Abstract
The toxicity of plant proteins, later identified as ribosome-inactivating proteins (RIPs), was described more than a century ago and their enzymatic activity was established more than 30 years ago. However, their physiological role and related biological activities are still uncertain. Therefore, despite the body of literature, research on RIPs is ongoing. This review deals with new RIPs being purified, sequenced, characterized, and cloned, and an increasing number of 3D-structures that are determined at high resolution. This is the case of the five type 1 RIPs (PD-S1-3, PD-L1/2, PD-L3/4, dioicin 1, and dioicin 2) from seeds and leaves of the ombú tree (Phytolacca dioica L.), native of the grassy pampas of Argentina. The data collected so far will contribute to our understanding of important issues of RIP research: (1) identifying structural determinants responsible for new enzymatic activities such as the DNA cleaving activity; (2) glycosylation and its influence on the catalytic and biological activities; (3) cellular localization of endogenous RIPs and their physiological role(s).
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Ribosome-inactivating proteins (RIPs; rRNA N-β-glycosidases; EC 3.2.2.22) have been isolated from a number of higher plants; fungi, bacteria, and at least one alga (Girbés et al. 2004). The genus Phytolacca (Fam. Phytolaccaceae) has several tens of species of herbs, shrubs, and trees.Footnote 1 American pokeweed (Phytolacca americana L.)Footnote 2 and Indian pokeweed (Phytolacca esculenta Van Houtte)Footnote 3 contain antiviral proteins whose action was described long before their recognition as inhibitors of protein synthesis (Duggar and Armstrong 1925; Kassanis and Kleczkowski 1948). Moreover, the first type 1 RIP, an antiviral protein, was identified from American pokeweed (PAP; Dallal and Irvin 1978).
This chapter will focus on the isolation and characterization of type 1 RIPs from the ombú treeFootnote 4 (Phytolacca dioica Footnote 5 L.; Fig. 1). The plant is very useful for this type of research, because it produces new leaves for many months, except at the end of the winter, and can be propagated by seeds, thus allowing the monitoring of RIP expression under several experimental conditions.
(a) P. dioica tree growing in the Botanical Garden of the University of Naples Federico II, Italy. This 100-year-old plant provided leaves and seeds (b and c, respectively) for the purification of RIPs described in the chapter. P. dioica was introduced in Europe in 1768 and in Italy in 1840. It was propagated in the Botanical Garden of the University of Naples by seeds supplied by the French Botanist Aimé Bonplant
2 RIPs from P. dioica L.
The genus Phytolacca is a rich source of several highly conserved RIPs. The ability of PAP, isolated from P. americana leaves, to inhibit protein synthesis by enzymatically damaging ribosomes was initially reported by Obrig et al. (1973). Indeed, several members of this genus have been found to contain type 1 RIPs, such as PAP isoforms from P. americana seeds, leaves, and root cultures (Irvin 1975; Irvin et al. 1980; Rajamohan et al. 1999; Park et al. 2002), dodecandrins from leaves and cell cultures of Phytolacca dodecandra L’HeritFootnote 6 (Ready et al. 1984; Thomsen et al. 1991), heterotepalins from Phytolacca heterotepala H. Walter (Mexican pokeweed; Di Maro et al. 2007), insularin from Phytolacca insularis Nakai (Song et al. 2000), and PAP-icos isoforms from Phytolacca icosandra Footnote 7 L. (red ink plant).
The first paper on the presence of RIPs in P. dioica was published by Parente et al. (1993), reporting the purification to homogeneity of three type 1 RIPs from seeds. The major form, named PD-S2 (P. dioica-seeds 2), accounted for about 90% of the total protein synthesis inhibitory activity of the crude seed extract (Parente et al. 1993). After this first report, four type 1 RIPs were isolated and characterized both structurally and functionally from the leaves of the same plant (Di Maro et al. 1999; Parente et al. 2008).
2.1 Isolation of RIPs from Seeds and Leaves of P. dioica
Type 1 RIPs from seeds (PD-Ss; Parente et al. 1993), fully expanded leaves of adult P. dioica (PD-L1, PD-L2, PD-L3, and PD-L4; Di Maro et al. 1999; Parente et al. 2008), fully expanded leaves of young P. dioica (8–36 months old) and developing leaves of adult plants from 10 to 60-days old (dioicin 1 and dioicin 2; Parente et al. 2008) have been isolated and characterized. Preliminary data on RIP purification from the bark of summer shoots have shown the presence of the four RIPs isolated from fully expanded leaves of adult P. dioica PD-Ls1–4 (unpublished).
Experimental conditions employed for RIP isolation from P. dioica are reported in Table 1. The multi-step purification protocols provided samples with high purity, which facilitated structural studies (mass spectrometry analysis, sequence and pI determination, X-ray studies) and lent confidence to the enzymatic characteristics, especially the adenine polynucleotide glycosylase (APG) and DNA-cleaving activities. Indeed, some purification steps were essential for full separation of closely related RIPs (isoforms): in the case of the PD-S forms, with the same primary structure but different glycosylation pattern (see later), using the CM Sepharose resin for the second cation-exchange chromatography was essential not only for the complete separation of the three native forms, but also for nicked PD-S2 forms, one with a cut between Asn195 and Arg196 (Di Maro et al. 1995) and the other between Asp82 and Pro83 (Zacchia et al. 2009). The in vivo occurrence and the likely biological significance of these nicked seed forms are currently being investigated. Similarly, the third cation-exchange chromatography on S-Sepharose fast flow, after the CM-52 step, allowed the complete separation of PD-L2 from PD-L3. The yields of the CM-52 purified PD-L1, PD-L2, PD-L3, PD-L4, dioicin 1, and dioicin 2 were 1.54, 0.72, 2.48, 4.0, 0.08, and 0.61 mg/100 g leaves, respectively. In the case of PD-S1–3, the yields were 7.0, 86.0, and 3.4 mg/100 g seeds, respectively.
2.2 Basic Characteristics of RIPs from Seeds and Leaves of P. dioica
The main structural characteristics (Table 2) of the purified RIPs are well within the canonical parameters of type 1 RIPs: basic pI (range 7.5–9.5) and a ratio Lys + Arg/Asp + Glu higher than 1, Mr of the unglycosylated forms about 30 k, 261–266 amino acid residues, with an expected prevalence of basic amino acid residues, four half cysteines engaged in two S–S bridges (Di Maro et al. 2009; Parente et al. 2008), an average theoretical molar extinction coefficient at 280 nm 27,762.5 (corresponding to E0.1% = 1mg/mL = 0.941) and a GRAVY index in the range 0.343–0.559 (Table 2).
The amino acid sequencesFootnote 8 of PD-S1-3; PD-L1, PD-L2, PD-L3, PD-L4, and dioicin 2 were determined using a combined approach based on Edman degradation and mass spectrometry (Fig. 2a; Del Vecchio Blanco et al. 1997; Chambery et al. 2008; Parente et al. 2008; Di Maro et al. 2009). The sequence determination of dioicin 1 is ongoing. The comparative analysis of the amino acid sequences (with a consensus sequence of 278 amino acid residues) shows that PD-L1 and PD-L2 have identical primary structures, as is the case for PD-L3 and PD-L4. Hereafter, they will be reported as PD-L1/2 and PD-L3/4. Even PD-S RIPs were found to have the same amino acid sequence each other (Chambery et al. 2008). In the case of PD-L1/2, a microheterogeneity was found by mass spectrometry at position 20, with the alternative presence of Met or Leu. The identity/similarity matrix (Fig. 2b) shows that the highest identity (81.6%; 85.1% similarity) is between PD-L1/2 and PD-L3/4, while the lowest is between dioicin 2 and PD-L1/2 (36.2%; 46.4% similarity). The identity values of dioicin 2 with PS-Ss and PD-L3/4 is 39 and 41.3%, respectively.
(a) Multiple alignment of RIP sequences from P. dioica (PD-Ss, PD-L1/2, PD-L3/4, and dioicin 2 (accession numbers P34967, P84853, P84854, and P85208, respectively). Asterisk – identical, double dots – conserved and single dot – semiconserved amino acid residues. N-glycosylation sites have been shaded. Copyright indicates conserved amino acid residues found in the active site of Phytolaccaceae RIPs. (b) identity–similarity matrix of PD-Ss, PD-L1/2, PD-L3/4, and dioicin 2. Sequences were first aligned by the algorithm Clustal W2.0.11 and then analyzed by BOXSHADE
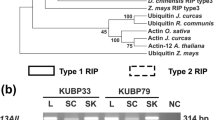
Figure 3a reports an identity/similarity matrix of RIP sequences from P. dioica, P. americana, P. acinosa, P. heterotepala, P. insularis, and P. icosandra.
(a) Identity/similarity matrix of type 1 RIP sequences from Phytolaccaceae. See Fig. 2 for P. dioica RIP sequences (PD-Ss, PD-L1/2, PD-L3/4, and dioicin 2). Sequences of RIPs other than from P. dioica (PAP-α, PAP, PAP-I, PAPII, PAP-R, PAP-S from P. americana; heterotepalin 4 from P. heterotepala; PIP 2 from P. insularis; Pap-aci and PapS1-aci from P. acinosa: PAP-icos and PAPII-icos from P. icosandra) were obtained from PubMed (Phytolaccaceae → taxonomy search). Sequences were first aligned by the algorithm Clustal W2.0.11 and then analyzed by BOXSHADE. (b) Unrooted phylogenetic tree of RIPs reported in (a). The tree shows the close relationship of dioicin 2 with PAP-II and PAPII-icos and their distance from other RIPs from P. americana and P. dioica, from both seeds and leaves. PD-S2 from seeds of P. dioica is more closely related to PAP-I or PAP-R from leaves or roots of P. americana, respectively, than to RIPs from seeds of P. americana (PAP, PAP-S, PAP-α) or PD-L4 from leaves of P. dioica. The neighbor-joining method was used with Poisson corrected distances. Scale bar, substitutions/site
Dioicin 2 has the lowest identity percentage (35.1%) with PD-L1/2 isolated from the same tissue of the same plant. The highest percentages are with PAPII-icos (86.1%) and PAP-II (81.6%). The identity values with the other RIPs are less than 41%.
The unrooted phylogenetic tree of the Phytolacca RIPs (Fig. 3b) clearly shows that dioicin 2, PAP II and PAPII-icos are located on a separate branch and may give rise to other RIPs. All other RIPs are grouped in four branches: (1) PAP-R; (2) PIP 2, PAP alpha, and PD-Ss; (3) PAP-icos, heterotepalin, Pap-aci, and PAP-I; (4) PD-L3/4, PAP, PD-L1/2, PapS1-aci, and PAP-S.
PD-L1/2 and PD-L3/4 from P. dioica leaves appear more closely related to RIPs from P. americana seeds than to PD-Ss from seeds of the same plant.
2.3 Differential Seasonal and Age Expression in Leaves
The expression of RIPs is developmentally regulated (Iglesias et al. 2008; Parente et al. 2008) and under transcriptional control (Kawade and Masuda 2009).
A differential seasonal expression was first found for PAP and PAP-II from P. americana, isolated from spring and summer leaves, respectively (Houston et al. 1983). This notion was later revisited by Rajamohan et al. (1999), reporting the expression of PAP-I (corresponding to PAP), PAP-II and PAP-III, in spring, early summer, and late summer leaves, respectively.
P. dioica has been very useful for studying RIP expression, because it produces new leaves for many months, except at the end of the winter, thus making it possible to monitor RIP activity during leaf ontogeny during most of the year. We have found that PD-L1, PD-L2, PD-L3, and PD-L4 RIPs from fully expanded leaves of adult P. dioica show differential seasonal and age expression. PD-L3 and PD-L4 are abundant in spring and summer leaves, decrease in autumn, and almost disappear in winter, when PD-L1 represents 80% of the RIP isoforms synthesized. On the contrary, the expression of PD-L2 remains constant throughout the year. PD-Ls 1–4 are not present in fully expanded leaves of young P. dioica (8–36 months old), where they appear to be replaced by two novel RIPs, dioicin 1 and dioicin 2. Furthermore, in developing leaves of adult plants (from 10- to 60- days old), PD-Ls 1–4 and dioicin 2 are always present, while dioicin 1 can be detected only at day 10 and 17. Dioicin 2 is also present in fully developed leaves of the adult plant; therefore, its expression is neither age- nor seasonally regulated.
The fact that P. dioica synthesizes and accumulates RIPs indirectly indicates that plant fitness benefits from these processes. In this view, the seasonal changes of RIP pattern could be due to the different and potentially adverse environmental conditions suffered, at least by the adult plant, in each season. Therefore, PD-L3 and PD-L4, mainly expressed in summer, could improve plant tolerance to drought and/or heat, whereas PD-L1, abundant in winter, may contribute to the avoidance of different abiotic stresses. Moreover, PD-Ls could be involved in the already known salt tolerance of P. dioica (Wheat 1977), thus conferring an adaptive, other than functional, role. As for the prevalence of diocin 1 and dioicin 2 in young plants, this may be due to the need to protect young and perhaps more susceptible tissues to pathogen attacks, e.g., by inactivating host ribosomes after virus challenge. The presence of diocin 1 only in developing leaves of adult plants could lend support to this hypothesis. Interestingly, treatment of tobacco and bean leaves with diocin 2 greatly reduced the infection of tobacco necrosis virus (TNV), an uncapped virus without the 5′ terminal m7GpppG cap, by inducing a slight H2O2 burst, but without activating any cell death phenomena (Faoro et al. 2008).
All the above suggested roles for P. dioica RIPs are also supported by the evidence of the newly reported enzymatic properties of RIPs other than N-glycosidase activity, such as RNAse, DNAse, superoxide dismutase (SOD), and phospholipase activities (reviewed in Park et al. 2004a, b). It must also be taken into account that senescence may induce RIP synthesis as well (Sawasaki et al. 2008). In this context, cytological changes activated by aging, as well as by pathogen infection, could alter the RIP compartmentalization, thus exposing the host ribosomes to the their action.
2.4 Cellular Localization
Diocin 1 and dioicin 2 from fully expanded leaves of young P. dioica were localized in the extracellular space, in the vacuole and in the Golgi apparatus of mesophyll cells (Parente et al. 2008). The presence of RIPs in the extracellular fluid was ascertained by western blot. For immunocytochemical localization studies, antidioicin 2 IgGs gave a positive signal mainly localized in the extracellular space of mesophyll cells, where they often aggregated forming amorphous and scarcely electron opaque deposits, intensely labeled by the gold probe. Some labeling was also found in the Golgi complex, indicating that the protein traffics via this route before being sorted into the cell vacuole or secreted.
Although double localization in the vacuole and apoplast is not unusual for type 1 RIPs (Carzaniga et al. 1994; Yoshinari et al. 1996, 1997), it is interesting to note that for the highly identical PAP-II (81.6%), an exclusive extracellular localization has been reported (Ready et al. 1986).
2.5 Glycosylation of P. dioica RIPs
The structure of glycan moieties present in PD-Ss and PD-Ls was determined by a fast and sensitive mass spectrometry-based approach, applying a precursor ion discovery mode on a Q-TOF mass spectrometer (Di Maro et al. 2009; Chambery et al. 2008). The MS analysis confirmed that PD-Ls 1–3 were glycosylated at different sites. In particular, PD-L1 contained three glycidic chains, with the well-known paucimannosidic structure (Man)3 (GlcNAc)2 (Fuc)1 (Xyl)1, linked to Asn10, Asn43, and Asn255. PD-L2 was glycosylated at Asn10 and Asn43, and PD-L3 was glycosylated only at Asn10. PD-L4, dioicin 1, and dioicin 2 were not glycosylated.
The standard plant paucimannosidic N-glycosylation pattern was found for PD-S1 and PD-S2 on Asn120, while in PD-S1 and PD-S3 Asn112 was shown to link an HexNAc residue, probably N-acetyl-d-glucosamine (GlcNAc) (Chambery et al. 2008). The glycosylation patterns of PD-Ss, PD-L1/2, and PD-L3/4 RIPs help explaining their different chromatographic behavior (Table 1).
Basic understanding of protein glycosylation is still an area of intense investigation. Several roles have been ascribed to N-linked glycans, such as prevention of proteolytic degradation, induction of the correct folding and influence on protein conformation, stability and biological activity, involvement in protein recognition, and cell–cell adhesion processes (Ceriotti et al. 1998; Elbein 1991; Lis and Sharon 1993; Sharon and Lis 1993; Varki 1993). Regarding the protein folding and stability, a direct contribution of N-glycans has also been related to the increase of protein solubility, the reduced tendency to aggregate, and to the presence of additional hydrogen bonds and hydrophobic interactions between the oligosaccharide and the protein (O’Connor and Imperiali 1996; Wyss and Wagner 1996). In this context, the four PD-Ls forms constitute an excellent experimental model suitable to further investigate the role of glycosylation in the modulation of the biological activity on different substrates.
The primary structure of PD-L1 and PD-L2 are identical, as well as those of PD-L3 and PD-L4 (see Sect. 10.2.2). Therefore, biological differences between each protein couple could be ascribed to the presence of the glycan moieties. The comparative modeling of PD-L1, PD-L2, PD-L3, and PD-L4 showed an overall high structural similarity, but also potential influences of the glycan chains on their APG activity on different substrates (Di Maro et al. 2009), possibly related to the bending of the glycan chain linked to Asn255. The observed catalytic activity decrease is much more evident with the poly(A) (41%), but it is repeatedly observed also with the rRNA (24%) and hsDNA (4%), suggesting that it is associated with an acquired impairment of adenine interaction with the enzyme when Asn255 is glycosylated. Indeed, the relatively lower activity on DNA and rRNA could be explained in terms of obvious lower frequency-abundance of adenines in these substrates. The same trend is also observed when the activities of PD-L4 are compared with those of PD-L3.
Of particular interest was the DNA cleaving activity shown by PD-L1 (and PD-L2), both native (with sugars) and recombinant (without sugars and likely without contaminating DNases), dioicin 1 and dioicin 2 on ds pBR322 DNA (see Sect. 10.3.5 later), while PD-L3/4 does not possess this activity. First, it was ascribed to differences in glycosylation; it has been later attributed solely to the differences of the protein sequences (see Sects. 3.5 and 4.3 below for PD-L1; Ruggiero et al. 2009).
3 Enzymatic and Biological Characteristics
3.1 N-β-Glycosidase and APG Activities
RIPs from P. dioica are N-β-glycosidases as shown by the appearance of the “aniline fragment” in the RNA from ribosomes treated with the RIPs (Parente et al. 1993, 2008). When assayed for the inhibition of protein synthesis on a cell-free system, they gave IC50 values in the pmolar range (Table 3), comparable to those of other type 1 RIPs. PD-S2 inhibited protein synthesis by cells at a much higher concentration (120 pM in the reticulocyte lysate against >3,310, 2,950, 6 and 90 nM for 3T3 fibroblasts, HeLa, NB 100, and BEWO cells, respectively; Parente et al. 1993). The maximum release of adenine from purified rat liver ribosomes in the case of PD-S2 was ∼0.5 mol/mol of ribosomes (as in the case of PAP-S). PD-S2 also inhibited phenylalanine polymerization by purified rat liver ribosomes. The inhibition was not complete, with a residual ∼40% of polymerization even at the highest concentrations of RIPs tested (Parente et al. 1993). This resistance was observed previously with abrin (Battelli et al. 1984) and with an RIP from Petrocoptis glaucifolia (Arias et al. 1992). These results suggest that part of the ribosomes escape inactivation, and this was confirmed by treating ribosomes with PD-S2 RIP, and incubating them again, after washing, with or without the same RIP. In this second incubation, pretreated ribosomes polymerized phenylalanine to the same extent (∼30% of controls) independently of the addition of PD-S2 RIP. Addition of supernatant from a rabbit reticulocyte lysate (22 μg of protein/sample) did not modify the inhibition. A similar incomplete inhibition was observed with ricin added to ribosomes at 1:1 molar ratio.
RIPs from P. dioica show APG activity, determined by measuring the adenine amount released from herring sperm DNA at 260 nm. This APG activity appears to be more variable among RIPs and related to amino acid residue(s) present in the active site other than the ones already known to be part of it (i.e. Tyr76, Tyr129, Glu186, Arg189, Trp220, numbering of the consensus sequence of P. dioica RIPs, Fig. 2a). Indeed, a conserved seryl residue identified by multiple sequence alignment analysis and located in the proximity of the catalytic tryptophan, appears to play a role in this activity. Its involvement in the enzymatic mechanism of RIPs was investigated in PD-L4 by site-directed mutagenesis (Chambery et al. 2007). The replacement of Ser211 (numbering of the PD-L4 sequence or Ser224, numbering of the aforementioned consensus sequence) with Ala apparently does not influence the inhibition of the protein synthesis (determined as IC50 in a cell-free system), but it reduces the APG activity, assayed spectrophotometrically on other substrates such as DNA, rRNA, and poly(A). The ability of PD-L4 to deadenylate polynucleotides appears more sensitive to the Ser211Ala replacement when poly(A) is used as substrate, as only 33% activity is retained by the mutant, while with more complex and heterogeneous substrates such as DNA and rRNA, its APG activity is 73% and 66%, respectively. While the mutated protein shows a conserved secondary structure by CD, it also exhibits a remarkably enhanced tryptophan fluorescence. This indicates that although the overall protein 3D structure is maintained, removal of the hydroxyl group locally affects the environment of a Trp residue. Modeling, docking analyses, and 3D structure (Sect. 10.4.2) confirmed the interaction between Ser211 and Trp207, which is located within the active site, thus likely affecting the PD-L4 APG activity (Chambery et al. 2007).
3.2 Toxicity to Mice
PD-S2 RIP was toxic to mice with an LD50 of 1.12 mg/kg of body wt (Parente et al. 1993). The pathology of dead animals was similar to that observed in mice poisoned with other RIPs (Battelli et al. 1990), with necrotic lesions in the liver and kidneys.
3.3 Immunotoxin
The PD-S2 RIP could be derivatized with 2-iminothiolane and subsequently linked to monoclonal antibodies retaining good inhibitory activity on protein synthesis by the reticulocyte lysate system (IC50 18 nM and 26 nM, respectively, after derivatization and after conjugation to the antibody). An immunotoxin prepared with the anti-CD30 monoclonal antibody and containing 2.5 mol of RIP per mol of antibody inhibited protein synthesis by target L540 cells with an IC50 < 50 pM. A similar immunotoxin made with a control LS3 antibody was much less toxic to the same cells (IC50 45 nM) (Parente et al. 1993). The toxicity of this immunotoxin to target cells was comparable to that of an immunotoxin prepared with the same antibody and saporin (Tazzari et al. 1992).
Consistent with the results obtained with other RIPs, the effects of PD-S RIPs on different cells were highly variable, BeWo and NB 100 being more sensitive than HeLa cells and fibroblasts (Battelli et al. 1992).
PD-S2 RIP is immunologically distinct from most RIPs and appears to be suitable for the preparation of immunotoxins. Thus PD-S2 RIP could be useful to overcome the immune reaction which would follow the administration of immunotoxins prepared with other RIPs.
3.4 Cross-Reactivity
The PD-S2 RIP gives a significant cross-reaction only with antibodies against dianthin 32 and PAP-R, and a weak or no cross-reactivity with antibodies against other RIPs, including saporin 6, momordin, momorcochin-S, and trichokirin (Parente et al. 1993). This immune-response pattern was somewhat unexpected, because of the many identities in the amino acid sequences of PD-S2 RIP and PAP-S (73.4% identity; Fig. 3a), and since RIPs from plants belonging to the same family often give a strong cross-reaction with the respective antisera (Strocchi et al. 1992).
Cross-reactivity data have also been obtained for dioicin 1 and dioicin 2. These two RIPs were localized, by immunoblot analysis, in the extracellular fluid proteins of fully expanded leaves of young P. dioica plants. Antidioicin 1-specific IgGs cross-reacted with dioicin 2, as they showed up both RIPs, while antidioicin 2-specific IgGs did not react with dioicin 1. When used for immunocytochemical localization studies, antidioicin 1 IgGs gave only faint or no staining, while antidioicin 2 IgGs gave a positive signal mainly localized in the extracellular space of mesophyll cells (Parente et al. 2008).
3.5 Activity on Double-Stranded pBR322 DNA
PD-L1/2, dioicin 1, and dioicin 2 purified on Red Sepharose® not only showed N-β-glycosidase and APG activity but cleaved supercoiled pBR322 dsDNA, generating relaxed and linear molecules. PD-L3/4, purified in the same way, did not produce the same effect. The DNA cleaving activity of PD-S2 could not be determined because of a very tight interaction with the substrate DNA (supercoiled pBR322 dsDNA), with the resulting complex migrating towards the cathode (Delli Bovi, personal communication). An extensive study has been performed with PD-L1, the most glycosylated P. dioica RIP isoform. This RIP produced both free 3′-OH and 5′-P termini randomly distributed along the DNA molecule, as suggested by labeling experiments with [α-32P]dCTP and [γ-32P]dATP. Moreover, when the reaction was carried out under low-salt conditions, cleavage was observed mainly at a specific site, located downstream of the ampicillin resistance gene (close to position 3200), ending with the deletion of a fragment of approximately 70 nucleotides. This cleavage pattern is similar to that obtained under the same conditions with mung bean nuclease, a single-strand endonuclease. Furthermore, pBR322 DNA treated with PD-L1 showed reduced transforming activity with Escherichia coli HB101 competent cells in comparison to untreated control plasmid DNA.
Semiquantitative analysis of the effect of PD-L1/2, dioicin 1 and 2 showed that pBR322, pGem-3, PM2, and ΦX174 replication form DNA were cleaved under standard experimental conditions (50 mM Tris–Cl, 12.5 μM EDTA, pH 7.5, 37°C), producing linearized and relaxed forms. However, an extensive study, using several experimental conditions and methodologies, was performed with PD-L1 on pBR322 dsDNA. These include (1) analyzing the effect of temperature, salt, and divalent metal ions; (2) mapping the preferential RIP cleavage site; (3) performing “nick-translation-like” experiments; (4) assessing the endonucleolytic activity, under low-salt conditions, on pBR322 DNA by low amounts of S1, DNase I, or mung bean endonucleases to map the pBR322 linearization cleavage site; (5) performing substrate competition experiments with pBR322-oligonucleotides and transformation assays.
Overall, the results suggest that PD-L1 RIP from P. dioica leaves induce the cleavage of phosphodiester bond(s) on pBR322 DNA. This action is similar to that previously reported for either type 1 or type 2 RIPs extracted from different sources (Ling et al. 1994; Roncuzzi and Gasperi-Campani 1996). The experiments have shown that the nicking activity on supercoiled pBR322 DNA results in the production of predominantly circular and linear forms. Furthermore, the nicking activity and the linearization cleavage(s) were dependent on (1) temperature, (2) ionic strength, with inhibition on increasing the NaCl concentration, and (3) divalent cations, such as Mg2+, Mn2+, Zn2+, and even more so Co2+. Their presence under standard conditions potentiated the capacity to produce linear and circular pBR322 forms. However, the activity was completely abolished in the presence of 10 mM EDTA. This result suggests that PD-L1 could be endowed with or contaminated by cation-dependent endonuclease activity because, as reported for the most well-known endonucleases, the addition of a chelating agent blocks the activity, even though PD-L1 does not seem to be strictly dependent on the presence of the divalent cations tested so far. However, the recombinant PD-L1 (rPD-L1; Ruggiero et al. 2009) exhibits the same activity. We can hypothesize that the addition of EDTA abolishes the activity because it might chelate metal ions present in trace amounts that are important for maintenance of the structure. It cannot be excluded that the activity is abolished merely because the ionic strength of 10 mM EDTA is high enough to compact the DNA structure and dramatically reduce any endonucleolytic activity on it.
Under low-salt conditions, PD-L1 exerted its nicking activity predominantly at a major site. In fact, the experiments in which the linearized and circular pBR322 forms, obtained after RIP incubation in the absence of salts, were digested with various restriction enzymes, showed fragments of different sizes according to the preferential cleavage downstream of the ampicillin resistance gene of the plasmid. It has already been reported that this region is sensitive to the single-strand-specific mung bean endonuclease (MBN; Sheflin and Kowalski 1985). Moreover, the linearization cleavage by the PD-L1 nicking activity under low-salt conditions generates ligatable blunt termini and a deletion of an approximately 70-bp DNA fragment at a specific site, as suggested by the sequence analysis of the mutant clones. The same deletion was present in clones obtained from both blunted and unblunted linear pBR322 DNA produced by PD-L1 cleavage. Furthermore, once this region was eliminated, as in the case of mutant clones with the deletion, the nicking activity under low-salt conditions was mainly restricted to a second preferential site, which is another structurally unstable sequence, described as a preferential site for topoisomerase II (Amir-Aslani et al. 1995) and single-strand endonucleases such as S1 nuclease and MBN (Sheflin and Kowalski 1985). These mapped preferential sites are the only two regions rich in A–T sequences that could assume a hairpin conformation, as found after analysis of the complete pBR322 sequence by the PC-GENE software hairpin option. Furthermore, analysis of the entire pBR322 sequence by Web-Thermodyn (the sequence analysis software for profiling DNA helical stability; Huang and Kowalski 2003), revealed that these two regions require the lowest free energy to unwind and separate the strands of the double helix under our low-salt conditions at 37°C (http://wings.buffalo.edu/gsa/dna/dk/WEBTHERMODYN). As described earlier, these two regions are very rich in A–T sequences, and the stable unwinding of these regions may be important for single-strand-specific nuclease hypersensitivity (Kowalski et al. 1988). PD-L1 activity on supercoiled DNA produced free 3′-OH and 5′-P, as shown by labeling experiments, in common with other endonucleases. However, the endonucleolytic effect, as in the case of MBN (Sheflin and Kowalski 1985), is mainly dependent on the ionic environment. In fact, PD-L1 endonucleolytic activity on pBR322 DNA was more pronounced at low-salt concentrations and was almost absent under high-salt conditions. It has often been suggested that the nicking activity of RIPs can be attributed to contaminating endonucleases (Barbieri et al. 2000; Day et al. 1998). We addressed this question by incubating pBR322 DNA with endonucleases such as DNase I, S1 and MBN, using the same experimental conditions as for PD-L1. Our experiments performed with DNase I and S1 showed that in the presence of very low amounts of these enzymes, the cleaving action appeared to be limited and largely resulted in linear and circular plasmid forms, as obtained with PD-L1 treatment. However, it should be noted that in both cases this result was achieved with the addition of a minimum amount of divalent cations (Mg2+ for DNase I and Zn2+ for S1, not necessary for PD-L1) and that the linearized form was a consequence of random cleavage of pBR322 DNA. The pattern obtained with PD-L1, conversely, seemed to be very similar to that obtained with MBN. Thus, if a contaminant is present, it should be an endonuclease (the orthologue of MBN in P. dioica?) with functional and structural properties very similar to the well-known properties of MBN, even though its complete amino acid sequence has not yet been determined (Di Maro et al. 2008). If this is the case, the contaminating MBN-like protein should be present in very small amounts because, apart from PD-L1 RIP, no traces of other proteins were detected using the purification protocol reported in Table 1. It has also been suggested that the contamination could not be detected by the analytical procedures used. However, contamination from DNases could also be excluded on the basis of the following considerations: (1) our purification procedure for PD-Ls and dioicins (Table 1) includes steps that have been reported to be capable of removing the contaminating DNase activities (Barbieri et al. 2000); (2) the contaminating DNase activities should be present in almost all our RIP preparations eluted over a wide range of ionic strength (from 20 to 120 mM NaCl) necessary to elute proteins with high pI. Indeed, pI values for RIPs (>8.5) used in this study appear to be higher than those reported for commercial DNase I (pI 4.5) and S1 (theoretical pI 4.26); (3) contamination by DNase II (pI between pH 6.0 and 8.0) is not likely because this enzyme acts in the presence of high-salt concentration (Adams et al. 1986), while RIP endonucleolytic activity is inhibited under these conditions; (4) finally, the purification and storage conditions (low pH, water, and the absence of metal ions) would cause a loss in activity of such contaminating endonucleases. It is well known that MBN stability and activity are Zn2+-dependent at pH 5.0. In fact, its presence is essential during the purification procedure and over 90% of the activity can be lost or restored by zinc deprivation or its addition after dialysis at pH 5.0 in the absence of EDTA (Kowalski et al. 1976). Hence, if MBN endonucleolytic activity is greatly potentiated in the presence of Zn2+, it cannot be explained why PD-L1 is only slightly potentiated by this ion and can exert its activity without the addition of any divalent cations. Moreover, as mentioned above, our purification procedure involves steps (dialysis at pH 4.5; Di Maro et al. 1999) that could inactivate the MBN-like protein and exclude copurification of a protein with MBN endonuclease properties (i.e., stepwise elution at pH 7.2, which allows the elution of proteins with pI in the range 5.0–7.0, such as MBN; Kowalski et al. 1976). In conclusion, our results suggest that the activity of PD-L1 on DNA is an intrinsic property of this RIP form and is exerted mainly at low ionic strength, where secondary single-stranded structures may be formed and unpaired bases are present. Similar behavior is shown by other known single-strand-specific endonucleases (Desai and Shankar 2003). The competition experiments performed at various single- and double-stranded oligonucleotide/substrate excesses support this hypothesis and show that the nicking RIP activity persisted even at a 1,000-fold molar excess of the scavenger oligos over pBR322. This suggests that the nicking activity is dependent more on the single-stranded secondary structure with unpaired bases than on the occurrence of a specific sequence. Recent reports are in agreement with this hypothesis (Park et al. 2004a, b). Experiments showing a decrease in the transformation capacity of plasmid DNA after PD-L1 treatment suggest that damage to DNA occurred. This could be considered the mechanism responsible for the additional biological activity of RIPs according to results described in a different system (Nicolas et al. 1997). The fact that some RIPs may be endowed with enzyme activity against DNA, even though it may occur only at high concentrations, helps to explain some of their different biological properties. In fact, direct or indirect RIP activity against DNA has been reported in several papers hypothesizing different roles and biological significance: (1) RIP binding DNA (Hao et al. 2001); (2) nuclear DNA damage (Brigotti et al. 2002); (3) internucleosomal DNA fragmentation activity (Bagga et al. 2003); and (4) a role in transforming mammalian cells (Barbieri et al. 2003). These different capabilities may be necessary for these proteins to perform different biological roles: (1) resistance to pathogenic microorganisms or viruses; (2) implication in the mechanism of apoptosis and in metabolic regulation; and (3) activity as gene expression regulators. There remains a need to elucidate the mechanism of DNA cleavage exerted by PD-L1 and the structural determinants involved in this activity, questions not yet fully resolved for other RIPs showing the same activity, although it has been suggested that RIPs may act as DNA glycosylase/AP lyases (Wang et al. 1999a). This mechanism proposed for MAP30 is based on the presence of a lysyl residue (K195) close to a triptophanyl residue, the side-chain amino group of which would function as a nucleophile that attacks the C19 of the ribose of the abasic site (Wang et al. 1999b). However, PD-L4, which contains a lysyl residue corresponding to K195 of MAP 30 in the 3D structure, does not show nicking activity under the experimental conditions used here.
4 X-ray Crystal Structure of P. dioica RIPs
X-ray structures of RIPs from P. dioica have recently been determined (Table 4) (Ruggiero et al. 2007a, b, 2008, 2009). These structures describe three of the four RIPs isolated from fully expanded leaves of adult P. dioica leaves (PD-L1-4). Crystallographic studies of PD-L4, in its unliganded and adenine-bound states (Table 4), have provided atomic resolution structural information. As such, they constitute reference structures for this class of proteins (Ruggiero et al. 2008).
4.1 Atomic Resolution Studies of PD-L4: A Reference RIP Structure
Analogous to other RIPs (ricin A-chain, trichosanthin, PAP-I), P. dioica RIPs are composed of two domains and possess a well-defined secondary structure (Fig. 4a). Despite a structural similarity with pokeweed antiviral protein (PDB code 1qcg), with an average r.m.s.d. value after superposition of 261 equivalent Cα atoms of 0.65 Å, significant differences exist in the electrostatic potential surface of the two proteins (Fig. 4b, c). These differences are particularly evident in the putative RNA binding site cleft, where the electrostatic potential surface of PD-L4 is more negatively charged (Fig. 4b, c). This behavior is predictive of a different activity/specificity of the two proteins.
(a) Ribbon structure of PD-L4. N-terminal and C-terminal domains are drawn in green and magenta, respectively; adenine is drawn in blue. The figure has been drawn using MOLSCRIPT (Esnouf 1999). (b) Surface electrostatic potential distributions of PD-L4 and 8C9 of PAP. Positive (blue) and negative (red) electrostatic potential is mapped on the molecular surfaces by GRASP. PD-L4 residue numbering has been used
4.2 An Insight into the Active Site of PD-L4: Tyr72 as a Substrate Carrier Through π–π Stacking Interactions with Adenine
Although high resolution structures of various RIPs have been determined (Ago et al. 1994; Fermani et al. 2005; Hou et al. 2007; Kurinov and Uckun 2003; Kurinov et al. 1999; Savino et al. 2000; Touloupakis et al. 2006; Zeng et al. 2003), the mechanism by which they inhibit cell growth is still not fully understood. The currently accepted reaction mechanism involves the protonation of the adenine to be cleaved and the successive hydrolysis by a water molecule of the positively charged oxicarbonium intermediate. However, the residues which are involved in the protonation of the adenyl group are not known and contrasting hypotheses have been proposed (Guo et al. 2003; Huang et al. 1995; Ren et al. 1994).
The well-conserved active site residues of PD-L4 (Tyr72, Glu175, Arg178, Trp207) are located in the central part of the long concave presumed RNA-binding region (Fig. 4a). Analysis of the refined model of PD-L4 in the unliganded state shows that two well-defined conformations exist for the active site Tyr72 (Fig. 5a). In contrast, only one of the two Tyr72 conformations, the less abundant in the unliganded state (occupancy factor 0.15), is observed in the adenine complex. In this conformation, the phenoxy plane of Tyr72 is almost parallel with that of the adenine base (Fig. 5b). Many different protein systems, beside RIPs, employ Tyr residues in the recognition of the adenine ring, through a π–π stacking interaction (Boehr et al. 2002). Also, the orientation of Ade and Tyr72, with the Tyr OH group pointing at the N9 atom of adenine, has been found to be the most frequent in proteins; this has been attributed to the occurrence of favorable electrostatic interactions between the two rings (Boehr et al. 2002). The observed tight interaction of the Tyr72 ring with adenine (with a distance of 3.75 Å between their centroids) as well as the existence of two conformations in the unliganded PD-L4 are likely to be a requirement for the catalytic role of the adenine interaction, attributed to Tyr72 (Huang et al. 1995). It is worth noting that the A conformation of Tyr72 (Fig. 5a) is nearly superposable to that observed in the complex of inactive mutants of trichosanthin with AMP (Guo et al. 2003). Consistent with these findings, it has been proposed that Tyr72 in the A conformation (Fig. 5a) is devoted to the binding of the adenyl group in the early stages of the reaction mechanism. Modeling of AMP with Tyr72 in the A conformation shows that adenine is in the near proximity of Asp91, likely to be the protonating moiety (Huang et al. 1995). Consistently, the equivalent residue of Asp91 in trichosanthin (Glu85) has been shown to strongly affect catalysis (Guo et al. 2003). Most probably, after protonation, the adenyl group remains bound to the A conformation of Tyr72 (Fig. 5a) since it is unlikely that a positively charged adenine could be hydrogen bonded to the positively charged Arg178 (Fig. 5b). Finally, only when the N-glycosidic bond has been cleaved, the product adenine is accompanied to its final destination by Tyr72 (B conformation, Fig. 5a) and establishes the hydrogen-bonding interactions (to Arg178, Val73, Ser120) observed in the structures of the adenine complex of PD-L4 (Fig. 5b).
(a) (2Fo–Fc) electron density map, contoured at 2σ, of A and B conformations of Tyr72 in PD-L4. (b) (2Fo–Fc) electron density map, contoured at 2σ, of PD-L4 in complex with adenine. (c) (2Fo–Fc) electron density map, contoured at 2.5σ, of (A) PD-L4. (d) PD-L4 mutant S211A at the mutation site. (e) Superposition of residues close to the mutation site in PD-L4 (dark gray) and the PD-L4 mutant S211A (light gray). Figures have been drawn using BOBSCRIPT and MOLSCRIPT (Esnouf 1999)
Sequence alignment of all known RIPs clearly evidences that a high sequence conservation characterizes residues which are not traditionally considered as catalytically relevant, since they are located outside the active site cleft. As a part of a systematic study of the impact of these residues on function and structure of RIPs, PD-L4 was used as a model for mutational studies (Chambery et al. 2007). This study provided evidence that the mutation of the invariant Ser211 to Ala causes a significant decrease in APG activity, with the major extent of reduction for poly(A) substrates (Chambery et al. 2007). Structural bases of the reduced activity of the PD-L4 S211A mutant were obtained by determining the X-ray structure of this mutant both in its unliganded state and in complex with adenine, the major product of their enzymatic reaction (Table 4). In the crystal structure of the PD-L4 S211A mutant, the formation of a cavity formed by the lack of the Ser211 OH group is not compensated by a reorganization of the local enzyme structure, but the OH is replaced by a water molecule (Fig. 5c, d). This resulted in subtle conformational changes of residues which play a fundamental role in substrate binding, like Trp207 and Arg178 (Fig. 5e; Ruggiero et al. 2008). These data point to the importance of precise catalytic residue positioning for substrate binding. Furthermore, these studies show that subtle, albeit significant, structural changes are responsible for significant differences in the enzymatic activity. This highlights the importance of atomic resolution studies for the understanding of enzymatic properties (Schmidt and Lamzin 2002; Vrielink and Sampson 2003).
4.3 PD-L1 and PD-L4 – Two Homologous Proteins with Distinct Functional Properties
As previously reported, leaves of P. dioica express four type 1 RIPs, named PD-L1, PDL-2, PD-L3, and PD-L4. PD-L1–3 isoforms exhibit different degrees of glycosylation, whereas PD-L4 is not glycosylated (see Sect. 10.2.5). Despite the high sequence identity of these proteins, PD-L1 (and PD-L2) induce the cleavage of supercoiled double-stranded pBR322 DNA, whereas PD-L4 (and PD-L3) do not (see Sect. 10.3.5, Aceto et al. 2005). The structural basis of the different functional behavior of PD-L1 and PD-L4 was identified by determining the X-ray structure of native PD-L1 and by evaluating the role of glycosylation on DNA cleaving activity (Ruggiero et al. 2009). The crystallographic structure of PD-L1 evidenced that the protein catalytic cleft is not large enough to host double-strand DNA. This suggested that DNA cleavage occurs at unstable sites, where the double helix is locally unfolded. Consistently, regions of the E. coli plasmid pBR322 identified as PD-L1 cleavage sites, are rich in adenine and thymine (AT-rich) (Aceto et al. 2005), and therefore characterized by a lower thermal stability. It is likely that the stress present within supercoiled DNA destabilizes double helices in AT rich regions, thus making them accessible to the action of RIPs (Ak and Benham 2005; Benham and Bi 2004). The structure of PD-L1 provided evidence for the flexible nature of its glycan chains, a result which suggested that glycan chains provide little, if any, contribution to the formation and stabilization of the enzyme–substrate complex prior to catalysis. Consistently, DNA cleavage assays on the E. coli plasmid pBR322 clearly showed that native and recombinant (nonglycosylated) PD-L1 were able to cleave the plasmid pBR322, as linearized forms were clearly detectable for both proteins (Ruggiero et al. 2009). By contrast, no cleavage of pBR322 was observed upon treatment with PD-L4, purified by the same procedure as native PD-L1. Altogether, these data unambiguously showed that the different behavior of PD-L1, compared to its homologue PD-L4, is not due to the protein glycosylation, but to differences in their protein sequences. When the structures of PD-L1 and PD-L4 are compared, most significant structural variations are observed in loop regions. Among these, a conformational change of the loop including Asp91 (Fig. 6) was identified. Asp91 has been proposed to play an important role in catalysis, as the equivalent amino acid residue in trichosanthin (Glu85) has important implications for N-glycosidase activity (Guo et al. 2003). Compared to the PD-L4 structure, the entire loop embedding Asp91 is pulled back in the PD-L1 structure (Fig. 6). This conformational change, induced by the presence of arginine at position 97, a serine in PD-L4, opens the active site cleft by about 2.5 Å. (Fig. 6). Notably, this same loop conformation is observed in the structure of the PAP, which displays a similar ability to induce DNA cleavage. In addition, other RIPs exhibiting DNA cleaving activity, like saporin 6 (Savino et al. 2000) and dianthin 30 (Fermani et al. 2005), are characterized by a two-residue shorter loop. The observed catalytic cleft opening may allow the binding of regions of the to-be-cleaved supercoiled DNA, whose binding is hampered by the obstructing loop in PD-L4. Following DNA binding and deadenylation by PD-L1, a spontaneous breakage of phosphodiester bonds was proposed. Consistently, thermodynamic studies have shown that abasic sites impact the stability, conformation, and melting behavior of a DNA duplex (Vesnaver et al. 1989). Consequently, phosphodiester bonds in extensively deadenylated regions of supercoiled DNA likely become liable because of the existence of tension in supercoiled DNA.
Overall, structural studies on PD-L1 confirmed that DNA cleaving activity is not to be attributed to nuclease contaminations during RIP preparation, as previously proposed (Ruggiero et al. 2009). In this framework, DNA cleavage is proposed to be a consequence of PD-L1 catalytic action, although not directly catalyzed by the enzyme. This interpretation is in line with evidences that (1) various RIPs which exhibit DNA relaxing activity are also able to depurinate supercoiled double-stranded DNA (Wang et al. 1999) and that (2) mutants of PAP that inhibit N-glycosidase activity also inhibit the cleavage of supercoiled double-stranded DNA (Bagga et al. 2003). In this study, PD-L1 and PD-L4, which share a sequence identity of 81.5%, offered a good opportunity for understanding the structural basis of DNA cleavage, given the limited number of diverse residues in their sequences.
5 Concluding Remarks
Structure and function studies of type 1 RIPs from P. dioica have provided fundamental knowledge on these plant toxins. These include (1) phylogenetic relationships among Phytolacca RIPs; (2) confirmation that RIP expression is developmentally regulated; (3) double localization, both in the extracellular spaces and in the cell vacuoles of leaf tissues. This offers a view of the physiological role of RIPs. Furthermore, detailed structural information obtained by high resolution X-ray studies, contribute to our knowledge of the role of single amino acid residues and of the glycan moieties, shedding new light on the discussed DNA cleaving activity reported for RIPs. However, there is still much to be learned about this family of enzymes: such as gene number and organization; complete screening of plant tissues; detection of the factor(s) regulating RIP expression; biosynthesis and characterization of their biological activity in plant. An even better knowledge on the intracellular trafficking and protein target(s) upon intoxication of these type 1 RIPs will extend the utility of theses enzymes for better targeted biotechnological applications.
Notes
- 1.
The genus Phytolacca is suspected to contain a toxic saponin, which causes enteritis with vomiting, abdominal pain and diarrhea. The illness may be fatal (cfr. Saunders Comprehensive Veterinary Dictionary, 3rd edn. © 2007 Elsevier, Inc.). No information is reported of a likely involvement in the symptoms of ribosome-inactivating proteins. Poisoning of cattle and chickens from Phytolacca dioica L. (packalacca) or Phytolacca dodecandra l'Herit were reported (Storie et al. 1992; Mugera 1970).
- 2.
Synonym: P. decandra L.
- 3.
Synonyms: Phytolacca acinosa Maxim. and Phytolacca kaempferi (A. Gray).
- 4.
The ombú tree was introduced in Italy from South America. The plant (also called umbú tree) grows to a height and spread of 60 ft (20 m) or more, often with multiple trunks developing from an enormous base resembling a giant pedestal. The huge base may be 3–6 ft tall (1–2 m) and 95 ft (30 m) in circumference. Ombú tree is native of the grassy pampas of Argentina, usually widely spaced and the only trees for miles. It is dioecious, and the female tree produces large quantities of white, fleshy fruits. It is a salt-resistant species, often planted near the sea.
- 5.
Synonyms and common names. Synonyms (from www.hear.org/pier/species/phytolacca_dioica.htm): Phytolacca arborea Moq., Phytolacca populifolia Salisb., Pircunia dioica Moq., Sarcoca dioica Rafin. Common names (English language): belhambra, packalacca (also trade name) and phytolacca; (Spanish language) bella sombra tree, belombra, ombú and umbú (the last two also trade names).
- 6.
Synonym: Phytolacca abissynica Hoffm.
- 7.
Synonym: Phytolacca octandra L.
- 8.
The protein sequence data of P. dioica RIPs have been deposited in the UniProtKB with accession numbers P34967 for PD-S2, P84853 for PD-L1/2, P84854 for PD-L3/4 and P85208 for dioicin 2.
References
Aceto S, Di Maro A, Conforto B, Siniscalco GS, Parente A, Delli Bovi P, Gaudio L (2005) Nicking activity on pBR322 DNA of ribosome-inactivating proteins from Phytolacca dioica L. leaves. Biol Chem 386:307–317
Adams RLP, Knowler JT, Leader DP (1986) Degradation and modification of nucleic acids. In: The biochemistry of the nucleic acids, 10th edn. Chapman and Hall, London, p 87
Ago H, Kataoka J, Tsuge H, Habuka N, Inagaki E et al (1994) X-ray structure of a pokeweed antiviral protein, coded by a new genomic clone, at 0.23 nm resolution. A model structure provides a suitable electrostatic field for substrate binding. Eur J Biochem 225:369–374
Ak P, Benham CJ (2005) Susceptibility to superhelically driven DNA duplex destabilization: a highly conserved property of yeast replication origins. PLoS Comput Biol 1:e7
Amir-Aslani A, Mauffret O, Bittoun P, Sourgen F, Monnot M, Lescot E, Fennandjian S (1995) Hairpins in a DNA site for topoisomerase II studied by 1H- and 31P-NMR. Nucleic Acids Res 23:3850–3857
Arias FJ, Rojo MA, Ferreras MJ, Iglesias R, Muñoz R, Rocher A, Mendez E, Barbieri L, Girbés T (1992) Isolation and partial characterization of a new ribosome-inactivating protein from Petrocoptis glaucifolia (Lag.) Boiss. Planta 186:532–540
Bagga S, Seth D, Batra JK (2003) The cytotoxic activity of ribosome-inactivating protein saporin-6 is attributed to its rRNA N-glycosidase and internucleosomal DNA fragmentation activities. J Biol Chem 278:4813–4820
Barbieri L, Valbonesi P, Righi F, Zuccheri G, Monti G, Gorini P, Samorì B, Stirpe F (2000) Polynucleotide:adenosine glycosidase is the sole activity of ribosome-inactivating proteins on DNA. J Biochem (Tokyo) 128:883–889
Barbieri L, Brigotti M, Perocco P, Carnicelli D, Ciani M, Mercatali L, Stirpe F (2003) Ribosome-inactivating proteins depurinate poly(ADP-ribosyl)ated poly(ADP-ribose) polymerase and have transforming activity for 3T3 fibroblasts. FEBS Lett 538:178–182
Battelli MG, Lorenzoni E, Stirpe F, Cella R, Parisi B (1984) Differential effect of ribosome-inactivating proteins on plant ribosomes activity and plant cells growth. J Exp Bot 155:882–889
Battelli MG, Barbieri L, Stirpe F (1990) Toxicity of, and histological lesions caused by, ribosome-inactivating proteins, their IgG-conjugates, and their homopolymers. Acta Pathol Microbiol Immunol Scand 98:585–593
Battelli MG, Montacuti V, Stirpe F (1992) High sensitivity of cultured human trophoblasts to ribosome-inactivating proteins. Exp Cell Res 201:109–112
Benham CJ, Bi C (2004) The analysis of stress-induced duplex destabilization in long genomic DNA sequences. J Comput Biol 11:519–543
Boehr DD, Farley AR, Wright GD, Cox JR (2002) Analysis of the pi–pi stacking interactions between the aminoglycoside antibiotic kinase APH(3′)-IIIa and its nucleotide ligands. Chem Biol 9:1209–1217
Brigotti M, Alfieri R, Sestili P, Bonelli M, Petronini PG, Guidarelli A, Barbieri L, Stirpe F, Sperti S (2002) Damage to nuclear DNA induced by Shiga toxin 1 and ricin in human endothelial cells. FASEB J 16:365–372
Carzaniga R, Sinclair L, Fordharm-Skelton AP, Harris N, Croy RDR (1994) Cellular and subcellular distribution of saporins, type-1 ribosome-inactivating proteins, in soapwort (Saponaria officinalis L.). Planta 194:461–470
Ceriotti A, Duranti M, Bollini R (1998) Effects of N-glycosylation on the folding and structure of plant proteins. J Exp Bot 49:1091–1103
Chambery A, Pisante M, Di Maro A, Di Zazzo E, Ruvo M, Costantini S, Colonna G, Parente A (2007) Invariant Ser211 is involved in the catalysis of PD-L4, type I RIP from Phytolacca dioica leaves. Proteins 67:209–218
Chambery A, Di Maro A, Parente A (2008) Primary structure and glycan moiety characterization of PD-Ss, type 1 ribosome-inactivating proteins from Phytolacca dioica L. seeds, by precursor ion discovery on a Q-TOF mass spectrometer. Phytochemistry 69:1973–1982
Dallal JA, Irvin JD (1978) Enzymatic inactivation of eukaryotic ribosomes by the pokeweed antiviral protein. FEBS Lett 89:257–259
Day PJ, Lord JM, Roberts LM (1998) The deoxyribonuclease activity attributed to ribosome-inactivating proteins is due to contamination. Eur J Biochem 258:540–545
Del Vecchio Blanco F, Bolognesi A, Malorni A, Sande MJ, Savino G, Parente A (1997) Complete amino-acid sequence of PD-S2, a new ribosome-inactivating protein from seeds of Phytolacca dioica L. Biochim Biophys Acta 1338:137–144
Desai NA, Shankar V (2003) Single-strand-specific nucleases. FEMS Microbiol Rev 26:457–491
Di Maro A, Del Vecchio Blanco F, Savino G, Parente A (1995) Isolation and characterization of a nicked form of the single-chain ribosome inactivating protein from seeds of Phytolacca dioica L. In: First European symposium of the protein society, vol 4, Protein Science, Davos, Switzerland, p 128 (com 495)
Di Maro A, Valbonesi P, Bolognesi A, Stirpe F, De Luca P et al (1999) Isolation and characterization of four type-1 ribosome-inactivating proteins, with polynucleotide:adenosine glycosidase activity, from leaves of Phytolacca dioica L. Planta 208:125–131
Di Maro A, Chambery A, Daniele A, Casoria P, Parente A (2007) Isolation and characterization of heterotepalins, type 1 ribosome-inactivating proteins from Phytolacca heterotepala leaves. Phytochemistry 68:767–776
Di Maro A, Di Giovannantonio L, Delli Bovi P, De Andrés SF, Parente A (2008) N-terminal amino acid sequences of intact and cleaved forms of mung bean nuclease. Planta Med 74:588–590
Di Maro A, Chambery A, Carafa V, Costantini S, Colonna G et al (2009) Structural characterization and comparative modeling of PD-Ls 1–3, type 1 ribosome-inactivating proteins from summer leaves of Phytolacca dioica L. Biochimie 91:352–363
Duggar BM, Armstrong JK (1925) The effect of treating the virus of TMV with juices of various plants. Ann Missouri Bot Garden 12:359–366
Elbein AD (1991) The role of N-linked oligosaccharides in glycoprotein function. Trends Biotechnol 9:346–352
Esnouf RM (1999) Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr D Biol Crystallogr 55:938–940
Faoro F, Conforto B, Di Maro A, Parente A, Iriti M (2009) Activation of plant defence response contributes to the antiviral activity of diocin 2 from Phytolacca dioica. IOBC/wprs Bull 44:53–57
Fermani S, Falini G, Ripamonti A, Polito L, Stirpe F et al (2005) The 1.4 angstroms structure of dianthin 30 indicates a role of surface potential at the active site of type 1 ribosome inactivating proteins. J Struct Biol 149:204–212
Girbés T, Ferreras JM, Arias FJ, Stirpe F (2004) Description, distribution, activity and phylogenetic relationship of ribosome-inactivating proteins in plants, fungi and bacteria. Mini Rev Med Chem 4:461–476
Guo Q, Zhou W, Too HM, Li J, Liu Y et al (2003) Substrate binding and catalysis in trichosanthin occur in different sites as revealed by the complex structures of several E85 mutants. Protein Eng 16:391–396
Hao Q, Peumans WJ, Van Damme EJ (2001) Type-1 ribosome-inactivating protein from iris (Iris hollandica var. Professor Blaauw) binds specific genomic DNA fragments. Biochem J 357:875–880
Hou X, Chen M, Chen L, Meehan EJ, Xie J et al (2007) X-ray sequence and crystal structure of luffaculin 1, a novel type 1 ribosome-inactivating protein. BMC Struct Biol 7:29
Houston LL, Ramakrishnan S, Hermodson MA (1983) Seasonal variations in different forms of pokeweed antiviral protein, a potent inactivator of ribosomes. J Biol Chem 258:9601–9604
Huang Y, Kowalski D (2003) Web-Thermodyn: sequence analysis software for profiling DNA helical stability. Nucleic Acids Res 31:3819–3821
Huang Q, Liu S, Tang Y, Jin S, Wang Y (1995) Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins. Biochem J 309:285–298
Iglesias R, Pérez Y, Citores L, Ferreras JM, Méndez E, Girbés T (2008) Elicitor-dependent expression of the ribosome-inactivating protein beetin is developmentally regulated. J Exp Bot 59:1215–1223
Irvin JD (1975) Purification and partial characterization of the antiviral protein from Phytolacca americana which inhibits eukaryotic protein synthesis. Arch Biochem Biophys 169:522–528
Irvin JD, Kelly T, Robertus JD (1980) Purification and properties of a second antiviral protein from Phytolacca americana which inactivates eukaryotic ribosomes. Arch Biochem Biophys 200:418–425
Kassanis B, Kleczkowska I (1948) The isolation and some properties of a virus-inhibiting protein from Phytolacca esculenta. J Gen Microbiol 2:143–153
Kawade K, Masuda K (2009) Transcriptional control of two ribosome-inactivating protein genes expressed in spinach (Spinacia oleracea) embryos. Plant Physiol Biochem 47:327–334
Kowalski D, Kroeker WD, Laskowski MSR (1976) Mung bean nuclease I. Physical, chemical, and catalytical properties. Biochemistry 15:4457–4463
Kowalski D, Natale DA, Eddy MG (1988) Stable DNA unwinding, not ‘breathing’, accounts for single-strand-specific nuclease hypersensitivity of specific A + T-rich sequences. Proc Natl Acad Sci USA 85:9464–9468
Kurinov IV, Uckun FM (2003) High resolution X-ray structure of potent anti-HIV pokeweed antiviral protein-III. Biochem Pharmacol 65:1709–1717
Kurinov IV, Myers DE, Irvin JD, Uckun FM (1999) X-ray crystallographic analysis of the structural basis for the interactions of pokeweed antiviral protein with its active site inhibitor and ribosomal RNA substrate analogs. Protein Sci 8:1765–1772
Ling J, Liu WY, Wang TP (1994) Cleavage of supercoiled double-stranded DNA by several ribosome-inactivating proteins in vitro. FEBS Lett 345:143–146
Lis H, Sharon N (1993) Protein glycosylation. Structural and functional aspects. Eur J Biochem 218:1–27
Mugera GM (1970) Phytolacca dodecandra l'Herit toxicity in livestock in Kenya. Bull Epizoot Dis Afr 18:41–43
Nicolas E, Gooyer ID, Taraschi TF (1997) An additional mechanism of ribosome-inactivating protein cytotoxicity: degradation of extrachromosomal DNA. Biochem J 327:413–417
O’Connor SE, Imperiali B (1996) Modulation of protein structure and function by asparagine-linked glycosylation. Chem Biol 3:803–812
Obrig TG, Irvin JD, Hardesty B (1973) The effect of an antiviral peptide on the ribosomal reactions of the peptide elongation enzymes, EF-I and EF-II. Arch Biochem Biophys 155:278–289
Parente A, De Luca P, Bolognesi A, Barbieri L, Battelli MG, Abbondanza A, Sande JWM, Gigliano SG, Tazzari PL, Stirpe F (1993) Purification and partial characterization of single-chain ribosome-inactivating proteins from the seeds of Phytolacca dioica L. Biochim Biophys Acta 1216:43–49
Parente A, Conforto P, Di Maro A, Chambery A, De Luca P, Bolognesi A, Iriti M, Faoro F (2008) Type 1 ribosome-inactivating proteins from Phytolacca dioica L. leaves: differential seasonal and age expression, and cellular localization. Planta 228:963–975
Park S-W, Lawrence CB, Linden JC, Vivanco JM (2002) Isolation and characterization of a novel ribosome-inactivating protein from root cultures of pokeweed and its mechanism of secretion from roots. Plant Physiol 130:164–178
Park S-W, Vepachedu R, Owens RA, Vivanco JM (2004a) The N-glycosidase activity of the ribosome-inactivating protein ME1 targets single-stranded regions of nucleic acids independent of sequence or structural motifs. J Biol Chem 279:34165–34174
Park S-W, Vepachedu R, Sharma N, Vivanco JM (2004b) Ribosome inactivating proteins in plant biology. Planta 219:1093–1096
Rajamohan F, Venkatachalam TK, Irvin JD, Uckun FM (1999) Pokeweed antiviral protein isoforms PAP-I, PAP-II, and PAP-III depurinate RNA of human immunodeficiency virus (HIV)-1. Biochem Biophys Res Commun 260:453–458
Ready MP, Adams RP, Robertus JD (1984) Dodecandrin, a new ribosome-inhibiting protein from Phytolacca dodecandra. Biochim Biophys Acta 791:314–319
Ready MP, Brown DT, Robertus JD (1986) Extracellular localization of pokeweed antiviral protein. Proc Natl Acad Sci USA 83:5053–5056
Ren J, Wang Y, Dong Y, Stuart DI (1994) The N-glycosidase mechanism of ribosome-inactivating proteins implied by crystal structures of alpha-momorcharin. Structure 2:7–16
Roncuzzi L, Gasperi-Campani A (1996) DNA-nuclease activity of the single-chain ribosome-inactivating proteins dianthin 30, saporin 6 and gelonin. FEBS Lett 392:16–20
Ruggiero A, Chambery A, Di Maro A, Pisante M, Parente A et al (2007a) Crystallization and preliminary X-ray diffraction analysis of PD-L4, a ribosome inactivating protein from Phytolacca dioica L. leaves. Protein Pept Lett 14:97–100
Ruggiero A, Chambery A, Di Maro A, Mastroianni A, Parente A et al (2007b) Crystallization and preliminary X-ray diffraction analysis of PD-L1, a highly glycosylated ribosome inactivating protein with DNase activity. Protein Pept Lett 14:407–409
Ruggiero A, Chambery A, Di Maro A, Parente A, Berisio R (2008) Atomic resolution (1.1 A) structure of the ribosome-inactivating protein PD-L4 from Phytolacca dioica L. leaves. Proteins 71:8–15
Ruggiero A, Di Maro A, Severino V, Chambery A, Berisio R (2009) Crystal structure of PD-L1, a ribosome inactivating protein from Phytolacca dioica L. leaves with the property to induce DNA cleavage. Biopolymers 91:1135–1142
Savino C, Federici L, Ippoliti R, Lendaro E, Tsernoglou D (2000) The crystal structure of saporin SO6 from Saponaria officinalis and its interaction with the ribosome. FEBS Lett 470:239–243
Sawasaki T, Nishihara M, Endo Y (2008) RIP and RALyase cleave the sarcin/ricin domain, a critical domain for ribosome function, during senescence of wheat coleoptiles. Biochim Biophys Res Commun 370:561–565
Schmidt A, Lamzin VS (2002) Veni, vidi, vici – atomic resolution unravelling the mysteries of protein function. Curr Opin Struct Biol 12:698–703
Sharon N, Lis H (1993) Carbohydrates in cell recognition. Sci Am 268:82–89
Sheflin LG, Kowalski D (1985) Altered DNA conformation detected by mung bean nuclease occur in promoter and terminator regions of supercoiled pBR322 DNA. Nucleic Acids Res 13:6137–6154
Song SK, Choi Y, Moon YH, Kim SG, Choi YD, Lee JS (2000) Systemic induction of a Phytolacca insularis antiviral protein gene by mechanical wounding, jasmonic acid, and abscisic acid. Plant Mol Biol 43:439–450
Stirpe F, Battelli MG (2006) Ribosome-inactivating proteins: progress and problems. Cell Mol Life Sci 63:1850–1866
Storie GJ, McKenzie RA, Fraser IR (1992) Suspected packalacca (Phytolacca dioica) poisoning of cattle and chickens. Aust Vet J 69:21–22
Strocchi P, Barbieri L, Stirpe F (1992) Immunological properties of ribosome-inactivating proteins and a saporin immunotoxin. J Immunol Methods 155:57–63
Tazzari PL, Bolognesi A, de Totero D, Falini B, Lemoli RM, Soria MR, Pileri S, Gobbi M, Stein H, Flenghi L et al (1992) Ber-H2 (anti-CD30)-saporin immunotoxin: a new tool for the treatment of Hodgkin's disease and CD30+ lymphoma: in vitro evaluation. Br J Haematol 81:203–211
Thomsen S, Hansen HS, Nyman U (1991) Ribosome-inhibiting proteins from in vitro cultures of Phytolacca dodecandra. Planta Med 57:232–236
Touloupakis E, Gessmann R, Kavelaki K, Christofakis E, Petratos K et al (2006) Isolation, characterization, sequencing and crystal structure of charybdin, a type 1 ribosome-inactivating protein from Charybdis maritima agg. FEBS J 273:2684–2692
Varki A (1993) Biological roles of oligosaccharides: all of the theories are correct. Glycobiology 3:97–130
Vesnaver G, Chang CN, Eisenberg M, Grollman AP, Breslauer KJ (1989) Influence of abasic and anucleosidic sites on the stability, conformation, and melting behavior of a DNA duplex: correlations of thermodynamic and structural data. Proc Natl Acad Sci USA 86:3614–3618
Vrielink A, Sampson N (2003) Sub-angstrom resolution enzyme X-ray structures: is seeing believing? Curr Opin Struct Biol 13:709–715
Wang P, Tumer NE (1999a) Pokeweed antiviral protein cleaves double-stranded supercoiled DNA using the same active site required to depurinate rRNA. Nucleic Acids Res 27:1900–1905
Wang YX, Neamati N, Jacob J, Palmer I, Stahl SJ, Kaufman JD, Huang PL, Huang PL, Winslow HE, Pommier Y et al (1999b) Solution structure of anti-HIV-1 and antitumor protein MAP30: structural insights into multiple functions. Cell 99:433–442
Wheat D (1977) Successive cambia in the stem of Phytolacca dioica. Am J Bot 64:1209–1217
Wyss DF, Wagner G (1996) The structural role of sugars in glycoproteins. Curr Opin Biotechnol 7:409–416
Yoshinari S, Yokota S, Sawamoto H, Koresawa S, Tamura M, Endo Y (1996) Purification, characterization and subcellular localization of a type-1 ribosome-inactivating protein from the sarcocarp of Cucurbita pepo. Eur J Biochem 242:585–591
Yoshinari S, Koresawa S, Yokota S, Sawamoto H, Tamura M, Endo Y (1997) Gypsophilin, a new type 1 ribosome-inactivating protein from Gypsophila elegans: purification, enzymatic characterization, and subcellular localization. Biosci Biotechnol Biochem 61:324–331
Zacchia E, Tamburino R, Di Maro A, Parente A (2009) Isolamento e caratterizzazione di forme tagliate di una proteina inattivante i ribosomi da semi di Phytolacca dioica L. Giornate Scientifiche della SUN, VIS-1. http://www.gsa.unina2.it/index.php?option=com_wrapper&Itemid=42
Zeng ZH, He XL, Li HM, Hu Z, Wang DC (2003) Crystal structure of pokeweed antiviral protein with well-defined sugars from seeds at 1.8 Å resolution. J Struct Biol 141:171–178
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2010 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Parente, A., Berisio, R., Chambery, A., Di Maro, A. (2010). Type 1 Ribosome-Inactivating Proteins from the Ombú Tree (Phytolacca dioica L.). In: Lord, J., Hartley, M. (eds) Toxic Plant Proteins. Plant Cell Monographs, vol 18. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-12176-0_5
Download citation
DOI: https://doi.org/10.1007/978-3-642-12176-0_5
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-12175-3
Online ISBN: 978-3-642-12176-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)