–antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Research, 33, 966–976.
Park, J. H., Yamaguchi, Y., & Inouye, M. (2011). Bacillus subtilis MazF-bs (EndoA) is a UACAU-specific mRNA interferase. FEBS Letters, 585, 2526–2532.
Pearce, M. J., Mintseris, J., Ferreyra, J., Gygi, S. P., & Darwin, K. H. (2008). Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science, 322, 1104–1107.
Poulsen, C., Akhter, Y., Jeon, A. H., Schmitt-Ulms, G., Meyer, H. E., Stefanski, A., et al. (2010). Proteome-wide identification of mycobacterial pupylation targets. Molecular System Biology, 6, 386.
Ramage, H. R., Connolly, L. E., & Cox, J. S. (2009). Comprehensive functional analysis of Mycobacterium tuberculosis toxin–antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genetics, 5, e1000767.
Randall, L. L., & Hardy, S. J. (2002). SecB, one small chaperone in the complex milieu of the cell. Cellular & Molecular Life Sciences, 59, 1617–1623.
Ribeiro-Guimaraes, M. L., & Pessolani, M. C. (2007). Comparative genomics of mycobacterial proteases. Microbial Pathogenesis, 43, 173–178.
Roberts, R. C., & Helinski, D. R. (1992). Definition of a minimal plasmid stabilization system from the broad-host-range plasmid RK2. Journal of Bacteriology, 174, 8119–8132.
Rustad, T. R., Harrell, M. I., Liao, R., & Sherman, D. R. (2008). The enduring hypoxic response of Mycobacterium tuberculosis. PLoS ONE, 3, e1502.
Sassetti, C. M., Boyd, D. H., & Rubin, E. J. (2003). Genes required for mycobacterial growth defined by high density mutagenesis. Molecular Microbiology, 48, 77–84.
Shah, D., Zhang, Z., Khodursky, A., Kaldalu, N., Kurg, K., & Lewis, K. (2006). Persisters: a distinct physiological state of E. coli. BMC Microbiology, 6, 53.
Sharp, J.D., Cruz, J.W., Raman, S., Inouye, M., Husson, R.N. and Woychik, N.A. (2012) Growth and translation inhibition through sequence specific RNA binding by a Mycobacterium tuberculosis VAPC toxin. Journal of Biological Chemistry, 287, 12835–12847.
Sherrid, A. M., Rustad, T. R., Cangelosi, G. A., & Sherman, D. R. (2010). Characterization of a Clp protease gene regulator and the reaeration response in Mycobacterium tuberculosis. PLoS ONE, 5, e11622.
Singh, R., Barry, C. E., 3rd, & Boshoff, H. I. (2010). The three RelE homologs of Mycobacterium tuberculosis have individual, drug-specific effects on bacterial antibiotic tolerance. Journal of Bacteriology, 192, 1279–1291.
Smollett, K. L., Fivian-Hughes, A. S., Smith, J. E., Chang, A., Rao, T., & Davis, E. O. (2009). Experimental determination of translational start sites resolves uncertainties in genomic open reading frame predictions—application to Mycobacterium tuberculosis. Microbiology, 155, 186–197.
Stewart, G. R., Wernisch, L., Stabler, R., Mangan, J. A., Hinds, J., Laing, K. G., et al. (2002). Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology, 148, 3129–3138.
Stinear, T. P., Seemann, T., Harrison, P. F., Jenkin, G. A., Davies, J. K., Johnson, P. D., et al. (2008). Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Research, 18, 729–741.
Tailleux, L., Waddell, S. J., Pelizzola, M., Mortellaro, A., Withers, M., Tanne, A., et al. (2008). Probing host pathogen cross-talk by transcriptional profiling of both Mycobacterium tuberculosis and infected human dendritic cells and macrophages. PLoS ONE, 3, e1403.
Tian, Q. B., Ohnishi, M., Tabuchi, A., & Terawaki, Y. (1996). A new plasmid-encoded proteic killer gene system: cloning, sequencing, and analyzing hig locus of plasmid Rts1. Biochemical & Biophysical Research Communications, 220, 280–284.
Van Melderen, L. (2010). Toxin–antitoxin systems: why so many, what for? Current Opinion in Microbiology, 13, 781–785.
Vesper, O., Amitai, S., Belitsky, M., Byrgazov, K., Kaberdina, A. C., Engelberg-Kulka, H., et al. (2011). Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell, 147, 147–157.
Voskuil, M. I., Schnappinger, D., Visconti, K. C., Harrell, M. I., Dolganov, G. M., Sherman, D. R., et al. (2003). Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. Journal of Experimental Medicine, 198, 705–713.
Wang, X., Kim, Y., Hong, S. H., Ma, Q., Brown, B. L., Pu, M., et al. (2011). Antitoxin MqsA helps mediate the bacterial general stress response. Nature Chemical Biology, 7, 359–366.
Winther, K. S., & Gerdes, K. (2011). Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proceedings of the National Academy of Sciences of the United States of America, 108, 7403–7407.
Wozniak, R. A., & Waldor, M. K. (2009). A toxin–antitoxin system promotes the maintenance of an integrative conjugative element. PLoS Genetics, 5, e1000439.
Yamaguchi, Y., & Inouye, M. (2011). Regulation of growth and death in Escherichia coli by toxin–antitoxin systems. Nature Reviews Microbiology, 9, 779–790.
Yamaguchi, Y., Park, J. H., & Inouye, M. (2011). Toxin–antitoxin systems in bacteria and archaea. Annual Review of Genetics, 45, 61–79.
Yang, M., Gao, C., Wang, Y., Zhang, H., & He, Z. G. (2010). Characterization of the interaction and cross-regulation of three Mycobacterium tuberculosis RelBE modules. PLoS ONE, 5, e10672.
Yoshizumi, S., Zhang, Y., Yamaguchi, Y., Chen, L., Kreiswirth, B. N., & Inouye, M. (2009). Staphylococcus aureus YoeB homologues inhibit translation initiation. Journal of Bacteriology, 191, 5868–5872.
Zhang, Y., Zhu, L., Zhang, J., & Inouye, M. (2005). Characterization of ChpBK, an mRNA interferase from Escherichia coli. Journal of Biological Chemistry, 280, 26080–26088.
Zhu, L., Phadtare, S., Nariya, H., Ouyang, M., Husson, R. N., & Inouye, M. (2008). The mRNA interferases, MazF-mt3 and MazF-mt7 from Mycobacterium tuberculosis target unique pentad sequences in single-stranded RNA. Molecular Microbiology, 69, 559–569.
Zhu, L., Sharp, J. D., Kobayashi, H., Woychik, N. A., & Inouye, M. (2010). Noncognate Mycobacterium tuberculosis toxin–antitoxins can physically and functionally interact. Journal of Biological Chemistry, 285, 39732–39738.
Zhu, L., Zhang, Y., Teh, J. S., Zhang, J., Connell, N., Rubin, H., et al. (2006). Characterization of mRNA interferases from Mycobacterium tuberculosis. Journal of Biological Chemistry, 281, 18638–18643.
Zuber, B., Chami, M., Houssin, C., Dubochet, J., Griffiths, G., & Daffe, M. (2008). Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. Journal of Bacteriology, 190, 5672–5680.
Acknowledgments
We thank Virginie Calderon and all the members of the Genevaux laboratory for insightful discussions. This work was supported by a French MENRT fellowship to AS and an ATIP-CNRS grant to PG.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Sala, A., Bordes, P., Fichant, G., Genevaux, P. (2013). Toxin-Antitoxin Loci in Mycobacterium tuberculosis . In: Gerdes, K. (eds) Prokaryotic Toxin-Antitoxins. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-33253-1_17
Download citation
DOI: https://doi.org/10.1007/978-3-642-33253-1_17
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-33252-4
Online ISBN: 978-3-642-33253-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)