Abstract
The major targets for direct-acting antivirals (DAAs) are the NS3/4A protease, the NS5A protein, and the NS5B polymerase. The latter enzyme offers several target sites: the catalytic domain for nucleoside/nucleotide analogs and different allosteric sites for non-nucleoside inhibitors. Two protease inhibitors have already been approved and more than 40 new NS3/4A, NS5A, or NS5B inhibitors are in development pipeline. Not only these agents can achieve very high cure rates when combined with PEG-IFN and RBV, but have also started to provide promising results when combined in IFN-free, all-oral combinations. In addition to the more canonical drug targets, new alternative viral targets for small molecule drug development are emerging, such as p7 or NS4B. Current research is focusing on defining the most efficacious DAA combination regimens, i.e., those which provide the highest rates of viral eradication, broadest spectrum of action, minimal or no clinical resistance, shortest treatment duration, and good tolerability.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
1 Introduction
With an estimated prevalence of HCV infection of 2 % worldwide (Shepard et al. 2005), chronic infection with hepatitis C virus (HCV) represents a major public health threat and a significant challenge for the physician and the patient. Until recently, the therapeutic regimens used to treat chronic infection have been based on the use of different forms of pegylated interferon-α (PEG-IFN) combined with the broad-spectrum antiviral ribavirin (RBV). These regimens not only are poorly tolerated and contraindicated in a high number of patients, but their effectiveness in eradicating the infection is limited to a fraction of the individuals who are eligible for therapy (see chapter “Treatment of Chronic Hepatitis C: Current and Future” by Pawlotsky, this volume, for further details). Therefore, there is an obvious and urgent need to develop more effective and tolerated treatments.
In order to address this need, new direct-acting antiviral agents (DAAs) are being developed that target specific HCV enzymes and proteins. With the recent approval of the first two NS3/4A oral protease inhibitors [boceprevir (Chang et al. 2012) and telaprevir (Forestier and Zeuzem 2012)], for the treatment of HCV genotype 1 infections in combination with a PEG-IFN/RBV backbone, we have witnessed a tremendous advance in the pharmacotherapy of chronic hepatitis C. With this new triple therapy regimen, the patient cure rates for HCV genotype 1 infections have increased from around 20–30 % to around 70–80 %, while significantly reducing treatment duration (Lee et al. 2012). However, these DAA-containing regimens still exhibit severe limitations: (1) they increase the spectrum of serious side effects associated with anti-HCV therapy; (2) they are associated with a huge pill-burden and complex dosing schedule; (3) they are limited to genotype 1 infections; (4) selection of DAA-resistant viral variants does occur in patients who respond poorly to the PEG-IFN/RBV component of the triple therapy (for further clinical details see chapter “Treatment of Chronic Hepatitis C: Current and Future” by Pawlotsky, this volume).
In order to overcome these limitations, a number of other DAAs are in development with the objective to develop therapeutic regimens that are more efficacious and convenient, better tolerated, active on all viral genotypes, and with a negligible likelihood to develop viral resistance. These are primarily targeted at the NS3/4A protease, NS5A protein, or NS5B RNA-dependent RNA polymerase. Additionally, other, less-studied viral proteins, such as the ion channel formed by p7 or the replication complex protein NS4B have been recently demonstrated to be “druggable”, providing potential new targets for pharmacological intervention (further details about the molecular properties of HCV proteins are given in chapter “Hepatitis C Virus Proteins From Structure to Function”by Moradpour and Penin, this volume). The ultimate goal of the drug discovery effort in HCV research will be that of developing a highly efficacious, IFN-free all-oral therapy and—to this aim—several clinical trials, combining only oral antivirals, have started to show very promising results.
In this chapter, we summarize the progress toward the development of novel HCV-targeted antiviral agents targeting the NS3/4A serine protease, the NS5A protein, the NS5B RNA-dependent RNA polymerase, the p7 ion channel, or the NS4B protein, with particular emphasis on those compound classes or combinations that have shown the most encouraging antiviral activity in the clinic.
2 Inhibitors of the NS3/4A Serine Protease
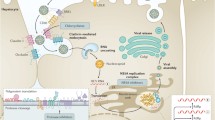
NS3 is a multifunctional protein that contains a serine protease domain in its ~180 N-terminal amino acids. The remainder of the protein encompasses an RNA helicase. The NS3 serine protease domain is, in turn, one component of a heterodimeric serine protease that requires the noncovalently associated viral protein NS4A for optimal catalytic activity (Failla et al. 1994). Accordingly, this viral enzyme is currently referred to as the NS3/4A protease (detailed in the chapter “Hepatitis C Virus Proteins: From Structure to Function” by Moradpour and Penin, this volume). The activity of the NS3/4A protease is responsible for the proteolytic cleavage of the viral polyprotein at the four junctions NS3-NS4A, NS5A-NS5B, NS4A-NS4B, and NS4B-NS5A (Tomei et al. 1993). The protease cofactor, NS4A, is a relatively small protein, consisting of only 54 residues. The first ~20 residues of NS4A are hydrophobic and form a transmembrane α-helix that is thought to anchor the NS3/4A protease/helicase complex to the outer membrane of the endoplasmic reticulum. A short, central domain of NS4A, amino acids 21–34, are directly implicated in the interaction with the NS3 protease domain and critically required for full enzymatic activity (Failla et al. 1995; Lin et al. 1995). Similar to other trypsin-like serine proteases, NS3/4A is made of two domains both composed of a ß-barrel and two short α-helices with the catalytic triad located in a crevice formed at the interface between the two domains (Kim et al. 1996; Love et al. 1996; Yan et al. 1998) (Fig. 1a). The central region of NS4A is an integral part of the protease domain. It forms one strand of an eight-stranded ß-barrel and increases the enzymatic activity of the protease by stabilizing the N-terminal domain of NS3 and by contributing to the formation of the substrate recognition site (De Francesco and Steinkuhler 2000).
Three-dimensional structures of the main targets for the development of HCV-specific direct-acting antivirals. a NS3/4A protease domain [PDB entry: 3SU3]. NS3 is depicted in green color. The NS4A cofactor is shown in red. The structural Zn++ ion is modeled in gray. Vaniprevir, a P4-P2 macrocyclic noncovalent NS4/4A protease inhibitor, is bound in the enzyme active site. The amino acids corresponding to the main resistance mutations are highlighted and numbered. b Two alternative dimeric conformations of NS5A protein Domain I [PDB entry: 1ZH1 (left) and 3FQM (right)]. The structural Zn++ ion is modeled in gray. Amino acids corresponding to main resistance mutations are highlighted and numbered. Substitutions of Y93 represent primary mutations, whereas mutations of position P58 act as a secondary mutation (Fridell et al. 2010). Other positions that are mutated in NS5A-resistant HCV variants (i.e., M28, Q30, L31, and P32) are not visible in the Domain I crystallographic structures. c Structure of the HCV RNA-dependent RNA polymerase and binding sites for nucleoside/nucleotide and non-nucleoside polymerase inhibitors. The palm, fingers, and thumb domains are colored green, red, and red blue, respectively. Mn++ ions in the active site are modeled in gray. Amino acids corresponding to resistance mutations in different regions have a different color: pink for P495, P496, and V499 (Thumb I site), light blue for L419, M423, and I482 (Thumb II site), yellow for N411, M414, Y448, and Y452 (Palm I site/beta-hairpin loop), and green for C316 and S365 (Palm II site)
The NS3/NS4A-dependent cleavage sites of the HCV polyprotein have the consensus sequence Asp/Glu-(Xaa)4-Cys/Thr↓Ser/Ala-(Xaa)2-Leu/Trp/Tyr, with the most efficient cleavage occurring after cysteine. The primary specificity of a protease is defined by the side chain of the amino acid that precedes the scissile bond (i.e., the P1 position; we follow the nomenclature of Schechter and Berger (Schechter and Berger 1967) in designating the cleavage sites as P3-P2-P1 … P1′-P2′-P3′, with the cleaved peptide bond between P1 and P1′ and the C-terminus of the substrate on the prime site). The specificity for the P1 amino acid is imposed by the features of the complementary S1 pocket on the enzyme. The S1 pocket of the NS3/4A protease is shallow and closed at its bottom by a bulky phenylalanine residue, explaining the preference of the NS3 protease for Cys in the substrate P1 position. In general, the substrate binding site of the NS3/4A serine protease appears to be relatively flat and featureless. Because of the absence of well-defined cavities in the substrate recognition site, the substrate sequence is specifically recognized owing to a series of weak molecular interactions distributed along a rather large surface. This mechanism of substrate recognition, more reminiscent of a protein-protein interaction compared to that typical for enzyme small molecule-substrate docking, has represented a very difficult starting point for the development of small molecule inhibitors of the enzyme. In spite of the formidable challenges encountered in the discovery of inhibitors of this target, a number of highly potent and efficacious active site inhibitors have now been developed.
From a chemical point of view, NS3/4A protease inhibitors can be divided into three main categories (Fig. 2): (1) linear peptidomimetics with an alpha-ketoamide group that bind the enzyme active site covalently but reversibly, (2) linear noncovalent peptidomimetic inhibitors, and (3) macrocyclic noncovalent peptidomimetic inhibitors. Macrocyclic inhibitors can be further classified as P3-P1 macrocycles or P4-P2 macrocycles. The clinical proof of concept for NS3/4A protease inhibitors was first achieved with ciluprevir (BILN 2061), a P3-P1 macrocyclic inhibitor that showed substantial antiviral activity in patients with HCV genotype 1 (Lamarre et al. 2003). However, ciluprevir was not further developed due to serious cardiotoxicity observed in a preclinical model (Goudreau and Llinas-Brunet 2005). The first NS3/4A inhibitors that received approval for use in chronic hepatitis C, telaprevir and boceprevir, both belong to the linear covalent inhibitor class. In the scientific community, telaprevir and boceprevir are also often referred to as “first-wave” NS3/4A protease inhibitors in order to distinguish them from the second-wave and/or second-generation of agents being developed.
2.1 First-Generation NS3/4A HCV Protease Inhibitors
First-generation NS3/4A protease inhibitors are defined as agents that (1) display potent antiviral activity on HCV genotype 1, but (2) oppose a low genetic barrier to selection of resistant viral variants, and (3) have a restricted spectrum of action on other viral genotypes, especially genotype 3. First-generation NS3/4A inhibitors can be in turn classified as belonging to the “first wave” (linear covalent inhibitors) or to the “second wave” (linear noncovalent or macrocyclic inhibitors).
2.1.1 First-Wave NS3/4A HCV Protease Inhibitors: Boceprevir and Telaprevir
Several groups have derived covalent serine protease inhibitors from known substrates by replacing the scissile amide bond with an electrophilic moiety. As a consequence of the catalytic action of the enzyme, such a molecular “warhead” is able to form a covalent adduct with the catalytic serine residue (Edwards and Bernstein 1994). Compounds with this mechanism of action are also referred to as “serine-trap inhibitors”. Several biopharmaceutical groups have reported a series of substrate-based, electrophile-based peptidic or peptidomimetic inhibitors of the NS3/4A protease, with electrophilic groups that included aldehydes, boronates, lactams, α-keto amides, and α-keto acids [reviewed in (Chen and Tan 2005)]. Here, we will focus on α-keto amides, since this is the warhead of telaprevir and boceprevir.
One very important mechanistic feature of α-keto amide protease inhibitors is that, in contrast to some other classes of covalent serine traps, the covalent bond formed with the enzyme catalytic serine is entirely reversible. Briefly, this compound class binds to the enzyme active site via a 2-step mechanism, wherein the initial transient formation of a noncovalent enzyme-inhibitor complex is followed by a slow rearrangement to a covalently bound hemiketal complex. The covalent complex undergoes then a very slow dissociation step. Unlike noncovalent inhibitors, covalently bound inhibitor-NS3/4A complexes are thus very long-lived, with half-lives ranging from several minutes to hours. This particular property might at least in part explain the striking antiviral efficacy associated with some of the compound in this class in spite of relatively weak binding affinities.
The evolution of α-ketoamide-based inhibitors of the NS3/4A protease, described in detail in a number of reviews (Chen and Tan 2005; Lin et al. 2006), has culminated in the development of boceprevir (Chang et al. 2012) and telaprevir (Forestier and Zeuzem 2012) (Fig. 2a). In May 2011, the NS3/4A protease inhibitors telaprevir and boceprevir gained approval by the US Food and Drug Administration for the treatment of patients chronically infected with HCV genotype 1, in combination with PEG-IFN and RBV. Both agents greatly improve rates of sustained viral response (SVR) among treatment-naïve as well as treatment-experienced patients. While addition of either one of these new agents to the standard PEG-IFN/RBV therapy has pushed SVR rates to unprecedented levels, these drugs still have a number of liabilities, including serious side effects (rash for telaprevir, dysgeusia for boceprevir, and anemia for telaprevir and boceprevir), suboptimal pharmacokinetic properties (dosing every 8 h is required), and low barrier to resistance (Aghemo et al. 2012) (described in more detail in the chapter “Treatment of Chronic Hepatitis C: Current and Future” by Pawlotsky, this volume). Mutations associated with in vitro as well as clinical resistance to telaprevir or boceprevir were identified at several positions close to the NS3 protease active site (Fig. 1a), including V36A/M/L, T54A/S, R155K/M/S/T, A156S (all four conferring low- to medium-level resistance), and A156T/Y (conferring high-level resistance) (Kieffer et al. 2007; Kwo et al. 2010; Sarrazin et al. 2007; Susser et al. 2009). Additionally, these agents have a very limited spectrum of action with respect to the different viral genotypes. For example, in clinical trials, telaprevir displayed activity against HCV genotype 2, but not genotype 3 (Foster et al. 2011a). Moreover, a small, randomized trial enrolling genotype 4 patients showed that addition of telaprevir to PEG-IFN/RBV only had a modest effect on SVR rates (Benhamou et al. 2009).
2.1.2 Second-Wave NS3/4A Protease Inhibitors
A number of so-called second-wave NS3/4A protease inhibitors are currently being studied in Phase II or III clinical trials. They include: linear noncovalent inhibitors faldaprevir/BI 201335 (White et al. 2010), asunaprevir/BMS-650032 (McPhee et al. 2012b), sovaprevir/ACH-1625 (Agarwal et al. 2012), and GS-9451 (Sheng et al. 2012a); P3-P1 macrocyclic inhibitors simeprevir/TMC435 (Lin et al. 2009), danoprevir/RG7227/ITMN-191 (Seiwert et al. 2008), ABT-450 (Wagaw et al. 2009), and GS-9256 (Sheng et al. 2012b); and P4-P2 macrocyclic inhibitor vaniprevir/MK-7009 (Liverton et al. 2010) (Fig. 2b–d). These agents are characterized by very potent activity on HCV genotype 1 [typically, low-nM EC50 in the replicon system (chapter “Cell Culture Systems for Hepatitis C Virus” by Steinmann and Pietschmann, this volume)] as well as antiviral efficacy on HCV genotype 1 patients similar to that of boceprevir or telaprevir (Lee et al. 2012; Sarrazin et al. 2012; Schaefer and Chung 2012). They differ from their first-wave counterpart in that they do not have the chemical reactivity required to make a covalent bond to their target and are therefore expected to display fewer- and less severe side effects. In addition, these agents have substantially improved pharmacokinetic profiles, which allows for less frequent dosing, typically once a day [qd: simeprevir (Reesink et al. 2010), faldaprevir (Manns et al. 2011), sovaprevir (Agarwal et al. 2012), ABT-450 (Poordad et al. 2012a), and GS-9451 (Dvory-Sobol et al. 2012)] or twice a day [bid: vaniprevir, danoprevir (Forestier et al. 2011), asuprenavir (Pasquinelli et al. 2012), and GS-9256 (Zeuzem et al. 2012a)]. It should be pointed out that low-dose ritonavir boosting is used with danoprevir (Reddy et al. 2012) and ABT-450 (Lawitz et al. 2010a) in order to decrease dosing frequency and to reduce drug-associated side effects.
Although second wave-first generation NS3/4A HCV protease inhibitors tend to have a significantly broader spectrum of action on the different HCV genotypes compared to the inhibitor of the first wave, these agents cannot be considered pan-genotypic antivirals since they do not exhibit the same potency across all viral genotypes. For example, simeprevir and faldaprevir have been reported to have a rather broad spectrum of action in biochemical assays, with variable in vitro activity against NS3/4A protease of HCV genotypes 1–6 (Reesink et al. 2010; White et al. 2010). For both compounds, the lowest inhibitory potency was observed against genotype 3. Simeprevir has also been investigated in patients infected with genotypes 2–6 viruses. In line with the in vitro data, medium to high antiviral activities (over −2.0 log10 IU/ml in all instances) were observed against genotypes 2, 4, 5, and 6, whereas the drug had no effect in patients infected with HCV genotype 3 (Moreno et al. 2012).
Along with the restricted genotype coverage, the genetic barrier posed to resistance by all first-generation NS3/4 protease inhibitors is low and extensive cross-resistance is observed between the different compounds or compound classes (Sarrazin and Zeuzem 2010). First-wave and second-wave NS3/4A protease inhibitors show slightly different resistance mutation patterns. In particular, mutations of Val 36 (V36A/M) or Thr 54 (T54S/A) have been exclusively observed in association with covalent, linear inhibitors of the first wave. Mutations at position Arg 155 (R155K/T/Q) have been shown to confer broad cross-resistance to all first-generation inhibitors. Conversely, mutations of Asp 168 (D168/E/G/H/T/Y) are specifically found to confer mutation to noncovalent peptidomimetic inhibitors, regardless whether of linear or macrocyclic structure (Dvory-Sobol et al. 2012; Lagace et al. 2012; Lenz et al. 2010; Lin et al. 2004; McPhee et al. 2012a; Sarrazin and Zeuzem 2010). This is believed to occur because of the interaction between the very large P2 substituent, common to all these inhibitors, with D168 in the enzyme active site (Romano et al. 2012). Notably, D168 is one of the few active site residues not entirely conserved among HCV genotypes, being replaced by glutamine in isolates belonging to genotype 3. This could partly explain why HCV genotype 3 is “naturally resistant” to virtually all first-generation, second-wave protease inhibitors (Trozzi et al. 2003). Agents belonging to this second-wave of first-generation NS3/4A protease inhibitors have showed high rates of SVR in HCV genotype 1 patients, when used in combination with PEG-IFN and RBV, of the same order of magnitude or higher than those reported for boceprevir or telaprevir triple-combination regimens (Lee et al. 2012; Schaefer and Chung 2012). Simeprevir and faldaprevir are currently in Phase III clinical evaluation for their use in PI/PEG-IFN/RBV combination to treat HCV genotype 1 chronic infection.
2.2 Second-Generation NS3/4A Protease Inhibitors
Second-generation NS3/4A protease inhibitors are defined as agents that (1) pose a high barrier to the development of viral resistance, (2) retain activity against the viral variants that are resistant to first-generation compounds, and (3) are active across the different genotypes, including HCV genotype 3.
MK-5172 (Fig. 2d) is a second-generation NS3/4A protease inhibitor with pan-genotype antiviral activity and improved resistance profile (Summa et al. 2012). Importantly, in biochemical assays, this agent maintained potent antiviral activity against the predominant amino acid substitutions known to confer resistance to first-generation protease inhibitors. Thus, MK-5172 remained highly active against HCV variants harboring mutations of Arg 155 or Asp 168. A recent crystallographic study analyzing the molecular basis of drug resistance against NS3/4A protease inhibitors revealed that telaprevir, danoprevir, and vaniprevir interact directly with sites that confer resistance upon mutation, whereas MK-5172 interacts in a unique conformation with the catalytic triad (Romano et al. 2012). This novel mode of MK-5172 binding explains its retained potency against the two multidrug-resistant variants, R155K and D168A. Results from early clinical trials are essentially in line with its in vitro properties. Thus, no viral breakthrough has been observed in HCV genotype 1-infected patients who received this drug alone for 7 days and antiviral activity persisted for several days beyond the treatment period (Brainard et al. 2010), suggesting a higher barrier to resistance compared to first-generation inhibitors. In the same trial, patients infected with HCV genotype 3 showed a robust antiviral response at the higher drug doses. Longer drug administration periods will be required to draw final conclusions regarding the in vivo resistance profile and genotype 3 efficacy of MK-5172.
ACH-2684, a P3-P1 macrocyclic inhibitor, is another second-generation HCV protease inhibitor reported to be in Phase I clinical trial. ACH-2684 showed potent biochemical activity against genotype 1–6 viruses and against known resistant variants (Huang et al. 2010). Additional examples of second-generation HCV protease inhibitors have started to fill the preclinical research space [see for example (Kazmierski et al. 2012)].
3 NS5A Inhibitors
HCV NS5A is a multifunctional protein that is essential for HCV RNA replication complex and required for virion assembly (see chapters “Hepatitis C Virus RNA Replication” by Lohmann and “Virion Assembly and Release” by Lindenbach, this volume). It is a phosphoprotein expressed in basally and hyperphosphorylated forms (p. 56 and p. 58, respectively) (Huang et al. 2007). The NS5A protein structure consists of three domains: Domain I (aa 1–213), Domain II (aa 250–342), and Domain III (aa 356–447). While Domains II and III have not been structurally characterized, the crystal structure of Domain I has been crystallized in alternative dimer forms (Love et al. 2009; Tellinghuisen et al. 2005), both containing zinc- and RNA-binding motifs (Fig. 1b). NS5A has been shown to interact with a variety of host proteins (Macdonald and Harris 2004), including a critical interaction with phosphatidylinositol 4-kinase IIIα (PI4KIIIα), a cellular lipid kinase that is absolutely required for HCV replication (Reiss et al. 2011). Not being associated to any measurable enzymatic activity, NS5A has been considered “not druggable” for a very long time. In recent years, however, compounds acting on NS5A have emerged as efficacious inhibitors of HCV replication, with specific examples displaying in vitro anti-HCV activity in the low pM range. The first NS5A inhibitors were discovered by replicon-based high-throughput screening (Conte et al. 2009; Lemm et al. 2010). The initial lead compounds (Fig. 3a) had moderate potency and narrow anti-HCV activity, mainly on genotype 1b. Subsequent medicinal chemistry efforts (Lemm et al. 2011) resulted in the discovery of extremely potent compounds characterized by a very peculiar, highly symmetrical dimeric structure (Fig. 3b). The most studied of this “palindromic” NS5A inhibitor class is daclatasvir/BMS-790052 (Gao et al. 2010), a highly optimized NS5A inhibitor, which exhibited 50 % effective concentration (EC50) values of 50 and 9 pM against replicon of genotype 1a and 1b, respectively. Moreover, daclatasvir exhibited low picomolar activity against replicons harboring NS5A from a broad range of HCV genotypes (McPhee et al. 2012b), thus indicating its pan-genotypic potential.
It should be pointed out that the precise mechanism of action of the so-called “NS5A-inhibitors” is not completely understood. They were initially claimed to be NS5A inhibitors mainly based on the selection of specific resistant mutations that mapped in NS5A Domain I (Conte et al. 2009; Lemm et al. 2010). In particular, changes corresponding to variants of NS5A Tyr93 (Y93H/C/N) were found by different groups to be most common mutations conferring broad resistance to this class of antivirals. Interestingly, Tyr93 is found near the protein dimer interface (Fig. 1b), leading to speculate that NS5A inhibitors might act by modulating NS5A monomer/dimer equilibrium (Conte et al. 2009). However, the palindromic topology of daclatasvir and related compounds rather suggests a binding interaction with NS5A in which the inhibitor interacts across the dimer interface, making simultaneous contacts to both protein monomers (Belda and Targett-Adams 2012). This could at least partly explain the extraordinary potency observed for the palindromic inhibitor series. Among the reported properties of NS5A inhibitors are the ability to inhibit NS5A hyperphosphorylation (Lemm et al. 2010) and to cause the redistribution of NS5A from the HCV membranous web to different subcellular localizations, thus impeding the formation of new replication complexes (Qiu et al. 2011; Targett-Adams et al. 2011) (see also chapter “Hepatitis C Virus RNA Replication” by Lohmann, this volume). NS5A inhibitors were also shown to interfere with the accumulation of phosphatidylinositol 4-phosphate (PI4P) in the membranous HCV replication compartment, the so-called membranous web (Berger et al. 2012; Reghellin et al. 2012). Membranous web PI4P is produced, in HCV-infected or replicon cells, by PI4KIIIα, a phosphatidylinositol-specific kinase that is specifically recruited and activated by the interaction with NS5A (Reiss et al. 2011). These new data indirectly suggest that interaction of these antiviral agents with NS5A might also interfere with the recruitment and/or activation of PI4KIIIα by HCV NS5A.
Clinically, inhibition of NS5A has been associated with steep reductions in HCV RNA in monotherapy (Gao et al. 2010) and enhanced SVR rates when combined with PEG-IFN and RBV (Pol et al. 2012). When patients with chronic HCV genotype 1 infection were administered single doses of 1, 10, and 100 mg of daclatasvir, viral load dropped by up to 3.6 log10 IU/ml. Strikingly, the drop in viremia observed at the highest dose was sustained for 144 h post-dose in two patients infected with genotype 1b virus (Gao et al. 2010). In spite of the striking antiviral activity, the genetic barrier to resistance for this drug is low, and resistant variants are selected readily in vitro as well as in patients, with the more relevant substitutions found at NS5A residues 28, 30, 31, and 93 for genotype 1a and residues 31 and 93 for genotype 1b (Fridell et al. 2011). A recent Phase II trial highlighted the efficacy of daclatasvir in combination with PEG-IFN/RBV. All subjects received 48 weeks of triple therapy. SVR rates for 10 and 60 mg doses were 83 %, whereas the standard therapy only achieved an SVR of 25 % (Pol et al. 2012) (for further details see chapter “Treatment of Chronic Hepatitis C: Current and Future” by Pawlotsky, this volume). Daclatasvir is currently being evaluated in IFN-free regimens in combination with sofosbuvir (nucleotide polymerase inhibitor), asunaprevir (protease inhibitor), and/or BMS791325 (non-nucleoside polymerase inhibitor) (Everson et al. 2012; Sulkowski et al. 2012a).
GS-5885 (Lawitz et al. 2012) is a more recently reported NS5A inhibitor with EC50 values of 34 pM against genotype 1a and 4 pM against genotype 1b replicons. Antiviral activity of once-daily dosing (1 to 90 mg) of GS-5885 for 3 days in patients with chronic genotype 1 HCV infection resulted in median maximal reductions in viral RNA ranging from 2.3 to 3.3 log10 IU/ml. HCV RNA sequencing following GS-5885 dosing revealed a pattern of resistance mutations virtually identical to what observed with daclatasvir. Like daclatasvir, GS-5885 is also evaluated in IFN-free regimens in combination with sofosbuvir (nucleotide polymerase inhibitor) and ribavirin (Gane et al. 2012b).
Other NS5A inhibitors in clinical development include: ABT-267, GSK2336805, PPI-561 (Phase II) and ACH-2928, ACH-3102, PPI-668, IDX719, and MK-8742 (Phase I) (Belda and Targett-Adams 2012). It is worth pointing out that ACH-3102 and MK-8742 are two early-stage NS5A inhibitors that (1) show a relatively higher barrier to resistance and (2) retain substantial levels of potency against resistance mutations that affect early NS5A inhibitors, such as daclatasvir and GS-5885 (Liu et al. 2012; Yang et al. 2012). In analogy with the evolution of NS3/4A protease inhibitors, these novel agents can be viewed as “second-generation” NS5A inhibitors (Belda and Targett-Adams 2012).
4 Inhibitors of the NS5B Polymerase
The HCV NS5B RNA-dependent RNA polymerase (RdRp) catalyzes the synthesis of a complementary minus-strand RNA, using the (incoming) RNA genome as a template, and subsequently the synthesis of new progeny genomic plus-strand RNA from the minus-strand RNA template (detailed in chapter “Hepatitis C Virus RNA Replication” by Lohmann, this volume). Similar to other nucleic acid polymerases, NS5B has the typical right-hand polymerase structure, consisting of a thumb domain and a fingers domain, both encircling the enzyme active site located within the palm domain (Bressanelli et al. 1999; Lesburg et al. 1999) (Fig. 1c). The evolutionarily very well-conserved active site includes a GDD motif, implicated in coordinating two Mg++ ions required to bind the nucleotide substrate(s) and to assist the enzymatic reaction (Fig. 1c). Inhibitors of the NS5B RdRp are classified into nucleoside (NI), nucleotide, and non-nucleoside (NNI) inhibitors (Membreno and Lawitz 2011) (Fig. 4).
The nucleoside/nucleotide inhibitors are mimics of the natural polymerase substrates and bind to the NS5B active site, causing chain termination. Importantly, because of the active site conservation, they have similar efficacy across all HCV genotypes and isolates (Ludmerer et al. 2005). For the same reason, nucleoside/nucleotide inhibitors are associated with a high barrier to development of drug resistance (McCown et al. 2008). NNIs bind to one of at least four, less conserved, allosteric sites, resulting in the inhibition of enzyme conformational changes that are necessary for the enzyme catalytic function (Tomei et al. 2003; Tomei et al. 2004). In contrast to NIs, these molecules have shown a restricted spectrum of activity against the various HCV genotypes (Ludmerer et al. 2005) and present a very low barrier to emergence of resistance (McCown et al. 2008). So far, NNIs have targeted four main allosteric sites in the NS5B polymerase: thumb domains I and II and palm domains I and II (depicted in Fig. 1c).
4.1 Nucleoside Inhibitors
HCV-specific nucleoside inhibitors (NI) are ribonucleoside analogs that need to be converted to their corresponding 5′-triphosphate by cellular kinases in the cytoplasm of infected cells, thereby mimicking natural polymerase substrates (Carroll and Olsen 2006). NIs with anti-HCV activity possess a free 3′-hydroxyl function and may therefore not be considered as obligate chain terminators. Virtually, all nucleoside/-tide inhibitors in development contain modifications at the sugar 2′-position. It is thus conceivable that these compounds may act as virtual chain terminators because of steric hindrance exerted by 2′-C-methyl or 2′-fluoro groups. The primary mutation identified to be associated with decreased susceptibility to a broad range of 2′-modified nucleoside/-tide analogs is NS5B S282T (Le Pogam et al. 2006a; Migliaccio et al. 2003; Pawlotsky et al. 2012) (Fig. 1c). This mutation dramatically reduces HCV replication capacity, explaining at least in part the high barrier to resistance posed by these agents (Pawlotsky et al. 2012).
Valopicitabine (NM283) is an oral prodrug (3′-O-valine ester) of 2′-C-methylcytidine (NM107) (Pierra et al. 2006) and the first nucleoside analog inhibitor of HCV that entered clinical trials. In Phase II clinical trials in HCV-infected patients, valopicitabine was combined for 48 weeks of treatment with PEG-IFN-α. The decline in viral load was not significantly different when compared to the standard of care group, i.e., PEG-IFN and RBV (Afdhal et al. 2007). Whereas HCV replicons resistant to 2′-C-methylcytidine carry the S282T mutation, no selection of resistant viruses was observed in patients treated with this nucleoside analog (Lawitz et al. 2006). This is consistent with the low antiviral effect exerted by this drug in the clinical trials. Based on the low-benefit profile observed in clinical trials, the development of valopicitabine was stopped.
Following 2′-C-methylcytidine, various other 2′-modified nucleoside analogs targeting the HCV NS5B polymerase have been reported to inhibit HCV replication in vitro: these include 2′-O-methylcytidine (Carroll et al. 2003), 2′-C-methyladenosine (Carroll et al. 2003), 2′-C-methylguanosine (Migliaccio et al. 2003), 7-deaza-2′-C-methyladenosine (Olsen et al. 2004), 2′-deoxy-2′-fluoro-2′-C-methylcytidine (PSI-6130) (Stuyver et al. 2006), and 2′-deoxy-2′-spirocyclopropylcytidine (TMC647078) (Berke et al. 2011). Except for PSI-6130, the parent drug of RG7128, none of these nucleoside analogs are further developed.
Mericitabine/RG7128 (Fig. 4a) is a di-isobutyl ester prodrug of PSI-6130, a cytidine nucleoside analog (β-D-2′-deoxy-2′-fluoro-2′-C-methylcytidine) (Stuyver et al. 2006). PSI-6130 is metabolized intracellularly to the 5′-triphosphate of β-D-2′-deoxy-2′-fluoro-2′-C-methylcytidine and also, following deamination, to the triphosphate of β-D-2′-deoxy-2′-fluoro-2′-C-methyluridine (PSI-6206) (Ma et al. 2007). Both triphosphates are incorporated as nonobligate chain terminators into RNA synthesized by purified NS5B (Murakami et al. 2007). Replicons resistant to PSI-6130 carry the S282T mutation, as is the case for valopicitabine-resistant replicons. Interestingly, this mutation results only in a low level (three to fourfold) of in vitro resistance and emergence of resistance is slower than for other classes of DAAs (Pawlotsky et al. 2012). Mericitabine has been shown to be active against genotype 1 and 4 HCV and is currently in Phase II clinical development in triple combination with PEG-IFN and RBV (Le Pogam et al. 2010). Mericitabine is also being evaluated in early-stage clinical trials as a component of IFN-free, all-oral regimen in combination with the NS3/4A protease inhibitor danoprevir (Gane et al. 2010; Pawlotsky et al. 2012). In line with the high barrier to resistance observed in vitro, no evidence of genotypic resistance to mericitabine has been detected by population or clonal sequence analysis in any baseline or on-treatment samples collected from >600 patients enrolled in Phase I/II trials of mericitabine administered as monotherapy, in combination with PEG-RBV, or in combination with danoprevir (Pawlotsky et al. 2012) (see also chapter “Treatment of Chronic Hepatitis C: Current and Future” by Pawlotsky, this volume).
4.2 Nucleotide Inhibitors
Nucleotide polymerase inhibitors are liver-targeted prodrugs designed to enhance formation of its active triphosphate in the liver, while minimizing systemic exposure of the nucleotide drug and its nucleoside metabolite. Such prodrugs of nucleotide analogs (i.e., nucleoside 5′-monophosphates) are preferentially cleaved by hepatic enzymes, thereby efficiently releasing their nucleoside monophosphate in liver cells. The rate-limiting step for metabolic activation of nucleoside analogs, the initial phosphorylation to a nucleoside monophosphate, is bypassed, resulting in higher levels of nucleoside triphosphates in the cell.
Sofosbuvir/GS-7977 (Fig. 4a) is currently the most advanced NS5B polymerase inhibitor in clinical development (Phase III). It is a chirally pure isomer of PSI-7851, a phosphoramidate prodrug of a uridine nucleotide analog (beta-D-2′-deoxy-2′-fluoro-2′-C-methyluridine monophosphate) (Murakami et al. 2008). It was previously shown that PSI-7851 exists as a 1:1 mixture of two diastereoisomers, PSI-7976 and PSI-7977, at the phosphorus center of the phosphoramidate moiety. PSI-7977, the Sp isomer, was found to inhibit HCV replication more effectively than PSI-7976 (Rp isomer) in replicon-based assays. The S282T mutation was the common resistance mutation emerging during resistance selection in vitro (Lam et al. 2012). While this mutation conferred resistance to sofosbuvir in genotype 1 replicons, it only caused a very minor shift in potency in genotype 2a, thus suggesting that additional mutations in genotype 2a NS5B are required for the resistant phenotype (Lam et al. 2012). Phase II clinical studies revealed that addition of sofosbuvir to PEG-IFN and RBV for 12 weeks resulted in SVR rates in excess of 90 % for naïve genotype 1 HCV infected patients (Kowdley et al. 2012b). Furthermore, IFN-free, all-oral sofosbuvir/RBV demonstrated consistent antiviral suppression across HCV genotypes (Gane et al. 2012b). Viral resistance was hardly observed in any clinical studies using sofosbuvir, confirming the high genetic barrier to resistance. Sofosbuvir is currently studied in IFN-free combinations with a number of other DAAs, including NS3/4A protease inhibitors (GS-938, simeprevir) and NS5A inhibitor (daclatasvir, GS-5885). Striking preliminary data have been reported from an ongoing Phase II study examining a 12 weeks course of therapy with sofosbuvir in combination with GS-5885 and ribavirin in patients with genotype 1 chronic HCV infection. In this trial, among treatment-naïve patients receiving this combination, 100 % (n = 25/25) remained HCV RNA undetectable 4 weeks after completing therapy (SVR4) (Gane et al. 2012a). The very high efficacy of the combination between sofosbuvir and an NS5A inhibitor was also demonstrated in a trial where 12 weeks viral elimination rates (SVR12) of greater than 93 % were observed in previously untreated patients infected with HCV genotype 1, 2, or 3 (Sulkowski et al. 2012b). Importantly, in this trial, the inclusion of RBV did not influence the virological outcome.
GS-938 (formerly PSI-938) is a prodrug of 2′-F-2′-C-methylguanosine monophosphate and was created to be an optimal partner DAA for pyrimidine nucleotide GS-7977 (Lam et al. 2011b). The nucleotides employ different prodrug cleavage pathways, largely independent phosphorylation pathways, competition with separate endogenous nucleotide pools (purine/pyrimidine), and complementary resistance profiles. No resistant genotype 1a and 1b replicons could be selected. In contrast, various mutations in NS5B were identified in resistant genotype 2a replicons. Phenotypic characterization showed that single mutations were not able to confer resistance to GS-938, but that a combination of three amino acid changes, S15G/C223H/V321I, was required (Lam et al. 2011a). Interestingly, no cross-resistance was observed with 2′-modified nucleoside/-tide analogs such as PSI-6130, GS-7977, BMS-986094, and IDX-184. Conversely, GS-938 remained fully active against both the S96T and S282T replicons (Lam et al. 2011b). The potential of the combination of GS-938 and GS-7977 was studied in the NUCLEAR clinical trial (Lawitz et al. 2011). The 14 days proof-of-concept study demonstrated excellent safety/tolerability, lack of antagonism (pharmacokinetic or antiviral), and no virologic breakthrough. However, in the subsequent Phase IIb Quantum study all treatment arms containing GS-938 were discontinued because abnormal liver function tests were observed in some patients receiving GS-938 (Press release Pharmasset, 16 December 2011).
A novel, double prodrug approach was used to synthesize nucleotide analog BMS-986094 (formerly INX-08189). A phosphoramidate ProTide motif and a 6-O-methoxy base moiety were combined to generate a lipophilic prodrug of the monophosphate of 2′-C-methyl guanosine (McGuigan et al. 2010). BMS-986094 is highly active in HCV replicon assays with an EC50 of 1–10 nM (genotypes 1a, 1b, 2a) (Vernachio et al. 2011), and replicons resistant to BMS-986094 carried the S282T mutation in NS5B, resulting in an approximately 10-fold reduction in sensitivity to the drug. Following 7 days of monotherapy, a median reduction of −4.25 log10 in HCV RNA levels was achieved (Rodriguez-Torres et al. 2011). Clinical development of BMS-986094 was stopped, however, due to adverse effects involving heart and kidney toxicity.
IDX184 is a different nucleotide prodrug also designed to deliver high concentrations of 2′-C-methyl guanosine 5′-monophosphate (MP) in hepatocytes (Zhou et al. 2011). In spite of the relatively encouraging clinical results, development of this drug was significantly slowed down in light of the severe toxicity associated with BMS-986094, with which IDX184 shares the parent nucleotide (2′-C-methyl guanosine MP). Other nucleotide polymerase inhibitors are currently studied in preclinical, Phase I and II clinical trials including PSI-353661 (prodrugs of 2′-C-methylguanosine monophosphate), GS-6620 (nucleotide prodrug of C-nucleoside analog), and ALS-2200 (structure unknown).
4.3 Non-nucleoside Inhibitors
Screening of inhibitor libraries has resulted in the discovery of several classes of allosteric non-nucleoside inhibitors (NNIs). The hallmark of all allosteric HCV NNIs described so far is that, in contrast to active site nucleoside inhibitors, they are noncompetitive with NTP substrates and inhibit the polymerase at a stage preceding the elongation reaction (Gu et al. 2003; Tomei et al. 2003; Tomei et al. 2004). To date, at least four distinct allosteric binding sites are known to be targeted by NNIs; two of them, dubbed “thumb I” and “thumb II”, are located on the polymerase thumb domain, whereas the other two sites, “palm I” and “palm II”, are close to the active site cavity and involve primarily amino acids from the palm domain (Fig. 1c).
4.3.1 Thumb I Inhibitors
The thumb I NNI binding site is located in the thumb domain 30 Å away from the active site (Fig. 1c). Several structurally related inhibitors have been shown to bind to this site (Beaulieu 2006). This class of inhibitors interrupts the intramolecular contacts between the thumb and the finger-tip loop, thereby preventing the formation of a productive enzyme complex for RNA synthesis (Di Marco et al. 2005; Kukolj et al. 2005). For this reason, these agents are also known as “finger-loop” inhibitors and are characterized by having a common benzimidazole or indole chemical core (Fig. 4b). The in vitro antiviral efficacy of finger-loop inhibitors was originally demonstrated in HCV subgenomic replicons (Kukolj et al. 2005; Tomei et al. 2003), where drug resistant variants carrying mutations at positions P495, P496, and T389 could be readily selected (Delang et al. 2012; Kukolj et al. 2005; Tomei et al. 2003) (Fig. 1c). Optimization of cellular permeability by conversion of the benzimidazole scaffold to the more lipophilic indole scaffold provided further enhancement of the antiviral activity, leading eventually to anti-HCV clinical activity (Beaulieu et al. 2012; Brainard et al. 2009; Devogelaere et al. 2012; Larrey et al. 2009; Narjes et al. 2011). Notably, agents belonging to this class of NNIs display reduced activity against genotype 1a HCV compared to genotype 1b (Brainard et al. 2009; Erhardt et al. 2009; Larrey et al. 2009). Several such indole-based inhibitors (BI 207127, TMC647055, and BMS791325) are currently being investigated in clinical trials (Beaulieu et al. 2012; Devogelaere et al. 2012; Larrey et al. 2012; Larrey et al. 2009). The most advanced thumb site I-targeting NNI is BI 207127 (Fig. 4b). It is currently in Phase II clinical development. In line with the in vitro findings, BI 207127-resistant mutants that encode P495 substitutions were observed in 11 % of patients who received a 5 days monotherapy (Larrey et al. 2009). BI 207127 in combination with PEG-IFN and RBV demonstrated strong antiviral activity with a favorable safety and tolerability profile (Larrey et al. 2012). In a Phase IIb IFN-free clinical trial (Zeuzem et al. 2012b), BI 207127 was studied in combination with the NS3/4Aprotease inhibitor BI 201335 together with RBV for 28 weeks. This IFN-free combination, which included 362 treatment-naïve patients with genotype-1 HCV infection, resulted in virus elimination with 70 % of patients overall, compared with 85 % seen in the GT-1b patient subgroup. More strikingly, a 12 weeks IFN-free and RBV-free triple therapy with daclatasvir, asunaprevir, and BMS-791325, another thumb I NNI, achieved 94 % (n = 15/16) 4 weeks post-treatment response (SVR4) in treatment-naïve patients infected with HCV genotype 1 viruses (Everson et al. 2012). Importantly, although preliminary, these results demonstrate the potential for thumb I NNIs to be successfully utilized in an IFN-free all-oral regimen, at least for selected patients.
4.3.2 Thumb II Inhibitors
Thiophene carboxylic acid derivatives were reported to inhibit NS5B RdRp activity and HCV RNA replication in the replicon cell culture system. Crystallographic studies revealed the existence of a hydrophobic cavity located at the base of the thumb domain of NS5B (Wang et al. 2003). Like thumb I NNIs, thiophene carboxylic acid derivatives are noncompetitive with nucleotide incorporation and inhibit an initiation step of RNA synthesis by interfering with conformational changes that are likely required for processive elongation of the replicating strand. Thiophene-based inhibitors were found to select for M423, L419, and I482 resistant mutants in replicon cell culture experiments (Le Pogam et al. 2006b). Lomibuvir/VX-222 (Fig. 4b) exhibits low micromolar antiviral activity against HCV 1a and 1b isolates in replicon assays. Upon 3 days of monotherapy, lomibuvir achieved a mean reduction in HCV RNA ranging from 3.1 to 3.4 log10 with similar activity in genotype 1a- and 1b-infected patients (Rodriguez-Torres et al. 2010). Lomibuvir is also studied in combination with protease inhibitor telaprevir. In particular, combining lomibuvir and telaprevir with PEG-IFN + RBV resulted in overall cure rates of up to 90 % (Nelson et al. 2011) with no on-treatment viral breakthroughs observed. A back-up drug for lomibuvir, VX-759, resulted in a significant reduction in viral load in treatment-naïve genotype 1 infected patients during a proof-of-concept study (Cooper et al. 2009). On-treatment rebound of viremia suggested the emergence of resistant strains. Genotypic analysis of clinical isolates confirmed mutations at positions L419, M423, and I482. VX-759 is currently being evaluated in Phase II clinical trials.
In addition to thiophene-derived carboxylic acids, other classes of molecules have been identified to target this allosteric site. Filibuvir/PF-868554 (Fig. 4b), a dihydropyranone derivative, is a potent and selective HCV inhibitor in vitro (Li et al. 2009). It exerts strong in vitro antiviral activity against the HCV 1b replicon (EC50 of 0.075 μM), and reduced activity against the 1a replicon (EC50 of 0.39 μM). The predominant in vitro resistance mutation is M423T (Shi et al. 2009). Other amino acid mutations, M426T and I482T, were detected at a much lower frequency. Interestingly, the resistance mutations at residues L419, M423, and I482 had different effects on the level of inhibition of RNA synthesis by filibuvir or by lomibuvir (Yi et al. 2012). It was therefore hypothesized that the binding sites of filibuvir and lomibuvir are partially overlapping, but not identical. In treatment-naïve genotype 1 patients, addition of filibuvir to PEG-IFN and RBV was associated with a maximal mean reduction in HCV RNA of 4.7 log10 IU/mL at day 28 (Jacobson et al. 2009). However, a high relapse rate was observed, probably due to the short treatment duration. Sequence analysis of the NS5B coding region identified residue M423 as the predominant site of mutation (Wagner et al. 2011). Currently, filibuvir is in Phase II of clinical development.
Another class of non-nucleoside polymerase inhibitors that interacts with thumb domain II is the class of the pyranoindole derivatives (i.e., HCV-371 and a follow-up compound HCV-086). It entered clinical development, but failed to demonstrate significant efficacy and development was therefore discontinued (Howe et al. 2004; Howe et al. 2006). Recently, substituted N-phenylbenzene-sulphonamides were also shown to inhibit HCV genotype 1a and 1b replication in vitro (May et al. 2012).
4.3.3 Palm I Inhibitors
Palm domain I is located at the junction of the palm and the thumb domain of NS5B and is in relatively close proximity to the catalytic site. The first class of palm domain I inhibitors was originally discovered by GlaxoSmithKline and is characterized by a benzothiadiazine scaffold. Akin to the thumb domain targeting compounds, benzothiadiazine-based compounds inhibit RNA synthesis before formation of an elongation complex. In vitro benzothiadiazines select for M414-resistant mutants (Nguyen et al. 2003).
The benzothiadiazine inhibitor setrobuvir/RG7790 (formerly ANA598; Fig. 4c) exhibits low nanomolar potency against genotype 1 HCV replicons (Thompson et al. 2009). In Phase I studies, RG7790, dosed for 3 days as monotherapy, resulted in a median viral load decline of 2.4 log10 in treatment-naïve genotype 1 infected patients (Lawitz et al. 2009). No patient showed evidence of viral rebound while on setrobuvir. A Phase II clinical trial in which treatment-naïve genotype-1 HCV infected patients were treated with a combination of PEG-IFN, RBV, and RG7790 revealed that addition of RG7790 resulted in a more rapid viral clearance than PEG-IFN and RBV alone (Lawitz et al. 2010b). A low viral breakthrough rate of <2 % was observed.
Benzothiadiazine inhibitors ABT-333 and ABT-072 have been studied in vitro and in Phase I clinical trials. The most frequently observed in vitro resistance mutations selected by ABT-333 and ABT-072 were C316Y, M414T, Y448H/C, or S556G. Combinations of ABT-333 or ABT-072 with PEG-IFN and RBV both resulted in significantly greater antiviral activity in treatment-naïve patients than PEG-IFN and RBV alone (SVR of 63, 52, and 9 %, respectively) (Poordad et al. 2012b). Based on these results, ABT-333 was selected for further development in an IFN-free regimen. In the so-called Aviator Phase IIb trial, 12 weeks of treatment with the IFN-free multidrug combination of ABT-333, ritonavir-boosted protease inhibitor ABT-450, ABT-267 (NS5A inhibitor), and ribavirin resulted in a striking 97.5 % (n = 77/79) or 93.3 % (n = 42/45) SVR12 in genotype 1 treatment-naïve patients or previous null responders, respectively (Kowdley et al. 2012b).
Acylpyrrolidines are yet another class of palm site I binding compounds (Slater et al. 2007). The activity of acylpyrrolidines is affected by the M414T mutation, suggesting that these compounds bind within palm domain I (Pauwels et al. 2007). GSK625433 was advanced into Phase I clinical trials, but this study was halted because of adverse effects noted in long-term mouse carcinogenicity studies (Gray et al. 2007).
4.3.4 Palm II Inhibitors
The palm II NNI binding site partially overlaps with the palm I site and is located in proximity to the active site and the junction between the palm and thumb domain. A class of benzofurans were identified as potent inhibitor of in vitro HCV replication (Gopalsamy et al. 2006). These molecules select for resistant mutants at residues L314, C316, I363, S365, and M414 (Howe et al. 2008) (Fig. 1c). HCV-796 showed significant activity in early-stage clinical trials (Kneteman et al. 2009). The development of HCV-796 was halted, however, because elevated liver enzyme levels were detected in some patients treated with this agent.
A class of imidazopyridines exerts potent in vitro antiviral activity against HCV. Drug resistant variants carry mutations in palm domain II (C316Y) as well as in the β-hairpin loop (C445F, Y448H, Y452H) (Fig. 1c). This β-hairpin loop is located in close proximity to the catalytic active site and is believed to be involved in primer-independent initiation of RNA replication (Hong et al. 2001). The fact that the imidazopyridines, in contrast to other NNIs, do not inhibit the enzymatic activity of the purified RdRp, suggested that this class of molecules targets the enzyme via a unique mechanism. Recent data indeed revealed that upon intracellular activation by CYP1A, the resulting metabolite of tegobuvir, after forming a conjugate with glutathione, directly and specifically interacts with NS5B (Hebner et al. 2012). Within this class of imidazopyridines, tegobuvir/GS-9190 (Fig. 4c) has demonstrated antiviral activity in HCV-infected patients. In Phase II studies tegobuvir is evaluated in combination with GS-9256, a protease inhibitor, when used as: (1) a dual antiviral therapy; (2) a three-drug, all-oral regimen with RBV; or (3) a four-drug regimen with RBV and PEG-IFN. Median maximal reductions in HCV RNA were 4.1 log10 IU/mL for the dual therapy, 5.1 log10 IU/mL for the triple therapy, and 5.7 log10 IU/mL for the quadruple therapy during 28 days of treatment (Zeuzem et al. 2012b). No viral breakthroughs were observed in patients treated with a four-drug regimen, whereas it was observed in the dual and triple therapy arms. The highest rate of resistance mutations was detected in the dual therapy arm. Results from this study demonstrated that addition of RBV improved the antiviral activity, delayed resistance emergence, and resulted in a greater proportion of patients achieving an RVR. In another clinical study, treatment with an all-oral, IFN-free quad regimen containing tegobuvir, protease inhibitor GS-9451, NS5A inhibitor GS-5885 and RBV resulted in high SVR rates (Sulkowski et al. 2012b) with relapse and viral breakthrough being limited to GT1a infected patients.
5 Emerging Targets for DAA Development
5.1 The p7 ion Channel
The HCV p7 protein is a “viroporin” (virus encoded ion channel) that is critical for HCV virus infection (see chapters “Hepatitis C Virus Proteins From Structure to Function” by Moradpour and Penin and “Virion Assembly and Release” by Lindenbach, this volume). It is a small transmembrane protein that forms hexameric channels (Luik et al. 2009). The N and C termini are oriented toward the ER lumen, whereas the connecting loop faces the cytoplasm (Carrere-Kremer et al. 2002). P7 is critical for the release of infectious virions in vitro and in vivo (Sakai et al. 2003; Steinmann et al. 2007a). When its ion channel activity is blocked, virus production is significantly reduced (Steinmann et al. 2007b). A number of HCV p7 inhibitors have been identified.
Viroporin inhibitors were first approved 40 years ago for influenza A antiviral therapy. Adamantanes (amantadine and rimantadine) were shown to block influenza A infection by inhibition of the M2 ion channel activity. Interestingly, adamantanes are also able to inhibit H+ transport through p7 ion channels (Griffin et al. 2003). Binding of amantadine to the p7 ion channel prevents opening of the channel that is mediated by low pH. The antiviral effect of amantadine in a HCV-infected cell culture system was shown to be genotype-dependent (Griffin et al. 2008; Steinmann et al. 2007b). In contrast, rimantadine could inhibit virus infectivity of different gentoypes. Adamantane resistance is conferred by a L20F mutation in p7 (Foster et al. 2011b). Interestingly, this mutation is also observed in HCV infected patients unresponsive to PEG-IFN, RBV, and amantadine treatment. In 1997, it was reported that amantadine treatment could induce a sustained biochemical and virological response in HCV infected patients who previously failed to respond to IFN-α monotherapy (Smith 1997). Moreover, data from meta-analysis support a specific antiviral effect in vivo (Deltenre et al. 2004). However, the possible benefit of amantadine monotherapy, or in combination with PEG-IFN/RBV remains controversial since these findings could not be confirmed in some other studies (Pessoa et al. 2012; von Wagner et al. 2008). The lack of efficacy for adamantanes in vivo can possibly be explained by the proposed minimal genetic barrier to resistance, since adamantane resistance is conferred by a single mutation that has little consequence for viral replication fitness.
Inhibition of the p7 ion channel activity was also demonstrated with various long-alkylated iminosugar derivatives (Steinmann et al. 2007b), molecules that are also known to inhibit ER α-glucosidase I and II. Long-alkylated iminosugars prevent p7 channel oligomerization by intercalating between p7 protomers. Resistance to alkylated iminosugars is mediated by a F25A mutation in p7 (Foster et al. 2011b). In a Phase II clinical trial, iminosugar UT-231b did not demonstrate efficacy in HCV infected patients who previously failed standard therapy (United Therapeutics company website, clinical trial identifier NCT00069511).
Acylguanidine BIT225 (N-[5-(1methyl-1H-pyrazol-4-yl)-napthalene-2-carbonyl]-guanidine) was identified in a rational drug design program based on amiloride derivatives. It inhibits p7 ion channel activity in planar lipid bilayers and is active in vitro against bovine viral diarrhea (BVDV), an HCV-related virus (chapter “The Origin of Hepatitis C Virus” by Simmonds, this volume) that also encodes a p7 ion channel (Luscombe et al. 2010). BIT225 was shown to result in synergistic antiviral activity in combination with PEG-IFN + RBV and with nucleoside polymerase inhibitors in BVDV assays (Luscombe et al. 2010). In a Phase IIa clinical trial (Tanwandee et al. 2011), chronic HCV patients were administered BIT225 for 28 days in combination PEG-IFN plus RBV. Patients then continued on with SOC with PEG-IFN/RBV for a further 44 weeks. 87 % of patients receiving BIT225 achieved a complete early virological response [cEVR, defined as viral load below the level of detection (<50 IU/ml) at 12 weeks], versus 63 % for PEG-IFN and RBV alone. BIT225 also targets HIV-1 Vpu and could thus have potential in the treatment of HCV and HIV-1 coinfected patients (Khoury et al. 2010).
5.2 The NS4B Protein
HCV replication appears to be associated with intracellular membrane structures, the so-called membranous web (chapter “Hepatitis C Virus RNA Replication” by Lohmann, this volume). This structure is believed to be induced by the NS4B protein. NS4B is a highly hydrophobic protein consisting of an N-terminal part, a central part harboring four transmembrane segments and a C-terminal part (Gouttenoire et al. 2010). It is an integral membrane protein with the N- and C-terminal parts located on the cytosolic side of the ER membrane (chapter “Hepatitis C Virus Proteins From Structure to Function” by Moradpour and Penin, this volume). NS4B is also required to assemble the other viral nonstructural proteins within the apparent sites of RNA replication. NS4B and HCV RNA have been shown to colocalize to the membranous web, suggesting that NS4B is in intimate contact with viral RNA in the context of authentic viral RNA replication (El Hage and Luo 2003). NS4B binding to HCV RNA has a preference for the 3′-terminus of the HCV negative RNA strand (Einav et al. 2008). This region harbors a highly conserved secondary structure that is possibly recognized by HCV NS4B. Two classes of NS4B inhibitors have been recently identified: one class that inhibits the binding of NS4B to HCV RNA and a second class that disrupts interactions of NS4B with membranes [reviewed in (Rai and Deval 2011)].
Clemizole hydrochloride, a first-generation antihistamine, is the most advanced HCV NS4B inhibitor. Its ability to inhibit the binding of NS4B to HCV RNA was discovered during a screening with a microfluidic RNA-binding assay. It inhibits RNA binding with an IC50 of 24 nM and has a modest inhibitory effect on HCV RNA replication (EC50 for viral replication is ~8 μM for genotype 2a). In contrast, clemizole might not be active against HCV genotype 1 (EC50 > 20 μM) (Einav et al. 2010). Interestingly, the in vitro antiviral activity of clemizole was shown to be synergistic with HCV protease inhibitors, whereas combinations with IFN, ribavirin, or polymerase inhibitors were found to be additive (Einav et al. 2010). It is speculated that the observed synergy could be due to an interaction between HCV NS4B and NS3, possibly involving conformational changes. Clemizole resistant variants carry mutations at position W55 and R214 in the NS4B protein and mutations in the 3′-terminus of the negative strand HCV RNA (Einav et al. 2008). The combination of clemizole with a protease inhibitor decreased the emergence of resistance against the latter. A Phase I proof-of-concept study, evaluating the safety and efficacy of clemizole as a single agent therapy in treatment-naïve HCV infected patients, is ongoing. Preliminary data reveal that clemizole, while mostly not active in monotherapy, results, when combined with PEG-IFN and RBV, in a more efficacious reduction in viral load than PEG-IFN and RBV alone (Choong et al. 2010). Clemizole-related molecules in which the benzimidazole core is replaced by an indazole core have been reported to be more potent in vitro than clemizole (Einav et al. 2010).
During a large-scale screen for small molecules that bind to recombinant NS4B, different compound classes were identified that were able to inhibit HCV replication. One of these classes, the pyrazolopyrimidine family, was further studied. Anguizole, 7-[chloro(difluoro)methyl]-5-furan-2-yl-N-(thiophen-2-ylmethyl)pyrazolo[1,5a] pyrimidine-2-carboxamide, is active against genotype 1a and 1b, but is not active against genotype 2 (Bryson et al. 2010). The most commonly selected resistance mutation in vitro is H94R in NS4B, which results in a 37-fold resistance when reintroduced into a wild-type backbone. Anguizole treatment results in an altered subcellular localization pattern of NS4B. It specifically interacts with the N-terminal amphipathic helix AH2 of NS4B, and thereby inhibits the ability of AH2 to aggregate lipid vesicles. Another molecule that abrogates the ability of AH2 to aggregate lipid vesicles is C4 (5-(N-methyl-N-isobutyl)amiloride) (Cho et al. 2010). In contrast to anguizole, molecule C4 is active against HCV genotype 2a in vitro. Furthermore, as determined by atomic force microscopy, C4 inhibits AH2 oligomerization which is not observed for anguizole. Both AH2 inhibitors were not able to inhibit NS4B binding to HCV RNA.
6 Concluding Remarks
In the last few years, vast progress has been made in our understanding of the HCV replication cycle and in the development of small molecule-based therapeutics for the treatment of chronic hepatitis C. The ultimate goal is to develop a well-tolerated therapeutic regimen, with minimal contraindications, efficacious against all HCV genotypes and eradication of the virus with no or minimal risk of developing drug resistance. Ideally, such therapy would no longer require the use of PEG-IFN or RBV, which are associated with inconvenient dosing and numerous side effects. Results from recent clinical trials have firmly established the concept that a permanent cure can be achieved with IFN-free combinations of DAAs (Assis and Lim 2012; Ferenci 2012). While many novel DAAs are still being evaluated in combination with the PEG-IFN/RBV backbone, the race for IFN-free HCV therapies has started and several DAA-based, IFN-free drug combinations are being evaluated in Phase I and II clinical trials (Assis and Lim 2012; Chatel-Chaix et al. 2012). Drugs to be used in an IFN-free combination must have nonoverlapping resistance profiles. This is achievable because the DAAs under development target different viral proteins or even different drug-binding sites within the same protein. More than 40 new NS3/4A, NS5A, or NS5B inhibitors are in the development pipeline. Compelling preclinical as well as clinical evidence indicates that the current generation of nucleoside/nucleotide inhibitors of the NS5B polymerase has a broad spectrum of action on the different viral genotypes and that this is accompanied by a very high barrier to development of resistance. In light of these properties, nucleoside/nucleotide polymerase inhibitors have the potential to become the backbone of future DAA combination therapies. IFN-free combination of a nucleotide polymerase inhibitor with NS5A inhibitors have started to show viral eradication rates approaching 100 % in patients infected with different viral genotypes (Gane et al. 2012a; Sulkowski et al. 2012b). Although nucleotide polymerase inhibitors show the promise to become the cornerstone of future HCV therapy, we should also take note that—like is the case for IFN and RBV—inclusion of a nucleotide in a DAA combination regimen is not an absolute requirement. Recent clinical data have indicated that it is possible to achieve SVR in >90 % of patients by administering DAA cocktails that include an NS3/4A protease inhibitor, an NS5A inhibitor, and an NS5B NNI (Everson et al. 2012; Kowdley et al. 2012a). Second-generation NS3/4A protease inhibitors and second-generation NS5A inhibitors, with broader spectrum of action and high barrier to resistance, have now entered the development pipeline, thus expanding the number of possible ingredients for an effective combination regimen. Furthermore, the already-rich repertoire of HCV-specific DAAs is further widening, with the p7 ion channel and the NS4B protein emerging as potential drug targets. What DAA combination will turn out to be more successful and most convenient for which patients is the question that needs to be addressed in current and future research.
References
Afdhal N, O’Brien C, Godofsky E et al (2007) Valopicitabine (NM283), alone or with peginterferon, compared to peg interferon/ribavirin (pegIFN/RBV) retreatment in patients with HCV-1 infection and prior non-response to pegIFN/RBV: one-year results. J Hepatol 46:S5
Agarwal A, Zhang B, Olek E et al (2012) Rapid and sharp decline in hepatitis C virus upon monotherapy with NS3 protease inhibitor, ACH-1625. Antivirus Therapy, in press. doi:10.3851/IMP2359
Aghemo A, Degasperi E, Colombo M (2012) Directly acting antivirals for the treatment of chronic hepatitis C: unresolved topics from registration trials. Digistive and liver disease, in press. doi:10.1016/j.dld.2012.05.002
Assis DN, Lim JK (2012) New pharmacotherapy for hepatitis C. Clin Pharmacol Ther 92:294–305
Beaulieu PL (2006) The discovery of finger loop inhibitors of the hepatitis C virus NS5B polymerase: status and prospects for novel HCV therapeutics. IDrugs 9:39–43
Beaulieu PL, Bos M, Cordingley MG et al (2012) Discovery of the first thumb pocket 1 NS5B polymerase inhibitor (BILB 1941) with demonstrated antiviral activity in patients chronically infected with genotype 1 hepatitis C virus (HCV). J Med Chem 55:7650–7666
Belda O, Targett-Adams P (2012) Small molecule inhibitors of the hepatitis C virus-encoded NS5A protein. Virus Research, in press. doi:10.1016/j.virusres.2012.09.007
Benhamou Y, Moussalli J, Ratziu V et al (2009) Results of a proof-of-concept study (C210) of telaprevir monotherapy and in combination with peginterfern alfa-2a and ribavirin in treatment-naive genotype 4 HCV patients. J Hepatol 50(1):S6
Berger C, Romero-Brey I, Zayas M et al (2012) HCV NS5A inhibitors alter membranous web formation and reduce PI4P induction in cells expressing HCV non-structural proteins. In: Abstracts O23 of the 19th international symposium on hepatitis C virus and related viruses, Venice, Italy, 5–9 Octo 2012
Berke JM, Vijgen L, Lachau-Durand S et al (2011) Antiviral activity and mode of action of TMC647078, a novel nucleoside inhibitor of the hepatitis C virus NS5B polymerase. Antimicrob Agents Chemother 55:3812–3820
Brainard DM, Anderson MS, Petry A et al (2009) Safety and antiviral activity of NS5B polymerase inhibitor MK-3281, in treatment-naïve genotype 1a, 1b and 3 HCV-infected patients. Hepatology 50:1026A–1027A
Brainard DM, Petry A, Van Dyck K et al (2010) Safety and antiviral activity of MK-5172, a novel HCV NS3/4A protease inhibitor with potent activity against known resistance mutants in genotype 1 and 3 HCV-infected patients. Hepatology 52(1):706A–707A
Bressanelli S, Tomei L, Roussel A et al (1999) Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc Natl Acad Sci USA 96:13034–13039
Bryson PD, Cho NJ, Einav S et al (2010) A small molecule inhibits HCV replication and alters NS4B’s subcellular distribution. Antiviral Res 87:1–8
Carrere-Kremer S, Montpellier-Pala C, Cocquerel L et al (2002) Subcellular localization and topology of the p7 polypeptide of hepatitis C virus. J Virol 76:3720–3730
Carroll SS, Olsen DB (2006) Nucleoside analog inhibitors of hepatitis C virus replication. Infect Disord Drug Targets 6:17–29
Carroll SS, Tomassini JE, Bosserman M et al (2003) Inhibition of hepatitis C virus RNA replication by 2′-modified nucleoside analogs. J Biol Chem 278:11979–11984
Chang MH, Gordon LA, Fung HB (2012) Boceprevir: a protease inhibitor for the treatment of hepatitis C. Clin Ther 34:2021–2038
Chatel-Chaix L, Germain MA, Gotte M, Lamarre D (2012) Direct-acting and host-targeting HCV inhibitors: current and future directions. Curr Opin Virol 2:588–598
Chen SH, Tan SL (2005) Discovery of small-molecule inhibitors of HCV NS3-4A protease as potential therapeutic agents against HCV infection. Curr Med Chem 12:2317–2342
Cho NJ, Dvory-Sobol H, Lee C et al (2010) Identification of a class of HCV inhibitors directed against the nonstructural protein NS4B. Sci Transl Med 2:15–16
Choong IC, Cory D, Glenn JS, Yang W (2010) Methods and compositions of treating a Flaviviridae family viral infection. WO/2010/107739
Conte I, Giuliano C, Ercolani C et al (2009) Synthesis and SAR of piperazinyl-N-phenylbenzamides as inhibitors of hepatitis C virus RNA replication in cell culture. Bioorg Med Chem Lett 19:1779–1783
Cooper C, Lawitz EJ, Ghali P et al (2009) Evaluation of VCH-759 monotherapy in hepatitis C infection. J Hepatol 51:39–46
De Francesco R, Steinkuhler C (2000) Structure and function of the hepatitis C virus NS3-NS4A serine proteinase. Curr Top Microbiol Immunol 242:149–169
Delang L, Froeyen M, Herdewijn P, Neyts J (2012) Identification of a novel resistance mutation for benzimidazole inhibitors of the HCV RNA-dependent RNA polymerase. Antiviral Res 93:30–38
Deltenre P, Henrion J, Canva V et al (2004) Evaluation of amantadine in chronic hepatitis C: a meta-analysis. J Hepatol 41:462–473
Devogelaere B, Berke JM, Vijgen L et al (2012) TMC647055, a potent nonnucleoside hepatitis C virus NS5B polymerase inhibitor with cross-genotypic coverage. Antimicrob Agents Chemother 56:4676–4684
Di Marco S, Volpari C, Tomei L et al (2005) Interdomain communication in hepatitis C virus polymerase abolished by small molecule inhibitors bound to a novel allosteric site. J Biol Chem 280:29765–29770
Dvory-Sobol H, Wong KA, Ku KS et al (2012) Characterization of resistance to the protease inhibitor GS-9451 in hepatitis C virus-infected patients. Antimicrob Agents Chemother 56:5289–5295
Edwards PD, Bernstein PR (1994) Synthetic inhibitors of elastase. Med Res Rev 14:127–194
Einav S, Gerber D, Bryson PD et al (2008) Discovery of a hepatitis C target and its pharmacological inhibitors by microfluidic affinity analysis. Nat Biotechnol 26:1019–1027
Einav S, Sobol HD, Gehrig E, Glenn JS (2010) The hepatitis C virus (HCV) NS4B RNA binding inhibitor clemizole is highly synergistic with HCV protease inhibitors. J Infect Dis 202:65–74
El Hage N, Luo G (2003) Replication of hepatitis C virus RNA occurs in a membrane-bound replication complex containing nonstructural viral proteins and RNA. J Gen Virol 84:2761–2769
Erhardt A, Deterding K, Benhamou Y et al (2009) Safety, pharmacokinetics and antiviral effect of BILB 1941, a novel hepatitis C virus RNA polymerase inhibitor, after 5 days oral treatment. Antiviral Ther 14:23–32
Everson GT, Sims KD, Rodriguez-Torres M et al (2012) An interferon-free, ribavirin-free 12 weeks regimen of daclatasvir (DCV), asunaprevir (ASV), and BMS-791325 yielded SVR4 of 94 % in treatment-naïve patients with genotype (GT) 1 chronic hepatitis C virus (HCV) infection. In: Abstracts LB-3 of the 63rd annual meeting of the american association for the study of liver diseases, Boston, Massachusetts, 9–13 Nov 2012
Failla C, Tomei L, De Francesco R (1994) Both NS3 and NS4A are required for proteolytic processing of hepatitis C virus nonstructural proteins. J Virol 68:3753–3760
Failla C, Tomei L, De Francesco R (1995) An amino-terminal domain of the hepatitis C virus NS3 protease is essential for interaction with NS4A. J Virol 69:1769–1777
Ferenci P (2012) Treatment of chronic hepatitis C: are interferons really necessary? Liver Int 32(1):108–112
Forestier N, Zeuzem S (2012) Telaprevir for the treatment of hepatitis C. Expert Opin Pharmacother 13:593–606
Forestier N, Larrey D, Guyader D et al (2011) Treatment of chronic hepatitis C patients with the NS3/4A protease inhibitor danoprevir (ITMN-191/RG7227) leads to robust reductions in viral RNA: a phase 1b multiple ascending dose study. J Hepatol 54:1130–1136
Foster GR, Hezode C, Bronowicki JP et al (2011a) Telaprevir alone or with peginterferon and ribavirin reduces HCV RNA in patients with chronic genotype 2 but not genotype 3 infections. Gastroenterology 141(881–889):e881
Foster TL, Verow M, Wozniak AL et al (2011b) Resistance mutations define specific antiviral effects for inhibitors of the hepatitis C virus p7 ion channel. Hepatology 54:79–90
Fridell RA, Qiu D, Wang C et al (2010) Resistance analysis of the hepatitis C virus NS5A inhibitor BMS-790052 in an in vitro replicon system. Antimicrob Agents Chemother 54:3641–3650
Fridell RA, Wang C, Sun JH et al (2011) Genotypic and phenotypic analysis of variants resistant to hepatitis C virus nonstructural protein 5A replication complex inhibitor BMS-790052 in humans: in vitro and in vivo correlations. Hepatology 54:1924–1935
Gane EJ, Roberts SK, Stedman CAM et al (2010) Oral combination therapy with a nucleoside polymerase inhibitor (RG7128) and danoprevir for chronic hepatitis C genotype 1 infection (INFORM-1): a randomised, double-blind, placebo-controlled, dose-escalation trial. Lancet 376:1467–1475
Gane E, Stedman C, Hyland RH et al (2012a) Once daily sofosbuvir (GS-7977) plus ribavirin in patients with HCV genotypes 1, 2, and 3: the electron trial. In: Abstracts 229 of the 63rd annual meeting of the american association for the study of liver diseases, Boston, Massachusetts, 9–13 Nov 2012
Gane EJ, Stedman CA, Hyland RH et al (2012b) Electron: once daily PSI-7977 plus RBV in HCV GT1/2/3. J Hepatol 56(2):S438–S439
Gao M, Nettles RE, Belema M et al (2010) Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature 465:96–100
Gopalsamy A, Aplasca A, Ciszewski G et al (2006) Design and synthesis of 3,4-dihydro-1H-[1]-benzothieno[2,3-c]pyran and 3,4-dihydro-1H-pyrano[3,4-b]benzofuran derivatives as non-nucleoside inhibitors of HCV NS5B RNA dependent RNA polymerase. Bioorg Med Chem Lett 16:457–460
Goudreau N, Llinas-Brunet M (2005) The therapeutic potential of NS3 protease inhibitors in HCV infection. Expert Opin Investig Drugs 14:1129–1144
Gouttenoire J, Penin F, Moradpour D (2010) Hepatitis C virus nonstructural protein 4B: a journey into unexplored territory. Rev Med Virol 20:117–129
Gray F, Amphlett E, Bright H et al (2007) GSK625433; a novel and highly potent inhibitor of the HCVNS5B polymerase. J Hepatol 46:S225
Griffin SD, Beales LP, Clarke DS et al (2003) The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, amantadine. FEBS Lett 535:34–38
Griffin S, StGelais C, Owsianka AM et al (2008) Genotype-dependent sensitivity of hepatitis C virus to inhibitors of the p7 ion channel. Hepatology 48:1779–1790
Gu B, Johnston VK, Gutshall LL et al (2003) Arresting initiation of hepatitis C virus RNA synthesis using heterocyclic derivatives. J Biol Chem 278:16602–16607
Hebner CM, Han B, Brendza KM et al (2012) The HCV Non-nucleoside inhibitor tegobuvir utilizes a novel mechanism of action to inhibit NS5B polymerase function. Plos One 7:e39163
Hong Z, Cameron CE, Walker MP et al (2001) A novel mechanism to ensure terminal initiation by hepatitis C virus NS5B polymerase. Virology 285:6–11
Howe AY, Bloom J, Baldick CJ et al (2004) Novel nonnucleoside inhibitor of hepatitis C virus RNA-dependent RNA polymerase. Antimicrob Agents Chemother 48:4813–4821
Howe AY, Cheng H, Thompson I et al (2006) Molecular mechanism of a thumb domain hepatitis C virus nonnucleoside RNA-dependent RNA polymerase inhibitor. Antimicrob Agents Chemother 50:4103–4113
Howe AY, Cheng H, Johann S et al (2008) Molecular mechanism of hepatitis C virus replicon variants with reduced susceptibility to a benzofuran inhibitor, HCV-796. Antimicrob Agents Chemother 52:3327–3338
Huang Y, Staschke K, De Francesco R, Tan SL (2007) Phosphorylation of hepatitis C virus NS5A nonstructural protein: a new paradigm for phosphorylation-dependent viral RNA replication? Virology 364:1–9
Huang M, Podos S, Patel D et al (2010) ACH-2684: HCV NS3 protease inhibitor with potent activity against multiple genotypes and known resistant variants. Hepatology 52(S1):1204A
Jacobson I, Pockros P, Lalezari J et al (2009) Antiviral activity of filibuvir in combination with pegylated interferon alfa-2a and ribavirin for 28 days in treatment naive patients chronically infected with HCV genotype 1. J Hepatol 50:S382–S383
Kazmierski WM, Hamatake R, Duan M et al (2012) Discovery of novel urea-based hepatitis C protease inhibitors with high potency against protease-inhibitor-resistant mutants. J Med Chem 55:3021–3026
Khoury G, Ewart G, Luscombe C et al (2010) Antiviral efficacy of the novel compound BIT225 against HIV-1 release from human macrophages. Antimicrob Agents Chemother 54:835–845
Kieffer TL, Sarrazin C, Miller JS et al (2007) Telaprevir and pegylated interferon-alpha-2a inhibit wild-type and resistant genotype 1 hepatitis C virus replication in patients. Hepatology 46:631–639
Kim JL, Morgenstern KA, Lin C et al (1996) Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell 87:343–355
Kneteman NM, Howe AYM, Gao TJ et al (2009) HCV796: a selective nonstructural protein 5B polymerase inhibitor with potent anti-hepatitis C virus activity in vitro, in mice with chimeric human livers, and in humans infected with hepatitis C virus. Hepatology 49:745–752
Kowdley KV, Lawitz E, Crespo I et al (2012a) Atomic: 97 % RVR for PSI-7977 + PEG/RBV X 12 weeks regimen in HCV GT1: an end to response-guided therapy? J Hepatol 56(2):S1
Kowdley KV, Lawitz E, Poordad F et al (2012b) A 12 weeks interferon-free treatment regimen with ABT-450/r, ABT-267, ABT-333 and ribavirin achieves SVR12 rates (observed data) of 99 % in treatment-naïve patients and 93 % in prior null responders with HCV genotype1 infection. In: Abstracts LB-1 of the 63rd annual meeting of the american association for the study of liver diseases, Boston, Massachusetts, 9–13 Nov 2012
Kukolj G, McGibbon GA, McKercher G et al (2005) Binding site characterization and resistance to a class of non-nucleoside inhibitors of the hepatitis C virus NS5B polymerase. J Biol Chem 280:39260–39267
Kwo PY, Lawitz EJ, McCone J et al (2010) Efficacy of boceprevir, an NS3 protease inhibitor, in combination with peginterferon alfa-2b and ribavirin in treatment-naive patients with genotype 1 hepatitis C infection (SPRINT-1): an open-label, randomised, multicentre phase 2 trial. Lancet 376:705–716
Lagace L, White PW, Bousquet C et al (2012) In vitro resistance profile of the hepatitis C virus NS3 protease inhibitor BI 201335. Antimicrob Agents Chemother 56:569–572
Lam AM, Espiritu C, Bansal S et al (2011a) Hepatitis C virus nucleotide inhibitors PSI-352938 and PSI-353661 exhibit a novel mechanism of resistance requiring multiple mutations within replicon RNA. J Virol 85:12334–12342
Lam AM, Espiritu C, Murakami E et al (2011b) Inhibition of hepatitis C virus replicon RNA synthesis by PSI-352938, a cyclic phosphate prodrug of {beta}-D-2′-deoxy-2′-{alpha}-fluoro-2′-{beta}-C-Methylguanosine. Antimicrob Agents Chemother 55:2566–2575
Lam AM, Espiritu C, Bansal S et al (2012) Genotype and subtype profiling of PSI-7977 as a nucleotide inhibitor of hepatitis C virus. Antimicrob Agents Chemother 56:3359–3368
Lamarre D, Anderson PC, Bailey M et al (2003) An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426:186–189
Larrey DG, Benhamou Y, Lohse AW et al (2009) Bi 207127 is a potent HCV RNA polymerase inhibitor during 5 days monotherapy in patients with chronic hepatitis C. Hepatology 50:1044A
Larrey D, Lohse AW, de Ledinghen V et al (2012) Rapid and strong antiviral activity of the non-nucleosidic NS5B polymerase inhibitor BI 207127 in combination with peginterferon alfa 2a and ribavirin. J Hepatol 57:39–46
Lawitz E, Nguyen T, Younes Z et al (2006) Valopicitabine (NM283) plus PEG-interferon in treatment-naive hepatitis C patients with HCV genotype-1 infection: HCV RNA clearance during 24 weeks of treatment. Hepatology 44:223A
Lawitz E, Rodriguez-Torres M, DeMico M et al (2009) Antiviral activity of ANA598, a potent non-nucleoside polymerase inhibitor, in chronic hepatitis C patients. J Hepatol 50:S384
Lawitz E, Gaultier I, Poordad F et al (2010a) 4 weeks virologic response and safety of ABT-450 given with low-dose ritonavir (ABT-450r) in combination with pegylated interferon alfa 2a and ribavirin after 3 days monotherapy in HCV-infected treatment-naive subjects. Hepatology 52(1):878A–979A
Lawitz E, Rodriguez-Torres M, Rustgi V et al (2010b) Safety and antiviral activity of ANA598 in combination with pegylated IFN alpha 2a plus ribavirin in treatment-naïve genotype-1 chronic HCV patients. Hepatology 52:334A–335A
Lawitz E, Rodriguez-Torres M, Denning JM et al (2011) Once dialy dual-nucleotide combination of PSI-938 and PSI-7977 provides 94 % HCV RNA < LOD at day 14: first purine/pyrimidine clinical combination data (the nuclear study). J Hepatol 54:S543
Lawitz EJ, Gruener D, Hill JM et al (2012) A phase 1, randomized, placebo-controlled, 3 days, dose-ranging study of GS-5885, an NS5A inhibitor, in patients with genotype 1 hepatitis C. J Hepatol 57:24–31
Le Pogam S, Jiang WR, Leveque V et al (2006a) In vitro selected Con1 subgenomic replicons resistant to 2′-C-methyl-cytidine or to R1479 show lack of cross resistance. Virology 351:349–359
Le Pogam S, Kang H, Harris SF et al (2006b) Selection and characterization of replicon variants dually resistant to thumb- and palm-binding nonnucleoside polymerase inhibitors of the hepatitis C virus. J Virol 80:6146–6154
Le Pogam S, Seshaadri A, Ewing A et al (2010) RG7128 alone or in combination with pegylated interferon-alpha 2a and ribavirin prevents hepatitis C virus (HCV) replication and selection of resistant variants in HCV-infected patients. J Infect Dis 202:1510–1519
Lee LY, Tong CY, Wong T, Wilkinson M (2012) New therapies for chronic hepatitis C infection: a systematic review of evidence from clinical trials. Int J Clin Pract 66:342–355
Lemm JA, O’Boyle D 2nd, Liu M et al (2010) Identification of hepatitis C virus NS5A inhibitors. J Virol 84:482–491
Lemm JA, Leet JE, O’Boyle DR 2nd et al (2011) Discovery of potent hepatitis C virus NS5A inhibitors with dimeric structures. Antimicrob Agents Chemother 55:3795–3802
Lenz O, Verbinnen T, Lin TI et al (2010) In vitro resistance profile of the hepatitis C virus NS3/4A protease inhibitor TMC435. Antimicrob Agents Chemother 54:1878–1887
Lesburg CA, Cable MB, Ferrari E et al (1999) Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat Struct Biol 6:937–943
Li H, Tatlock J, Linton A et al (2009) Discovery of (R)-6-Cyclopentyl-6-(2-(2,6-diethylpyridin-4-yl)ethyl)-3-((5,7-dimethyl-[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)methyl)-4-hydroxy-5,6-dihydropyran-2-on e (PF-00868554) as a potent and orally available hepatitis C virus polymerase inhibitor. J Med Chem 52:1255–1258
Lin C, Thomson JA, Rice CM (1995) A central region in the hepatitis C virus NS4A protein allows formation of an active NS3-NS4A serine proteinase complex in vivo and in vitro. J Virol 69:4373–4380
Lin C, Lin K, Luong YP et al (2004) In vitro resistance studies of hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061: structural analysis indicates different resistance mechanisms. J Biol Chem 279:17508–17514
Lin C, Kwong AD, Perni RB (2006) Discovery and development of VX-950, a novel, covalent, and reversible inhibitor of hepatitis C virus NS3-4A serine protease. Infect Disord Drug Targets 6:3–16
Lin TI, Lenz O, Fanning G et al (2009) In vitro activity and preclinical profile of TMC435350, a potent hepatitis C virus protease inhibitor. Antimicrob Agents Chemother 53:1377–1385
Liu R, Kong R, Mann P et al (2012) In vitro resistance analysis of HCV NS5A inhibitor MK-8742 demonstrates increased potency against clinical resistance variants ans higher resistance barrier. J Hepatol 56(2):S334–S335
Liverton NJ, Carroll SS, Dimuzio J et al (2010) MK-7009, a potent and selective inhibitor of hepatitis C virus NS3/4A protease. Antimicrob Agents Chemother 54:305–311
Love RA, Parge HE, Wickersham JA et al (1996) The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site. Cell 87:331–342
Love RA, Brodsky O, Hickey MJ et al (2009) Crystal structure of a novel dimeric form of NS5A domain I protein from hepatitis C virus. J Virol 83:4395–4403
Ludmerer SW, Graham DJ, Boots E et al (2005) Replication fitness and NS5B drug sensitivity of diverse hepatitis C virus isolates characterized by using a transient replication assay. Antimicrob Agents Chemother 49:2059–2069
Luik P, Chew C, Aittoniemi J et al (2009) The 3-dimensional structure of a hepatitis C virus p7 ion channel by electron microscopy. Proc Natl Acad Sci USA 106:12712–12716
Luscombe CA, Huang Z, Murray MG et al (2010) A novel hepatitis C virus p7 ion channel inhibitor, BIT225, inhibits bovine viral diarrhea virus in vitro and shows synergism with recombinant interferon-alpha-2b and nucleoside analogues. Antiviral Res 86:144–153
Ma H, Jiang WR, Robledo N et al (2007) Characterization of the metabolic activation of hepatitis C virus nucleoside inhibitor beta-D-2′-deoxy-2′-fluoro-2′-C-methylcytidine (PSI-6130) and identification of a novel active 5′-triphosphate species. J Biol Chem 282:29812–29820
Macdonald A, Harris M (2004) Hepatitis C virus NS5A: tales of a promiscuous protein. J Gen Virol 85:2485–2502
Manns MP, Bourliere M, Benhamou Y et al (2011) Potency, safety, and pharmacokinetics of the NS3/4A protease inhibitor BI201335 in patients with chronic HCV genotype-1 infection. J Hepatol 54:1114–1122
May MM, Brohm D, Harrenga A et al (2012) Discovery of substituted N-phenylbenzenesulphonamides as a novel class of non-nucleoside hepatitis C virus polymerase inhibitors. Antiviral Res 95:182–191
McCown MF, Rajyaguru S, Le Pogam S et al (2008) The hepatitis C virus replicon presents a higher barrier to resistance to nucleoside analogs than to nonnucleoside polymerase or protease inhibitors. Antimicrob Agents Chemother 52:1604–1612
McGuigan C, Madela K, Aljarah M et al (2010) Design, synthesis and evaluation of a novel double pro-drug: INX-08189. A new clinical candidate for hepatitis C virus. Bioorg Med Chem Lett 20:4850–4854
McPhee F, Friborg J, Levine S et al (2012a) Resistance analysis of the hepatitis C virus NS3 protease inhibitor asunaprevir. Antimicrob Agents Chemother 56:3670–3681
McPhee F, Sheaffer AK, Friborg J et al (2012b) Preclinical profile and characterization of the hepatitis C virus NS3 protease inhibitor asunaprevir (BMS-650032). Antimicrob Agents Chemother 56:5387–5396
Membreno FE, Lawitz EJ (2011) The HCV NS5B nucleoside and non-nucleoside inhibitors. Clin Liver Dis 15:611–626
Migliaccio G, Tomassini JE, Carroll SS et al (2003) Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J Biol Chem 278:49164–49170
Moreno C, Berg T, Tanwandee T et al (2012) Antiviral activity of TMC435 monotherapy in patients infected with HCV genotypes 2–6: TMC435-C202, a phase IIa, open-label study. J Hepatol 56:1247–1253
Murakami E, Bao H, Ramesh M et al (2007) Mechanism of activation of beta-D-2′-deoxy-2′-fluoro-2′-c-methylcytidine and inhibition of hepatitis C virus NS5B RNA polymerase. Antimicrob Agents Chemother 51:503–509
Murakami E, Niu CR, Bao HY et al (2008) The mechanism of action of beta-D-2′-deoxy-2′-fluoro-2′-C-methylcytidme involves a second metabolic pathway leading to beta-D-2′-deoxy-2-fluoro-2′-C-methyluridine 5′-triphosphate, a potent inhibitor of the hepatitis C virus RNA-dependent RNA polymerase. Antimicrob Agents Chemother 52:458–464
Narjes F, Crescenzi B, Ferrara M et al (2011) Discovery of (7R)-14-cyclohexyl-7-{[2-(dimethylamino)ethyl](methyl) amino}-7,8-dihydro-6H-indolo[1,2-e][1,5]benzoxazocine-11-carboxylic acid (MK-3281), a potent and orally bioavailable finger-loop inhibitor of the hepatitis C virus NS5B polymerase. J Med Chem 54:289–301
Nelson DR, Gane EJ, Jacobson IM et al (2011) VX-222/telaprevir in combination with peginterferon-alfa-2a and ribavirin in treatment-naive genotype 1 HCV patients treated for 12 weeks: zenith study, SVR12 interim analysis. Hepatology 54:1435A
Nguyen TT, Gates AT, Gutshall LL et al (2003) Resistance profile of a hepatitis C virus RNA-dependent RNA polymerase benzothiadiazine inhibitor. Antimicrob Agents Chemother 47:3525–3530
Olsen DB, Eldrup AB, Bartholomew L et al (2004) A 7-deaza-adenosine analog is a potent and selective inhibitor of hepatitis C virus replication with excellent pharmacokinetic properties. Antimicrob Agents Chemother 48:3944–3953
Pasquinelli C, McPhee F, Eley T et al (2012) Single- and multiple-ascending-dose studies of the NS3 protease inhibitor asunaprevir in subjects with or without chronic hepatitis C. Antimicrob Agents Chemother 56:1838–1844
Pauwels F, Mostmans W, Quirynen LMM et al (2007) Binding-site identification and genotypic profiling of hepatitis C virus polymerase inhibitors. J Virol 81:6909–6919
Pawlotsky JM, Najera I, Jacobson I (2012) Resistance to mericitabine, a nucleoside analogue inhibitor of HCV RNA-dependent RNA polymerase. Antivir Ther 17:411–423
Pessoa MG, Cheinquer H, Almeida PR et al (2012) Re-treatment of previous non-responders and relapsers to interferon plus ribavirin with peginterferon alfa-2a (40KD), ribavirin ± amantadine in patients with chronic hepatitis C: randomized multicentre clinical trial. Ann Hepatol 11:52–61
Pierra C, Amador A, Benzaria S et al (2006) Synthesis and pharmacokinetics of valopicitabine (NM283), an efficient prodrug of the potent anti-HCV agent 2′-C-methylcytidine. J Med Chem 49:6614–6620
Pol S, Ghalib RH, Rustgi VK et al (2012) Daclatasvir for previously untreated chronic hepatitis C genotype-1 infection: a randomised, parallel-group, double-blind, placebo-controlled, dose-finding, phase 2a trial. Lancet Infect Dis 12:671–677
Poordad F, Lawitz E, Kowdley KV et al (2012a) 12 weeks interferon-free regimen of ABT-450/R + ABT-333 + ribavirin achieved SVR12 in more than 90 % of treatment-naive HCV genotype-1-infected subjects and 47 % of previous non-responders. J Hepatol 56(2):S549–S550
Poordad F, Lawitz E, Kowdley KV et al (2012b) 12 weeks interferon-free regimen of ABT-450/R + ABT-333 + ribavirin achieved svr12 in more than 90 % of treatment-naive HCV genotype-1-infected subjects and 47 % of previous non-responders. J Hepatol 56:S549–S550
Qiu D, Lemm JA, O’Boyle DR 2nd et al (2011) The effects of NS5A inhibitors on NS5A phosphorylation, polyprotein processing and localization. J Gen Virol 92:2502–2511
Rai R, Deval J (2011) New opportunities in anti-hepatitis C virus drug discovery: targeting NS4B. Antiviral Res 90:93–101
Reddy MB, Chen Y, Haznedar JO et al (2012) Impact of low-dose ritonavir on danoprevir pharmacokinetics: results of computer-based simulations and a clinical drug-drug interaction study. Clin Pharmacokinet 51:457–465
Reesink HW, Fanning GC, Farha KA et al (2010) Rapid HCV-RNA decline with once daily TMC435: a phase I study in healthy volunteers and hepatitis C patients. Gastroenterology 138:913–921
Reghellin V, Fenu S, Bianco A et al (2012) HCV NS5A inhibitor BMS-790052 exerts its antiviral effects by reducing phosphatidylinositol 4-phosphate levels in the virus-induced membranous web. In: Abstracts LB-9 of the 19th international symposium on hepatitis C virus and related viruses, Venice, Italy, 5–9 Octo 2012
Reiss S, Rebhan I, Backes P et al (2011) Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe 9:32–45
Rodriguez-Torres M, Lawitz E, Conway B et al (2010) Safety and antiviral activity of the HCV non-nucleoside polymerase inhibitor VX-222 in treatment-naive genotype 1 HCV-infected patients. J Hepatol 52:S14
Rodriguez-Torres M, Lawitz E, Hazan L et al (2011) Antiviral activity and safety of INX-08189, a nucleotide polymerase inhibitor, following 7 days of oral therapy in naive genotype-1 chronic HCV patients. Hepatology 54:535A
Romano KP, Ali A, Aydin C et al (2012) The molecular basis of drug resistance against hepatitis C virus NS3/4A protease inhibitors. Plos Pathog 8:e1002832
Sakai A, St Claire MS, Faulk K et al (2003) The p7 polypeptide of hepatitis C virus is critical for infectivity and contains functionally important genotype-specific sequences. Proc Natl Acad Sci USA 100:11646–11651
Sarrazin C, Zeuzem S (2010) Resistance to direct antiviral agents in patients with hepatitis C virus infection. Gastroenterology 138:447–462
Sarrazin C, Kieffer TL, Bartels D et al (2007) Dynamic hepatitis C virus genotypic and phenotypic changes in patients treated with the protease inhibitor telaprevir. Gastroenterology 132:1767–1777
Sarrazin C, Hezode C, Zeuzem S, Pawlotsky JM (2012) Antiviral strategies in hepatitis C virus infection. J Hepatol 56(1):S88–S100
Schaefer EA, Chung RT (2012) Anti-hepatitis C virus drugs in development. Gastroenterology 142(1340–1350):e1341
Schechter I, Berger A (1967) On the size of the active site in proteases. I Papain Biochem Biophys Res Commun 27:157–162
Seiwert SD, Andrews SW, Jiang Y et al (2008) Preclinical characteristics of the hepatitis C virus NS3/4A protease inhibitor ITMN-191 (R7227). Antimicrob Agents Chemother 52:4432–4441
Sheng XC, Appleby T, Butler T et al (2012a) Discovery of GS-9451: an acid inhibitor of the hepatitis C virus NS3/4A protease. Bioorg Med Chem Lett 22:2629–2634
Sheng XC, Casarez A, Cai R et al (2012b) Discovery of GS-9256: a novel phosphinic acid derived inhibitor of the hepatitis C virus NS3/4A protease with potent clinical activity. Bioorg Med Chem Lett 22:1394–1396
Shepard CW, Finelli L, Alter MJ (2005) Global epidemiology of hepatitis C virus infection. Lancet Infect Dis 5:558–567
Shi ST, Herlihy KJ, Graham JP et al (2009) Preclinical characterization of PF-00868554, a potent nonnucleoside inhibitor of the hepatitis C virus RNA-dependent RNA polymerase. Antimicrob Agents Chemother 53:2544–2552
Slater MJ, Amphlett EM, Andrews DM et al (2007) Optimization of novel acyl pyrrolidine inhibitors of hepatitis C virus RNA-dependent RNA polymerase leading to a development candidate. J Med Chem 50:897–900
Smith JP (1997) Treatment of chronic hepatitis C with amantadine. Dig Dis Sci 42:1681–1687
Steinmann E, Penin F, Kallis S et al (2007a) Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. Plos Pathog 3:962–971
Steinmann E, Whitfield T, Kallis S et al (2007b) Antiviral effects of amantadine and iminosugar derivatives against hepatitis C virus. Hepatology 46:330–338
Stuyver LJ, McBrayer TR, Tharnish PM et al (2006) Inhibition of hepatitis C replicon RNA synthesis by beta-D-2′-deoxy-2′-fluoro-2′-C-methylcytidine: a specific inhibitor of hepatitis C virus replication. Antivir Chem Chemother 17:79–87
Sulkowski M, Rodriguez-Torres M, Lawitz E et al (2012a) High sustained virologic response rate in treatment-naïve HCV genotype 1a and 1b patients treated for 12 weeks with an interferon-free all-oral quad regimen: interim results. J Hepatol 56:S560
Sulkowski MS, Gardiner DF, Rodriguez-Torres M et al (2012b) High rate of sustained virologic response with the all-oral combination of daclatasvir (NS5A inhibitor) plus sofosbuvir (nucleotide NS5B inhibitor), with or without ribavirin, in treatment-naïve patients chronically infected with HCV genotype 1, 2, or 3. In: Abstracts LB-2 of the 63rd annual meeting of the american association for the study of liver diseases, Boston, Massachusetts, 9–13 Nov 2012
Summa V, Ludmerer SW, McCauley JA et al (2012) MK-5172, a selective inhibitor of hepatitis C virus NS3/4a protease with broad activity across genotypes and resistant variants. Antimicrob Agents Chemother 56:4161–4167
Susser S, Welsch C, Wang Y et al (2009) Characterization of resistance to the protease inhibitor boceprevir in hepatitis C virus-infected patients. Hepatology 50:1709–1718
Tanwandee T, Luscombe CA, Ewart G et al (2011) Antiviral activity and tolerability of BIT225 plus pegylated interferon alpha 2a or 2b and weight-based ribavirin for 28 days in HCV treatment-naïve monoinfected patients. Glob Antiviral J 7(1):64 HEP DART 2011
Targett-Adams P, Graham EJ, Middleton J et al (2011) Small molecules targeting hepatitis C virus-encoded NS5A cause subcellular redistribution of their target: insights into compound modes of action. J Virol 85:6353–6368
Tellinghuisen TL, Marcotrigiano J, Rice CM (2005) Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature 435:374–379
Thompson P, Patel R, Steffy K, Appleman J (2009) Preclinical studies of ANA598 combined with other Anti-HCV agents demonstrate potential of combination treatment. J Hepatol 50:S37
Tomei L, Failla C, Santolini E et al (1993) NS3 is a serine protease required for processing of hepatitis C virus polyprotein. J Virol 67:4017–4026
Tomei L, Altamura S, Bartholomew L et al (2003) Mechanism of action and antiviral activity of benzimidazole-based allosteric inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J Virol 77:13225–13231
Tomei L, Altamura S, Bartholomew L et al (2004) Characterization of the inhibition of hepatitis C virus RNA replication by nonnucleosides. J Virol 78:938–946
Trozzi C, Bartholomew L, Ceccacci A et al (2003) In vitro selection and characterization of hepatitis C virus serine protease variants resistant to an active-site peptide inhibitor. J Virol 77:3669–3679
Vernachio JH, Bleiman B, Bryant KD et al (2011) INX-08189, a phosphoramidate prodrug of 6-O-methyl-2′-C-methyl guanosine, is a potent inhibitor of hepatitis C virus replication with excellent pharmacokinetic and pharmacodynamic properties. Antimicrob Agents Chemother 55:1843–1851
von Wagner M, Hofmann WP, Teuber G et al (2008) Placebo-controlled trial of 400 mg amantadine combined with peginterferon alfa-2a and ribavirin for 48 weeks in chronic hepatitis C virus-1 infection. Hepatology 48:1404–1411
Wagaw S, Ravn M, Engstrom K et al (2009) Process for making macrocyclic oximyl hepatitis C protease inhibitors. WO/2009/073780
Wagner F, Thompson R, Kantaridis C et al (2011) Antiviral activity of the hepatitis C virus polymerase inhibitor filibuvir in genotype 1-infected patients. Hepatology 54:50–59
Wang M, Ng KK, Cherney MM et al (2003) Non-nucleoside analogue inhibitors bind to an allosteric site on HCV NS5B polymerase. Crystal structures and mechanism of inhibition. J Biol Chem 278:9489–9495
White PW, Llinas-Brunet M, Amad M et al (2010) Preclinical characterization of BI 201335, a C-terminal carboxylic acid inhibitor of the hepatitis C virus NS3-NS4A protease. Antimicrob Agents Chemother 54:4611–4618
Yan Y, Li Y, Munshi S et al (1998) Complex of NS3 protease and NS4A peptide of BK strain hepatitis C virus: a 2.2 A resolution structure in a hexagonal crystal form. Protein Sci 7:837–847
Yang G, Wiles J, Patel D et al (2012) Preclinical characteristics of of ACH-3102: a novel HCV NS5A inhibitor with imporved potency against genotype-1a virus and variants resistant to 1st generation of NS5A inhibitors. J Hepatol 56(2):S330
Yi G, Deval J, Fan B et al (2012) Biochemical study of the comparative inhibition of hepatitis C virus RNA polymerase by VX-222 and filibuvir. Antimicrob Agents Chemother 56:830–837
Zeuzem S, Buggisch P, Agarwal K et al (2012a) The protease inhibitor, GS-9256, and non-nucleoside polymerase inhibitor tegobuvir alone, with ribavirin, or pegylated interferon plus ribavirin in hepatitis C. Hepatology 55:749–758
Zeuzem S, Soriano A, Asselah T et al (2012b) Interferon (IFN)-free combination treatment with the HCV NS3/4A protease inhibitor BI 201335 and the non-nucleoside NS5B inhibitor BI 207127 ± ribavirin (R): final results of SOUND-C2 and predictors of response. In: Abstracts 232 of the 63rd annual meeting of the american association for the study of liver diseases, Boston, Massachusetts, 9–13 Nov 2012
Zhou XJ, Pietropaolo K, Chen J et al (2011) Safety and pharmacokinetics of IDX184, a liver-targeted nucleotide polymerase inhibitor of hepatitis C virus, in healthy subjects. Antimicrob Agents Chemother 55:76–81
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Delang, L., Neyts, J., Vliegen, I., Abrignani, S., Neddermann, P., De Francesco, R. (2013). Hepatitis C Virus-Specific Directly Acting Antiviral Drugs. In: Bartenschlager, R. (eds) Hepatitis C Virus: From Molecular Virology to Antiviral Therapy. Current Topics in Microbiology and Immunology, vol 369. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-27340-7_12
Download citation
DOI: https://doi.org/10.1007/978-3-642-27340-7_12
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-27339-1
Online ISBN: 978-3-642-27340-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)