Abstract
Ribosome-inactivating proteins (RIPs) are a group of proteins exhibiting N-glycosidase activity leading to an inactivation of protein synthesis. Thirteen predicted Jatropha curcas RIP sequences could be grouped into RIP types 1 or 2. The expression of the RIP genes was detected in seed kernels, seed coats, and leaves. The full-length cDNA of two RIP genes (26SK and 34.7(A)SK) were cloned and studied. The 34.7(A)SK protein was successfully expressed in the host cells while it was difficult to produce even only a small amount of the 26SK protein. Therefore, the crude proteins were used from E. coli expressing 26SK and 34.7(A)SK constructs and they showed RIP activity. Only the cell lysate from 26SK could inhibit the growth of E. coli. In addition, the crude protein extracted from 26SK expressing cells displayed the effect on the growth of MDA-MB-231, a human breast cancer cell line. Based on in silico analysis, all 13 J. curcas RIPs contained RNA and ribosomal P2 stalk protein binding sites; however, the C-terminal region of the P2 stalk binding site was lacking in the 26SK structure. In addition, an amphipathic distribution between positive and negative potential was observed only in the 26SK protein, similar to that found in the anti-microbial peptide. These findings suggested that this 26SK protein structure might have contributed to its toxicity, suggesting potential uses against pathogenic bacteria in the future.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Jatropha curcas (commonly known as the physic nut) belongs to the Euphorbiaceae family and is a drought-resistant plant distributed in tropical regions [1]. It has been exploited as an alternative source for biodiesel production because it accumulates relatively a high content of oil in the seed kernel [2]. The seed cake, a by-product after oil-extraction, is used as animal feedstock because of its high protein content. However, seeds of some J. curcas varieties contain toxic substances, for example, nitric oxide, phorbol ester, cyclic peptides, and ribosome-inactivating proteins (RIPs).
RIPs are proteins with rRNA N-glycosidase (EC 3.2.2.22). They catalyze the cleavage of the N-glycosidic bond at a specific adenine residue in the conserved sarcin/ricin loop of prokaryotic and eukaryotic rRNA [3]. Adenine depurination precludes the binding of elongation factors to ribosomes leading to a translation inhibition and finally to cell death [4,5,6,7]. RIPs are categorized into three types. Type 1 RIPs consist of a single polypeptide chain with a molecular weight of approximately 25–30 kDa. Type 2 RIPs contain two polypeptide chains connected by disulfide linkage. Chain A possesses enzymatic activity while chain B has lectin activity. The molecular weights of type 2 RIPs are in the range 60–65 kDa. Type 3 RIPs are produced as an inactive form with molecular mass approximately 56–63 kDa. Activation of type 3 RIPs occurs through proteolytic cleavage [8]. Cell entry has been proposed through binding of the chain B of RIPs type 2 to cell surface receptors, allowing the endocytosis of chain A into the cell [9]. However, the entry routes of RIPs type 1 and type 3 are still unknown.
J. curcas RIPs have been studied for more than a decade. Curcin is the well-studied type 1 RIP which initially was purified from the seed kernel. The molecular weight of curcin is approximately 28 kDa [4]. Biological activities of curcin have been reported including antimicrobial, anti-tumor, and pesticidal activities [10, 11]. In addition to curcin, other RIPs are also presented in J. curcas such as curcin-L [12], curcin 2 [13, 14], and curcin C [15]. Curcin-L is a type 1 RIP which shows a high sequence similarity to curcin but this protein was expressed only in the leaves of stressed plants [12]. Curcin 2 is another homolog of curcin. This protein has a molecular weight of approximately 32 kDa and the expression of curcin 2 was activated by fungal infections, cold stresses, and heat stresses [13]. The transgenic tobacco expressing curcin 2 displayed an increased tolerance against viral and fungal pathogens [14]. Additionally, a novel type 1 RIP, namely Jc-SCRIP, has been purified from the seed coat of J. curcas [16]. Jc-SCRIP has N-glycosidase activity, in vitro cytotoxicity to cancer cell lines and antimicrobial activity against human pathogens [16].
In addition to their biological activities, structural analysis of RIPs has been reported to provide the link between the functions and protein structure. The crystal structure of the ricin A chain (RTA) displayed key amino acid residues (Y80, Y123, E177, R180, and T211). These amino acids were hypothesized to form an RNA binding pocket and play a key role in ribosomal RNA binding [17]. Indeed, the crystal structure of the recombinant RTA with pteroic acid (the RTA substrate analog) showed that pteroic acid was bound with some amino acids including Y80 and R180. In addition, the binding of RTA resulted in a deficiency of RTA activity [18]. The RIPs not only bind to the ribosomal RNA but also to the ribosome. The ricin A chain forms a complex with the human ribosomal P2 stalk through hydrogen bonds with the amino acids (Y183, R235, F240, and I251) in the hydrophobic pocket [19].
The objectives of this work were to investigate the biological activities of J. curcas RIPs recombinant protein and to understand whether the protein structure was related to the protein toxicity, by considering in the developed structural model RNA binding site residues, ribosomal binding site residues, the electrostatic potential map, and hydrophobicity of J. curcas RIPs compared with the ricin A chain. This study might help to understand the correlation between the toxicity of J. curcas RIPs and their protein structures.
Methodology
Sequence retrieval, Phylogenetic Tree Analysis, and 3D Structure Prediction
The amino acid sequences of RIPs from J. curcas, other plant RIPs, and non-RIPs proteins were retrieved from the NCBI and Kazusa DNA research institute (version 4.515) (http://www.kazusa.or.jp/ jatropha) databases. These amino acid sequences were aligned, and a phylogenetic tree was generated using the MEGAX software (https://www.megasoftware.net). The RIP amino acid sequences were aligned by multiple sequence comparison by Log-expectation (MUSCLE) method [20]. Phylogenetic tree was calculated using neighbor joining statistic mode with the number of Bootstrap replications as 1000 (analyzed on 11/10/2018). The 3D structure, RNA binding site, ribosomal P2 binding site, molecular surface, and electrostatic potential were generated using the Swiss model (https://swissmodel.expasy.org/interactive) [21,22,23,24,25]. The searching models were the structure of the ricin A chain with pteroic acid as substrate analog (Protein Data Bank: 1BR6) [18] and the ricin A chain without substrate analog (Protein Data Bank: 2AAI) [26]. These predicted conformations were further analyzed using the software package BIOVIA Discovery Studio Visualizer (version 2017, R2). The reliability of predicted RIPs conformations was validated using the online Ramachandran plot analysis servers including RAMPAGE (http://mordred.bioc.cam. ac.uk/~rapper/rampage.php) [27] and PROCHECK (http://servicesn.mbi.ucla.edu/PROCHECK/) [28].
Expression Analysis of J. curcas RIP Genes
The expression profiles of RIP genes were analyzed in two J. curcas varieties KUBP33 and KUBP79. Both varieties were provided by the Suwanwajokkasikit Field Crops Research Station, Nakhon Ratchasima, Thailand. Reverse transcriptase-polymerase chain reaction (RT-PCR) was performed according to the manufacturer’s instructions. In brief, J. curcas trees have been cultivated under field conditions without temperature and humidity controls for 3–4 years. Young leaves, immature seed coats, and immature seed kernels were collected and total RNAs were extracted using a GF-1 total RNA extraction kit (Vivantis, USA). Subsequently, 2.5 µg of total RNAs were converted to cDNA using the ImProm-IITM reverse transcriptase system (Promega, USA). Then, 125 ng of cDNAs was amplified with gene-specific primers (Supplementary Table 1) using the following PCR conditions: pre-denaturing at 95 °C for 5 min, 25 cycles with denaturing at 95 °C for 30 s, annealing at 50–65 °C for 30 s, extension at 72 °C for 30 s, and final extension at 72 °C for 10 min. The internal control was 18S rRNA.
26SK and 34.7(A)SK Plasmid Construction
The coding sequences of 26SK and 34.7(A)SK were cloned using gene-specific primers consisting of: 26SK (forward) 5′-CGC CAT ATG AAA CGA GGA AAC ACG AAG-3′; 26SK (reverse) 5′-GTG CTC GAG TTA AGC-3′; 34.7(A)SK (forward) 5′-GCG CAT ATG ATG AAA GGT GGA AAC ATG AAG-3′; and 34.7(A)SK (reverse) 5′-GTG CTC GAG CTA AAG CAA TGG CAG CCA CTT-3′ using PCR conditions as followed: pre-denaturing at 95 °C for 5 min, 30 cycles with denaturing at 95 °C for 30 s, annealing at 60 °C for 30 s, extension at 72 °C for 30 s, and final extension at 72 °C for 10 min. The full-length cDNAs of 26SK and 34.7(A)SK were ligated into pET28a(+) (Novagen Merck KGaA, Germany) and pETDuet-1TM (Novagen Merck KGaA, Germany), respectively, by NdeI and XhoI restriction enzymes (Supplementary Table 1). Additionally, the plasmid harboring the full-length cDNAs of 26SK and 34.7(A)SK was analyzed using the fluorescence dye terminator sequencing system (Macrogen Inc., Korea).
Expression of 26SK and 34.7(A)SK Recombinant Proteins
The 26SK and 34.7(A)SK recombinant proteins were induced in the E. coli host cell strain Rosetta (DE3) pRARE. The starter cells of 26SK were inoculated in Luria-Bertani (LB) media supplemented with 25 µg/ml kanamycin and 34 µg/ml chloramphenicol, whereas 34.7(A)SK was inoculated in LB media supplemented with 100 µg/ml ampicillin and 34 µg/ml chloramphenicol, respectively. Subsequently, the starter cells were cultured at 37 °C for 16–18 h with agitation at 220 rpm. The starter cells were inoculated into new LB media supplemented with appropriate antibiotics and cultured further for 2–3 h at 37 °C in a shaking incubator until the optical density at 600 nm (OD600) reached 0.4–0.6. The 0.1 mM IPTG was added to induce recombinant protein production. The cells were incubated at 30 °C for 1 h in a shaking incubator at 220 rpm. After induction, the cell pellets were harvested using centrifugation at 3,000 rpm for 10 min and suspended in 1×phosphate buffer saline (PBS) containing 1 mM PMSF before proceeding to protein extraction. The crude protein concentrations of 26SK and 34.7(A)SK were measured using Lowry’s assay.
rRNA N-Glycosidase Activity
The rRNA N-glycosidase activity of J. curcas RIPs crude protein extract was determined according to the modified method [29]. In brief, rRNA substrate was extracted from rabbit reticulocyte lysate (Promega Corp., USA) and was further treated with 10 µg of crude protein samples in 100 μl of reaction buffer (25 mM Tris–HCl, pH 7.6, 25 mM KCl, and 5 mM MgCl2). The samples were incubated at 37 °C for 10 min. The RNA pellet was re-suspended in 20 μl of DEPC-treated water and approximately equal volumes of the reaction were placed into two microcentrifuge tubes. One tube was treated with 1 M aniline/0.8 M acetic acid pH 4.5 and the other was untreated. The aniline-treated and aniline-untreated products were analyzed using 7M urea 6% polyacrylamide gel electrophoresis.
Antimicrobial Activity of 26SK and 34.7(A)SK Expressing Cells
The crude protein samples from E. coli Rosetta (DE3) pRARE cells carrying pET28a(+)-26SK, pETDuet-34.7(A)SK, and empty vectors were tested for antimicrobial activity against E. coli host cells using the disc diffusion method [30]. The E. coli cells were cultured in LB media until the OD600 was approximately 0.5 (1×108 cfu/ml) and then the cells were thoroughly spread on separate LB agar plates. A sterile paper disc was immersed in 300 µg of each type of crude protein and placed on LB plates. The plates were incubated at 37 °C for 16 h. Ten micrograms of ampicillin was used as the positive control and 1×PBS with 1 mM PMSF was used as the negative control. The antimicrobial activities of each protein sample were examined and compared with the positive and negative controls by measuring the diameters of the inhibition zones to the nearest millimeter ± standard error.
Results
Phylogenetic Tree Analysis of Predicted RIP Proteins from Jatropha curcas
Thirteen predicted J. curcas RIP amino acid sequences were identified based on information available in the NCBI and Kazusa databases (Supplementary Table 1). The proteins were named in the current study based on their molecular weights, namely, 13All, 26SK, 28SK, 32All, 33curcin, 33SK, 34.7(A)SK, 34.7(B)SKL, 35curcin, 35NF, 35SKL, 50SK, and 62NF. The curcin (accession number: ACO53803.1) and curcin-L (accession number: EU195892) were renamed to 33curcin and 35curcin. Phylogenetic tree analysis was carried out to compare the similarity between these J. curcas RIPs. For comparison, amino acid sequences of RIP from other plants and non-RIP proteins were also included in the analysis. Type 1 RIP from other plants comprised cucurmosin (Curcubita moschata), karasurin-C (Tricosanthes kirilowii), alpha-trichosathin (Tricosanthes kirilowii), alpha-momorcharin (Momordica charantia), and luffin-B (Luffa aegyptiaca). The type 2 RIPs were ricin A chain (Ricinus communis) and Viscum album RIP, while Zea mays RIP and Dianthus chinensis RIPs were the type 3 RIPs. Ubiquitin and actin were used as outgroup proteins in this analysis. The phylogenetic tree analysis showed that 12 predicted J. curcas RIPs consisting of 13All, 26SK, 28SK, 32All, 33curcin, 33SK, 34.7(A)SK, 34.7(B)SKL, 35curcin, 35NF, 35SKL, and 50SK were categorized as type 1 RIP (Fig. 1a), whereas only 62NF belonged to type 2 RIP (Fig. 1a). No J. curcas type 3 RIPs were found in these current public sequences. In addition, ubiquitin and actin proteins were separated to another clad which was not related to J. curcas RIP.
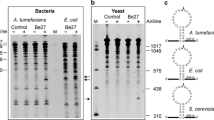
Phylogenetic tree and expression analysis of J. curcas RIPs. a Phylogenetic tree analysis of J. curcas RIPs, RIP homologs, and non-RIPs from different plants using Clustal Omega software. b Expression profiles of RIP genes in leaves (L), seed coats (SC) and seed kernels (SK) from J. curcas varieties KUBP33 and KUBP79. NC, negative control
RIP Genes Were Expressed in Various Tissues in Jatropha curcas
The expression of the predicted RIP genes was analyzed in various tissues (leaves, seed coat, and seed kernel) of the J. curcas varieties KUBP33 and KUBP79 using RT-PCR amplification. The expression pattern of the 13 RIP genes was similar in both cultivars. The expression patterns of the J. curcas RIP genes were categorized into four groups. In the first group, the RIP genes were expressed in all tissue types. These were 13All and 32All with the amplicons of approximately 314 bp and 861 bp, respectively (Fig. 1b). In the second group, the RIP genes were expressed in both leaves and seed kernel but not in the seed coat. These genes were 34.7(B)SKL and 35SKL with product bands at approximately 920 bp and 945 bp, respectively (Fig. 1b). In the third group, the RIP genes were expressed only in the seed kernel. The genes were 26SK, 28SK, 33curcin, 33SK, 34.7(A)SK, 35curcin, and 50SK with product bands at approximately 701 bp, 765 bp, 882 bp, 872 bp, 924 bp, 921 bp, and 1,368 bp, respectively (Fig. 1b). In the fourth group, there were two RIP genes (35NF and 62NF) and neither was expressed in any tissue type (Fig. 1b).
Some RIPs Lacked the C-Terminal Domain
It has been reported that there are 14 highly conserved amino acids in 165 angiosperm RIP sequences: Y46, F49, R54, Y130, Y185, L225, G226, L230, E280, A281, R283, F284, W324, and S328 (Supplementary Table 2) [31]. Thus, the presence of these amino acid was expected in the J. curcas RIPs. From the previous sequence analysis by Di Maro and colleagues, the methionine at position 1 of ricin A chain (PBD: 2AAI) was not included in their analysis [31]. Therefore, the first methionine of the ricin A chain (PDB: 1BR6) was also not included in the current analysis. Fig. 2 shows that the RIPs from J. curcas also contained these 14 conserved amino acids, indicating that these proteins might share some important characteristics. As the RIP enzymes function in the cleavage of ribosomal RNA, the RNA binding sites are crucial for their key function. Amino acid residues in the RNA binding site are also highly conserved. The Jatropha RIPs also contain these conserved amino acid residues. The conserved RNA binding sites of type 1 RIPs were Y, F/L, R, Y, Y/N, L/I, G, L, E, A, R, F/Y, W, and S (Fig. 2 and Supplementary Table 2). For type 2 RIPs, the amino acid residues in the RNA binding site consisted of Y, F, R, G, Y, L, G, L, E, A, R, F, W, and S (Fig. 2 and Supplementary Table 2). The conserved amino acid residues in the RNA binding sites among type 1 and type 2 RIPs were Y, R, Y, G, L, E, A, R, W, and S (Supplementary Table 2). Glutamic acid (E) and arginine (R) residues in the RNA binding site were conserved in many Jatropha curcas RIPs. These residues corresponded to positions E209 and R212 in 26SK, 34.7(A)SK, and 33curcin (Fig. 2 and Supplementary Table 2). However, these corresponding residues were not found in the 13All and 32All. Phenylalanine (F) was found in almost all RIPs from Jatropha curcas except for the 28SK and 33SK proteins (Supplementary Table 2). The sarcin-ricin loop of rRNA is well-known as a universal substrate for all RIPs. Proximity to an adenine was predicted for nine amino acids of 33curcin, namely Y118, V120, F131, N132, D133, L137, T156, G157, and S158 (Fig. 2). These amino acids were also present in other Jatropha RIPs (26SK, 34.7(A)SK, 34.7(B)SKL, and 35curcin) (Fig. 2 and Supplementary Table 3).
Amino acid sequence alignment of RIPs from J. curcas compared with ebulin (PDB: 1HWP) and ricin (PDB: 1BR6). Yellow color and blue color represent the key amino acid residues located in the RNA binding site and ribosome P2 binding site, respectively. Key amino acid residues related to specific adenine binding sites and acid-catalyzed depurination displayed by red and blue letters, respectively
Ribosomal proteins are involved in the susceptibility of rRNA to depurination by RIPs [32]. The RIP proteins bind to both rRNA and ribosome in order to function as N-glycosidase enzymes. The ricin A chain (PDB: 1BR6) has been proposed as recognizing and binding to the C-terminal region of P2 stalk protein through the key amino acid residues (Y183, L232, R235, and F240) [33, 34]. They were predicted to form three α-helices and two β-sheets (Fig. 2 and Supplementary Table 4). However, the 13All, 26SK, and 28SK proteins lack some parts of the α-helix and β-sheet where some of the key amino acids are located. This observation gave rise to the question of how these proteins could function as N-glycosidase enzyme (Fig. 2 and Supplementary Table 4). In addition, the P2 stalk protein recognition site residues were in the α-helix and β-sheet regions in almost all members of J. curcas RIPs. Differences in the key amino acids in the P2 stalk protein binding site were observed between J. curcas type 1 and type 2 RIPs. For the type 1 RIPs, the amino acids Y/S, L, E/P, and D/I/Y/K were predicted as the key amino acids functioned to bind the P2 binding protein while the amino acid residues Y, L, L, and N were predicted in the type 2 RIP (Fig. 2 and Supplement Table 4).
26SK and 34.7(A)SK Crude Proteins Possessed rRNA N-Glycosidase Activity
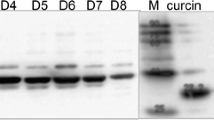
“Based on the phylogenetic and amino acid sequence analysis, some of the RIP proteins (13All, 26SK and 28SK) lacked their C-terminal domains which might be considered the truncated RIPs (Fig. 2). This raised the question of whether these proteins could function as RIPs. As shown in the phylogenetic analysis, 26SK and 34.7(A)SK proteins were the most similar among the predicted RIP family in Jatropha (Fig. 1a), thus, 26SK and 34.7(A)SK were chosen for further study. In addition, the 34.7(A)SK also showed high sequence similarity to the well-studied curcin (33curcin) (Fig. 2) making this protein interesting to dissect its functions.” The full-length cDNAs of 26SK and 34.7(A)SK were successfully cloned into the expression vectors. Several attempts have been made to express the 26SK recombinant protein in E. coli. We found that the growth of the host cells strikingly decreased after the induction. In contrast, induction of the 34.7(A)SK recombinant protein did not affect the growth of the host cells (Supplementary Fig. 1). The presence of 26SK and 34.7(A)SK recombinant proteins was determined in the crude extracted of the host cells after induction. The crude protein extract of his-tag containing protein, CPN60B1 (C. reinhardtii chloroplastic chaperonin proteins), served as positive control, while the crude extract from the cells containing pET28a(+) empty vector (EV) served as negative control. Both 26SK and 34.7(A)SK proteins were detected after induction by Western blot using anti-histidine antibody (Supplementary Fig. 2a and b). However, only small amount of 26SK protein was produced. Therefore, the crude extracts from E. coli cells harboring either 26SK or 34.7(A)SK recombinant plasmids were used for further studies.
In general, the enzymes classified as RIPs have rRNA N-glycosidase activity. Large ribosomal RNA treated with RIPs would release specific fragments, called Endo’s fragments, after the addition of an aniline in the test reaction indicating the presence of rRNA N-glycosidase activity [35]. Therefore, the rRNA N-glycosidase activity of the 26SK and 34.7(A)SK crude protein extracts was investigated (Fig. 3a and b). The enzyme activity was compared to the crude proteins from the empty vector, CPN60B1, and J. curcas seed kernel (Fig. 3a and b). The crude protein extract from the seed kernel of J. curcas contained natural curcin. It was used as a positive control while the crude proteins extracted from cells containing CPN60B1 and the empty vector served as negative controls. Specific Endo’s fragments were clearly seen after the rabbit reticulocyte lysate was treated with 10 µg of the crude protein extracts from 26SK, 34.7(A)SK, and seed kernel with the addition of aniline due to the rRNA N-glycosidase activity. Moreover, no rRNA fragment was released and detected without RIP treatment (Fig. 3a and b). Thus, the crude protein extracts from E. coli cells carrying 26SK and 34.7(A)SK plasmid possessed rRNA N-glycosidase activity.
N-glycosidase activity of recombinant RIPs. a N-glycosidase activity against rabbit reticulocyte lysate of crude extract from 26SK, empty vector (EV), CPN60B1 (CPN), and J. curcas seed kernel (SKC); b N-glycosidase activity against rabbit reticulocyte lysate of crude extract from 34.7(A)SK, empty vector (EV), CPN60B1 (CPN), and J. curcas seed kernel (SKC) when UT represented untreated rabbit reticulocyte lysate
Growth Inhibition of E. coli by 26SK
As the E. coli carrying the 26SK construct displayed growth inhibition after the induction of recombinant protein production, therefore, it was of interest whether this protein could indeed inhibit the growth of the E. coli host cells. Therefore the growth inhibition of 26SK and 34.7(A)SK crude protein lysate was tested against E. coli Rosetta (DE3) pRARE. The growth inhibition activity was determined by the presence of the growth inhibition zones. As shown in Fig. 4, the inhibition zones were clearly seen after treating E. coli cells with 300 µg of 26SK cell lysate with the diameters of the inhibition zones of 12.5±0.21 mm, while 10 µg ampicillin produced 16.2±0.15 mm inhibition zone. In contrast, 34.7(A)SK cell lysate did not inhibit the growth of bacteria, similar to the negative controls (empty vector cell lysate or 1xPBS containing 1 mM PMSF). This indicated that only the 26SK crude protein lysate could inhibit the growth of E. coli cells.
Inhibitory effect of the 26SK crude protein lysate to E. coli cells. Inhibition zones of E. coli Rosetta (DE3) pRARE cells treated with 300 µg 26SK cell lysate (a) or 300 µg 34.7(A)SK cell lysate (b). Treatments with 10 µg ampicillin (Amp), 300 µg empty vector (EV) cell lysate, and 1xPBS with 1 mM PMSF (PBS) were used positive and negative controls, respectively
26SK Crude Protein Exhibited In Vitro Antitumor Activity Against Triple Negative Human Breast Adenocarcinoma Cells
Many of the RIP proteins contain anti-tumor activity [10, 16, 29, 36, 37]; therefore, we assessed whether the 26SK protein could influence the growth of the cancer cell line. Cytotoxicity of 26SK protein on the MDA-MB-231 cells (triple-negative human breast adenocarcinoma) was assessed compared with the Vero cells (African green monkey’s kidney epithelial cells). The crude protein from E. coli with an empty vector (EV) was taken as a negative control, whereas doxorubicin (a chemotherapy drug) was used for a positive control. The viability of MDA-MB-231 was examined using an MTT assay. We found that the 26SK crude protein significantly inhibited the growth of MDA-MB-231 cells but did not affect the Vero cells (Fig. 5a and c). Compared with the negative control, EV crude protein showed no cytotoxic effect to both MDA-MB-231 and Vero cells (Fig. 5a and c). In addition, the IC50 values of 26SK crude protein on MDA-MB-231 cells after treatment for 72 h was 2,208±11.24 µg/ml, while the concentration of doxorubicin (a positive control) between 0.625 and 20 µg/ml (approximately 1.1–36.8 µM) displayed high cytotoxic effect against MDA-MB-231 (Fig. 5c) and Vero cells (Fig. 5d). These results suggested that 26SK crude protein lysate could inhibit the growth of triple-negative human breast adenocarcinoma cells.
Homology Modeling of the RNA Binding Site and Ribosome Binding Site of 26SK and 34.7(A)SK
The crude protein extracts from cells carrying 26SK and 34.7(A)SK displayed rather different characters. Both extracts had rRNA N-glycosidase activity but only the 26SK cell lysate could inhibit the growth of E. coli. We believed that there must be distinct factors that differed between the two proteins. With the sequence analysis, the 26SK protein was predicted to be smaller than for the 34.7(A)SK protein. The 26SK protein was predicted to have 233 amino acids while 309 amino acids were predicted for 34.7(A)SK. There might have been structural dissimilarities between the two proteins. Therefore, predicted models for 26SK and 34.7(A)SK were generated using the ricin A chain (RTA) from Ricinus communis (castor bean) with substrate analog (protein data bank: 1BR6) and without substrate analog (protein data bank: 2AAI) as templates. The percent similarity of J. curcas RIPs with 1BR6 ranged between 30.88 and 42.23, while 2AAI template had the percentage of similarity approximately 31.02–58.24 (Supplementary Table 5). The reliability of these protein structures was validated using a Ramachandran plot. The reliability of the models was considered based on the percentage of total amino acid residues in disallowed regions, which should be lower than two [28]. From the models generated using 1BR6 as a forced template, the amino acid residues arranged in disallowed regions ranged between 0–1.8% and 0–1.0% using RAMPAGE and PROCHECK, respectively (Supplementary Table 6). For the 2AAI forced template, the percentages of amino acid residues arranged in the outlier regions using RAMPAGE and PROCHECK were approximately 1.1–4.0% and 0.6–2.7%, respectively (Supplementary Table 6). This indicated that using the 1BR6 as the template generated a more appropriate RIP structure than the model generated with 2AAI.
Proper folding of proteins is crucial for protein functions within cells [38]. Therefore, 3D structure modelling of the representative proteins (26SK, 33curcin, and 34.7(A)SK) was generated and compared with the ricin A chain (RTA) using the Swiss-PdbViewer and BIOVIA Discovery Studio Visualizer software package. Based on the structure modelling, there was a variation in the C-terminal domain of the 26SK protein compared to the RTA, 33curcin, and 34.7(A)SK, reaffirming the notion of a truncated C-terminal domain in the 26SK protein (Fig. 6). The α-helix and two β-strands in the C-terminal domain were absent in the 26SK structure but present in the 33curcin and 34.7(A)SK structures (Fig. 6). This region represented the P2 stalk protein ribosome binding site (Fig. 2). The binding of RIP to the P2 stalk binding protein allows the access of RIP to the conserved sarcin-ricin loop leading to the translational inhibition.
Predicted conformation of 26SK, 33curcin, and 34.7(A)SK. The protein conformations were represented in green ribbons aligning with the ribosome P2 binding site of ricin A chain (blue ribbons). Ribosome P2 binding sites of 26SK, 33curcin, and 34.7(A)SK indicated by red circles and pteroic acid substrate analog are located in the center of the predicted conformations (red molecule). “RTA” represented the predicted conformation of ricin A chain (blue ribbons) which displayed the conserve RIP fold structure. “Merge” showed the similarity of RIP fold structure between 26SK, 34.7(A)SK, and 33curcin compared with RTA
3.7 26SK, 33curcin, and 34.7(A)SK Displayed Diverse Hydrophobicity and Electrostatic Potential on the Surfaces
As the structure model of the 26SK protein differed from those of 33curcin and 34.7(A)SK, it was of interest to determine whether this difference would influence other properties and activities of the proteins. Therefore, the predicted structure models of 26SK, 33curcin, and 34.7(A)SK were analyzed by comparing hydrophobicity and the electrostatic potential map using Swiss-PdbViewer and the BIOVIA Discovery Studio Visualizer software package. The hydrophobicity levels on the surfaces of 26SK, 33curcin, and 34.7(A)SK were indicated by the histogram spectrum ranging from −3 to 3 where blue color represented low hydrophobicity and brown color corresponded to high hydrophobicity of the proteins. As shown in Fig. 7, the overall hydrophobicity levels on the surface of 34.7(A)SK were similar to 33curcin though there were some differences between the surface hydrophobicity of 26SK to 33curcin and to 34.7(A)SK. Moderate and high hydrophobicity were detected near the substrate binding sites of 26SK, while the substrate binding sites of 33curcin and 34.7(A)SK had lower hydrophobicity. The other side of 33curcin and 34.7(A)SK proteins was masked with low hydrophobicity, whereas a high hydrophobicity patch was at the center of the 26SK protein (Fig. 7). The electrostatic potential map in Fig. 8 displays the overall charge distribution with red color indicating the negative potential and blue color representing the positive potential. The electrostatic potential maps of 26SK, 33curcin, and 34.7(A)SK were calculated and compared using default parameters (Supplementary Table 7). From in silico analysis, all the proteins had unique electrostatic potential maps. 26SK displayed striking distributions of positive and negative potentials (Fig. 8). Approximately half of the 26SK protein contained positive potential, while the other half showed negative potential. In contrast, the 33curcin had a negative potential map cloud condensed around the protein, whereas the positive potential was scattered and surrounded the outside of the protein. Interestingly, 34.7(A)SK presented a clearly different pattern of electrostatic potential mapping compared to the others, with its negative potential map dispersed around the 34.7(A)SK protein, while a positive potential map was rare (Fig. 8).
Discussion
RIP Expression Profiles in Jatropha curcas Varieties KUBP33 and KUBP79
Many publications have reported on RIP gene expression in various parts of J. curcas, such as curcin from the endosperm [39] and curcin 2 [13, 14], curcin-L from leaves [12], and curcin-C from cotyledons [15]. From the 13 RIP genes, expression of 12 genes was detected with four different expression patterns, while the expression of one gene could not be detected in the tissues under the conditions tested. The curcin protein (33curcin) was found in the endosperm [39]. Its expression was detected only in the seed kernel as expected.
26SK and 34.7(A)SK from J. curcas Belonged to Ribosome-Inactivating Proteins
Ribosome-inactivating proteins are defined as proteins with rRNA N-glycosidase activity [8]. Previous study reported that a purified curcin could interrupt a cell-free translation in rabbit reticulocyte lysate with an IC50 of 0.19 nmol/l (it is equivalent to 5.36 pg/ml) [29]. The purified Jc-SCRIP, another type 1 RIP from J. curcas, exhibited N-glycosidase activity against rabbit reticulocyte lysate at a concentration of 1 µg per reaction [16]. The crude protein lysates from E. coli harboring 26SK or 34.7(A)SK exhibited N-glycosidase activity similar to J. curcas seed kernel crude protein in which natural curcin was present. Therefore, 26SK and 34.7(A)SK were likely to function as ribosome-inactivating proteins in Jatropha.
Growth Inhibition of E. coli Cells by 26SK
The antimicrobial activity of various RIPs has been continuously reported. The minimum inhibitory concentration of a purified Jc-SCRIP that inhibited the growth of various types of Gram-positive and Gram-negative bacteria was in the range 0.20–12.85 µM (equivalent to 7.78–500.35 µg/ml) [16]. Antimicrobial activity has been reported of balsamin, a type 1 RIP isolated from Momordica balsamina. The diameters of inhibition zones using the various concentrations of purified balsamin against E. coli cells at 50, 100, and 200 μg/ml were 2.0 ± 0.1, 4.0 ± 0.3, and 6.0 ± 0.5 mm, respectively [36]. Another type 1 RIP from Momordica balsamina is MbRIP-1; this protein has displayed toxicity against E. coli cells, with the diameters of inhibition zones of 3, 5, and 10 μg/ml of purified MbRIP-1 being 3.0± 0.2, 5.0±0.5, and 8.0± 0.7 mm, respectively [40]. From our current work, 300-μg crude protein of 26SK cell lysate had a growth inhibition against E. coli strain Rosetta (DE3) pRARE. In contrast, no toxicity was observed for the 34.7(A)SK crude protein extract. Compared to previous reports, the effective concentrations of the purified Jc-SCRIP, balsamin, and MbRIP-1 were higher than for 26SK crude cell lysate. However, because the 26SK protein studied here was a crude protein, it is difficult to directly compare its effective concentration with other purified RIP proteins. However, the results indicated that even with the crude protein, 26SK still had an inhibitory effect on E. coli, suggesting that even a small amount of this protein was toxic to E. coli cells.
Antitumor Activity of 26SK Crude Proteins
The cytotoxicity of a purified curcin on various types of cancer cell lines has been reported. From the MTT assay, the IC50 of a purified curcin against SGC-7901, Sp2/0, and human hepatoma after treatment for 72 h were 0.23 µg/ml, 0.66 µg/ml, and 3.160 µg/ml, respectively. In contrast, curcin did not exhibit the cytotoxic effect on Hela cells and MRC normal cells [29]. The antitumor activity of other J. curcas type 1 RIP such as curcin-C and Jc-SCRIP have been also studied. Curcin-C, a highly conserved protein to curcin, had the IC50 against U20S cell line (osteosarcoma cells) of 0.019 µM (equivalent to 0.597 pg/ml) when IC50 of curcin was 0.27 µM (equivalent to 7.614 pg/ml) [15]. The IC50 concentrations of purified Jc-SCRIP against SW-620, MCF-7, and HepG2 after treatment for 24 h were 0.25 mM, 0.15 mM, and 0.40 mM, respectively (equivalent to 9.73 µg/ml, 5.84 µg/ml, and 15.58 µg/ml) [16]. From the MTT assay of 26SK crude protein, the IC50 of this crude protein against MDA-MB-231 after treatment for 72 h was 2,208±11.24 µg/ml. These results pointed that even though the 26SK crude protein presented lower cytotoxicity when compared with a purified Jc-SCRIP and curcin, but it still possessed inhibitory effect against one of an important cancer, a triple-negative breast cancer cell line with no toxicity to the Vero cells.
Secondary Structure of 26SK Might Be Related to Its Toxicity
The secondary structure of proteins plays a key role in protein stability and functions, particularly in enzymatic activity [38]. Structural analysis of curcin showed amino acids which were in proximity to adenine. These amino acids were Y118, V120, F131, N132, D133, L137, T156, G157, and S158. Among these residues, the amino acids Y118, F131, G157, and S158 were involved in the ricin-adenine complex [18, 41]. Ricin depurinated the specific adenine in the sarcin-ricin loop when the target adenine was inserted between 2 tyrosines in the catalytic region (Y80 and Y123). Furthermore, the residues E177 and R180 of ricin are important to the speed of the reaction and the breakage of N-glycosidic bonds [41]. The current study identified that the amino acid residues predicted to be arranged close to adenine were also conserved in J. curcas RIPs (Fig. 2 and Supplementary Table 3). The corresponding amino acids at the catalytic site were conserved in 26SK, 34.7(A)SK, and 33curcin suggesting that these proteins were likely to perform N-glycosidase activity in vitro. Indeed, this was true for the 26SK and 37.3(A)SK proteins (Fig. 4a and b).
RIP proteins function to cleave ribosomal RNAs, leading to translation inhibition. The proteins comprised of RNA binding sites and ribosome binding sites [42, 43]. The eukaryotic ribosomal stalk protein consists of the P0, P1, and P2 heterodimers to form a pentameric complex of the P-proteins [44]. The conserved C-terminal domain of the eukaryotic ribosome P stalk proteins is essential for the target recognition and the rRNA depurination [42, 45]. Specificity of RIPs to their rRNA substrate may be due to the sensitivity of the interaction between the RIPs and the ribosomal proteins [46]. The hydrophobic pocket of the ricin A chain (PDB: 1BR6) interacted with the ribosomal P2 stalk protein through the interaction formed by the following amino acid residues: Y183, L207, L232, F240, V242, I247, P250, and I251. Deficiencies in ribosome binding and translation inhibition were observed when the Y183 and R235 at the P2 stalk protein recognition sites of ricin were mutated by site-directed mutagenesis [19]. The conserved Y and R amino acid residues in the P2 stalk protein recognition sites were present in the J. curcas RIPs (Fig. 2), suggesting that the interaction between J. curcas proteins and a ribosome complex might occur by interaction through these amino acids.
In general, proteins designated as RIPs display a conserved “RIP fold” consisting of the N-terminal domain 1, domain 2 in the middle, and the C-terminal domain 3. Domain 1 consists of β-strands and α-helices. Domain 2 contains α-helices and the C-terminal domain encompasses two α-helices and two β-strands [31]. The current results revealed that a part of the C-terminal domain of the RIP fold was absent in the 26SK structure but was exhibited in 33curcin and 34.7(A)SK (Fig. 6). Of note, the 26SK protein was still able to perform N-glycosidase in vitro despite having only the partial C-terminal domain. This leads to the question of how important this domain is to rRNA binding and N-glycosidase activities. This region is probably responsible for enzyme function.
Several RIPs interact with the P proteins in vitro during depurination, such as the ricin A chain [33, 34, 43], pokeweed antiviral protein (PAP) [43], and trichosanthin [45]. The ricin A chain RIP was not able to depurinate the ribosomes without intact P stalk proteins [42], while PAP did not require the C-terminal domain to depurinate trypanosome [47] and yeast ribosomes [43]. This suggests that the interactions between the ribosomal P proteins and RIPs may not be a general feature of all RIPs. Thus, the 26SK protein may use different docking sites on the ribosome to access its target site.
Peptides with anti-microbial activity display some characteristics such as positive charge, amphipathic, and contain high amounts of cysteine in the sequence. Amphipathic nature is the character where the protein or peptide contains both hydrophilic and hydrophobic regions in the molecule. It has been proposed that this is involved with the penetration of the peptide into the cell wall of bacteria [48]. In addition, hydrophobic interactions between proteins are essential for protein stability, conformation, functions, and interactions [49]. Based on molecular surface analysis, 26SK, 34.7(A)SK, and 33curcin contained both hydrophobic and hydrophilic amino acids in their structures. Among them, there were no remarkable differences which might contribute to the toxicity of the 26SK protein to the E. coli cells.
Electrostatic potential has been applied as a tool to predict the interactions between antimicrobial peptides with the lipid bilayers of the bacterial cell wall. These interactions lead to the disruption of the bacterial cell wall [50]. The 26SK protein had an interesting electrostatic potential map, displaying a highly positive charge whereas a highly negative charge was observed in the 34.7(A)SK map (Fig. 8). As mentioned above, anti-microbial peptides usually carry a positive charge on their surface. This character was proposed to be involved with the binding of the peptides to the bacterial cell wall prior to cell penetration [48]. This was supported by the results in the current work where 26SK had anti-microbial activity while 34.7(A)SK did not. Luffin P1 was isolated and purified as a peptide from Luffa aegyptiaca. It was classified as the smallest RIP since the peptide had rRNA N-glycosidase activity, though without any sequence or structural similarity [51]. Luffin P1 also had anti-microbial activity [52]. Molecular surface analysis indicated some similarities among the electrostatic potentials of luffin P1, 26SK, and 33curcin (Supplementary Fig. 3). The three proteins displayed positive potential on their surfaces. Therefore, we hypothesized that 26SK might bind and disrupt the bacterial cell wall due to its surface characteristics of having a positive charge and an amphipathic nature. However, the mechanisms underlying its anti-microbial activity require further elucidation.
Conclusion
Phylogenetic tree analysis of predicted RIP protein sequences showed that most of the predicted J. curcas RIPs were categorized in type 1 RIP while only 62NF belonged to type 2 RIP. Moreover, the expression of predicted RIP genes was detectable in various tissues. From in silico analysis, we found that 26SK lacked the C-terminal domain which was the eukaryotic ribosome P stalk protein binding site such as 26SK. However, 26SK possessed rRNA N-glycosidase activity against substrate as well as 34.7(A)SK. Interestingly, 26SK exhibited inhibitory effects to E. coli and breast cancer cell line whereas 34.7(A)SK cell lysate did not exhibit this activity. Structure models of 26SK, 33curcin, and 34.7(A)SK displayed diverse hydrophobicity and electrostatic potential on their surfaces suggesting the functional differences among the Jatropha RIP proteins.
Data Availability
Not applicable.
Code Availability
Not applicable.
Abbreviations
- RIP:
-
Ribosome-inactivating protein
- rRNA:
-
Ribosomal RNA
- RTA:
-
Ricin A chain
- PMSF:
-
Phenylmethylsulfonyl fluoride
- PBS:
-
Phosphate buffer saline
References
Openshaw, K. (2000). A review of Jatropha curcas: An oil plant of unfulfilled promise. Biomass & Bioenergy, 19, 1–15.
King, A. J., He, W., Cuevas, J. A., Freudenberger, M., Ramiaramanana, D., & Graham, I. A. (2009). Potential of Jatropha curcas as a source of renewable oil and animal feed. Journal of Experimental Botany, 60, 2897–2905.
Endo, Y., & Tsurugi, K. (1988). The RNA N-glycosidase Activity of ricin A-chain. The characteristics of the enzymatic activity of ricin A-chain with ribosomes and with rRNA. Journal of Biological Chemistry, 263(18), 8735–8739.
Barbieri, L., Valbonesi, P., Gorini, P., Pession, A., & Stirpe, F. (1996). Polynucleotide: adenosine glycosidase activity of saporin-L1: Effect on DNA, RNA and poly(A). Biochemical Journal, 319, 507–513.
Park, S. W., Vepachedu, R., Owens, R. A., & Vivanco, J. M. (2004). The N-glycosidase activity of the ribosome-inactivating protein ME1 targets single-stranded regions of nucleic acids independent of sequence or structural motifs. Journal of Biological Chemistry, 279(33), 34165–34174.
Narayanan, S., Surendranath, K., Bora, N., Surolia, A., & Karande, A. A. (2005). Ribosome inactivating proteins and apoptosis. FEBS Letters, 579(6), 1324–1331.
Lancaster, L., Lambert, N. J., Maklan, E. J., Horan, L. H., & Noller, H. F. (2008). The sarcin-ricin loop of 23S rRNA is essential for assembly of the functional core of the 50S ribosomal subunit. RNA, 10, 1999–2012.
Stirpe, F. (2004). Ribosome-inactivating proteins. Toxicon, 44, 371–383.
Devappa, R. K., Makkar, H. P. S., & Becker, K. (2010). Nutritional, biochemical, and pharmaceutical potential of proteins and peptides from Jatropha. Journal of Agricultural and Food Chemistry, 58(11), 6543–6555.
Luo, M. J., Yang, X. Y., Liu, W. X., Xu, Y., Huang, P., Yan, F., & Chen, F. (2006). Expression, purification and anti-tumor activity of curcin. Acta Biochimica et Biophysica Sinica (Shanghai), 38(9), 663–668.
Kshirsagar, R. V. (2010). Insecticidal activity of Jatropha seed oil against Callosobruchus maculatus (Fabricius) infesting Phaseolus aconitifolius Jacq. The Bioscan, 5(3), 415–418.
Qin, X., Zheng, X., Shao, C., Gao, J., Jiang, L., Zhu, X., Yan, F., Tang, L., Xu, Y., & Chen, F. (2009). Stress-induced curcin-L promoter in leaves of Jatropha curcas L. and characterization in transgenic tobacco. Planta, 230, 387–395.
Qin, W., Huang, M. X., Xu, Y., Zhang, X. S., & Chen, F. (2005). Expression of a ribosome inactivating protein (curcin 2) in Jatropha curcas is induced by stress. Journal of Biosciences, 30, 351–357.
Huang, M. X., Hou, P., Wei, Q., Xu, Y., & Chen, F. (2008). A ribosome-inactivating protein (curcin 2) induced from Jatropha curcas can reduce viral and fungal infection in transgenic tobacco. Plant Growth Regulation, 54, 115–123.
Zhang, Y. X., Yang, Q., Li, C. Y., Ding, M. M., Lv, X. Y., Tao, C. Q., Yu, H. W., Chen, F., & Xu, Y. (2017). Curcin C, a novel type I ribosome-inactivating protein from the post-germinating cotyledons of Jatropha curcas. Amino Acids, 49, 1619–1631.
Nuchsuk, C., Wetprasit, N., Roytrakul, S., Choowongkomon, K., T-Thienprasert, N., Yokthongwattana, C., Arpornsuwan, T., & Ratanapo, S. (2013). Bioactivities of Jc-SCRIP, a type 1 ribosome-inactivating protein from Jatropha curcas seed coat. Chemical Biology & Drug Design, 82(4), 453–462.
Kim, Y. S., & Robertus, J. D. (1992). Analysis of several key active site residues of ricin A chain by mutagenesis and X-ray crystallography. Protein Engineering Design & Selection, 5, 775–779.
Yan, X. J., Hollis, T., Svinth, M., Day, P., Monzingo, A. F., Milne, G. W. A., & Robertus, J. D. (1997). Structure based identification of a ricin inhibitor. Journal of Molecular Biology, 266, 1043–1049.
Fan, X. J., Zhu, Y. W., Wang, C. Y., Niu, L. W., Teng, M. K., & Li, X. (2016). Structural insights into the interaction of the ribosomal P stalk protein P2 with a type II ribosome-inactivating protein ricin. Scientific Reports, 6, 1–10.
Kumar, S., Stecher, G., Li, M., Knyaz, C., & Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Molecular Biology and Evolution, 35, 1547–1549.
Guex, N., Peitsch, M. C., & Schwede, T. (2009). Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis, 30, 162–173.
Benkert, P., Biasini, M., & Schwede, T. (2011). Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics, 27, 343–350.
Bertoni, M., Kiefer, F., Biasini, M., Bordoli, L., & Schwede, T. (2017). Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Scientific Reports, 7, 1–15.
Bienert, S., Waterhouse, A., de Beer, T. A. P., Tauriello, G., Studer, G., Bordoli, L., & Schwede, T. (2017). The SWISS-MODEL Repository - New features and functionality. Nucleic Acids Research, 45, 313–319.
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., Heer, F. T., de Beer, T. A. P., Rempfer, C., Bordoli, L., Lepore, R., & Schwede, T. (2018). SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Research, 46(1), 296–303.
Rutenber, E., Katzin, B. J., Ernst, S., Collins, E. J., Mlsna, D., Ready, M. P., & Robertus, J. D. (1991). Crystallographic refinement of ricin to 2.5 A. Proteins, 10, 240–250.
Lovell, S. C., Davis, I. W., Arendall, W. B., III., de Bakker, P. I. W., Word, J. M., Prisant, M. G., Richardson, J. S., & Richardson, D. C. (2002). Structure validation by Cα geometry: φ/ψ and Cβ deviation. Proteins, 50, 437–450.
Laskowski, R. A., MacArthur, M. W., Moss, D. S., & Thornton, J. M. (1993). PROCHECK: A program to check the stereochemical quality of protein structures. Journal of Applied Crystallography, 26, 283–291.
Lin, J., Yan, F., Tang, L., & Chen, F. (2003). Antitumor effects of curcin from seeds of Jatropha curcas. Acta Pharmaceutica Sinica, 24(3), 241–246.
Bauer, A. W., Kirby, W. M. M., Sherris, J. C., & Turck, M. (1966). Antibiotic susceptibility testing by a standardized single disk method. American Journal of Clinical Pathology, 36, 493–496.
Di Maro, A., Citores, L., Russo, R., Iglesias, R., & Ferreras, J. M. (2014). Sequence comparison and phylogenetic analysis by the Maximum Likelihood method of ribosome-inactivating proteins from angiosperms. Plant Molecular Biology, 85(6), 575–588.
Tumer, N. E., & Li, X. P. (2012). Interaction of ricin and Shiga toxins with ribosomes. Current Topics in Microbiology and Immunology, 357, 1–18.
Zhao, Y. J., Li, X. P., Chen, B. Y., & Tumer, N. E. (2017). Ricin uses arginine 235 as an anchor residue to bind to P-proteins of the ribosomal stalk. Scientific Reports, 7(4291), 1–12.
Zhou, Y. J., Li, X. P., Kahn, J. N., McLaughlin, J. E., & Tumer, N. E. (2019). Leucine 232 and hydrophobic residues at the ribosomal P stalk binding site are critical for biological activity of ricin. Bioscience Reports, 39(10), 1–15.
Endo, Y., & Tsurugi, K. (1987). RNA N-glycosidase activity of ricin A-chain. Mechanism of action of the toxic lectin ricin on eukaryotic ribosomes. Journal of Biological Chemistry, 262(17), 8128–8130.
Ajji, P. K., Binder, M. J., Walder, K., & Puri, M. (2017). Balsamin induces apoptosis in breast cancer cells via DNA fragmentation and cell cycle arrest. Molecular and Cellular Biochemistry, 432(1–2), 189–198.
Lu, W. L., Mao, Y. J., Chen, X., Ni, J., Zhang, R., Wang, Y. T., Wang, J., & Wu, L. F. (2018). Fordin: A novel type I ribosome inactivating protein from Vernicia fordii modulates multiple signaling cascades leading to anti-invasive and pro-apoptotic effects in cancer cells in vitro. International Journal of Oncology, 53, 1027–1042.
Teilum, K., Olsen, J. G., & Kragelund, B. B. (2011). Protein stability, flexibility and function. Biochimica et Biophysica Acta, 1814, 969–976.
Stirpe, F., Brizzi-Pession, A., Lorenzoni, E., Strochi, P., Montanaro, L., & Sperti, S. (1976). Studies on the proteins from the seeds of Croton tigilium and Jatropha curcas. Biochemical Journal, 156, 1–6.
Kushwaha, G. S., Pandey, N., Sinha, M., Singh, S. B., Kaur, P., Sharma, S., & Singh, T. P. (2012). Crystal structures of a type-1 ribosome inactivating protein from Momordica balsamina in the bound and unbound states. Biochimica et Biophysica Acta, 1824, 679–691.
Srivastava, M., Gupta, S. K., Abhilash, P. C., & Singh, N. (2012). Structure prediction and binding sites analysis of curcin protein of Jatropha curcas using computational approaches. Journal of Molecular Modeling, 18(7), 2971–2979.
Chan, D. S. B., Chu, L. O., Lee, K. M., Too, P. H. M., Ma, K. W., Sze, K. H., Zhu, G., Shaw, P. C., & Wong, K. B. (2007). Interaction between trichosanthin, a ribosome inactivating protein, and the ribosomal stalk protein P2 by chemical shift perturbation and mutagenesis analyses. Nucleic Acids Research, 35(5), 1660–1672.
Chiou, J. C., Li, X. P., Remacha, M., Ballesta, J. P., & Tumer, N. E. (2008). The ribosomal stalk is required for ribosome binding, depurination of the rRNA and cytotoxicity of ricin A chain in Saccharomyces cerevisiae. Molecular Microbiology, 70, 1441–1452.
Krokowski, D., Boguszewska, A., Abramczyk, D., Liljas, A., Tchorzewski, M., & Grankowski, N. (2006). Yeast ribosomal P0 protein has two separate binding sites for P1/P2 proteins. Molecular Microbiology, 60, 386–400.
May, K. L., Li, X. P., Martínez-Azorín, F., Ballesta, J. P., Grela, P., Tchórzewski, M., & Tumer, N. E. (2012). The P1/P2 proteins of the human ribosomal stalk are required for ribosome binding and depurination by ricin in human cells. FEBS Journal, 279(20), 3925–3936.
McCluskey, A. J., Poon, G. M. K., Bolewska-Pedyczak, E., Srikumar, T., Jeram, S. M., Raught, B., & Gariepy, J. (2008). The catalytic subunit of Shiga-like toxin 1 interacts with ribosomal stalk proteins and is inhibited by their conserved C-terminal domain. Journal of Molecular Biology, 378, 375–386.
Ayub, M. J., Smulski, C. R., Ma, K. W., Levin, M. J., Shaw, P. C., & Wong, K. B. (2008). The C-terminal end of P proteins mediates ribosome inactivation by trichosanthin but does not affect the pokeweed antiviral protein activity. Biochemical and Biophysical Research Communications, 369, 314–319.
Tam, J. P., Wang, S. J., Wong, K. H., & Tan, W. L. (2015). Antimicrobial peptides from plants. Pharmaceuticals, 8, 711–757.
Loladze, V. V., Ermolenko, D. N., & Makhatadze, G. I. (2002). Thermodynamic consequences of burial of polar and non-polar amino acid residues in the protein interior. Journal of Molecular Biology, 320(2), 343–357.
La Rocca, P., Shai, Y., & Sansom, M. S. P. (1999). Peptide-bilayer interactions: simulations of dermaseptin B, an antimicrobial peptide. Biophysical Chemistry, 76, 145–159.
Li, F., Yang, X. X., Xia, H. C., Zeng, R., Hu, W. G., Li, Z., & Zhang, Z. C. (2003). Purification and characterization of Luffin P1, a ribosome-inactivating peptide from the seeds of Luffa cylindrica. Peptides, 24, 799–805.
Ng, Y. M., Yang, Y. H., Sze, K. H., Zhang, X., Zheng, Y. T., & Shaw, P. C. (2011). Structural characterization and anti-HIV-1 activities of arginine/glutamate-rich polypeptide Luffin P1 from the seeds of sponge gourd (Luffa cylindrica). Journal of Structural Biology, 174, 164–172.
Acknowledgements
The authors would like to thank Associate Professor Kittisak Yokthongwattana, Department of Biochemistry, Faculty of Science, Mahidol University, Bangkok, Thailand, for providing the plasmid, pETDuet-CPN60B1.
Funding
This work was supported in part by funding (v-t(d)45.54) to CY from the Kasetsart University Research and Development Institute (KURDI), Bangkok, Thailand.
Author information
Authors and Affiliations
Contributions
D.P. and C.Y. conceived and designed the experiments. D.P. performed the experiments. D.P., K.C., S.R., and C.Y. analyzed the data. D.P. and C.Y. wrote the manuscript with contributions from all authors. All authors read and commented on the manuscript before submission.
Corresponding author
Ethics declarations
Ethical Approval
Not applicable.
Consent to Participate
Not applicable.
Consent to Publish
Not applicable.
Human and Animal Participants
No such procedures were performed in studies.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.

Supplementary Fig. 1
The Growth rate of E. coli host cells expressing (a) 26SK and (b) 34.7(A)SK under inducing and non-inducing conditions. EV: E. coli carrying pET28a(+) or pETDuet-1 empty vector in (a) and (b) respectively; 26SK: E. coli containing pET28(a)+-26SK; 34.7(A)SK: E. coli harboring pETDuet-34.7(A)SK: CPN60B1: E. coli harboring pETDuet-CPN60B1; N: non-inducing condition; I: inducing condition (PNG 188 kb)

Supplementary Fig. 2
Recombinant RIPs production in E. coli cells. (a) SDS-PAGE (b) Western blot of pET28a(+) empty vector, CPN60B1, 26SK and 34.7(A)SK crude proteins under inducing and non-inducing conditions. M: Prestained SDS-PAGE standard broad range protein marker; EV: pET28a(+) empty vector; CPN60B1: E. coli harboring pETDuetTM-CPN60B1; 26SK: E. coli carrying pET28(a)+-26SK; 34.7(A)SK: E. coli containing pETDuetTM-34.7(A)SK; N: non- inducing condition; I: inducing condition. (Exposure time = 120 seconds) (PNG 1669 kb)

Supplementary Fig. 3
Electrostatic potential map of transmembrane helices located in 26SK, 33curcin, 34.7(A)SK and luffin P1. Predicted conformation of 26SK, 33curcin, and 34.7(A)SK represented by green ribbons and luffin P1 transmembrane helices and transmembrane helices of 26SK, 33curcin and 34.7(A)SK display by black ribbons. Histogram presents the charge of electrostatic potential from -1.8 (red) to 1.8 (blue). (PNG 1175 kb)
Supplementary Table 1
(DOCX 18 kb)
Supplementary Table 2
(DOCX 17 kb)
Supplementary Table 3
(DOCX 15 kb)
Supplementary Table 4
(DOCX 16 kb)
Supplementary Table 5
(DOCX 14 kb)
Supplementary Table 6
(DOCX 12 kb)
Supplementary Table 7
(DOCX 14 kb)
Rights and permissions
About this article
Cite this article
Pathanraj, D., Choowongkomon, K., Roytrakul, S. et al. Structural Distinctive 26SK, a Ribosome-Inactivating Protein from Jatropha curcas and Its Biological Activities. Appl Biochem Biotechnol 193, 3877–3897 (2021). https://doi.org/10.1007/s12010-021-03714-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-021-03714-6