Abstract
Vitamin B2 and arachidonic acid, both needed in food or feed, are produced by filamentous fungi on a scale of 1000 t/year. After 50 years of research and development, vitamin B2 production with Ashbya gossypii, a hemiascomycete, has won a competition with chemical synthesis by combining classical strain improvement, genetic engineering, and process optimization. Screening for antimetabolite resistance, overexpression, and disruption of genes were applied, resulting in a metabolically designed cellular factory. But although the genome of A. gossypii has been sequenced and analytical tools are highly developed, important questions are still open, e.g. concerning excretion of the vitamin. Arachidonic acid is a long-chain polyunsaturated fatty acid used for infant formula to support the development of the child’s brain. Mortierella alpina, a zygomycete, accumulates lipids rich in arachidonic acid when growth becomes limited, e.g. after consumption of the nitrogen source. The first steps in pathway engineering were done.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 I. Introduction
Vitamins are organic nutrients essential for the vital functions of humans or domestic animals because their own anabolism does not synthesize them adequately. Since they fulfil catalytic or hormone-like functions, an intake of small amounts is sufficient but because storage is limited they should be on the menu every day. Although at least in the so-called developed countries a more than sufficient and seasonally independent food supply allows adequate intake of all vitamins and vitamin-like compounds, up to 50% of the people tested in large scale studies were found to be short in vitamin B2, folate, vitamin A, vitamin D, vitamin E, and vitamin C (Aranceta et al. 2001). Between 1930 and 1950 all 13 vitamin groups were determined in their structure after extraction, purification, and chemical synthesis. Today the technical production of thousands of tonnes per year is performed by chemistry, extraction, fermentation or bioconversion, or combined processes (Table 11.1).
Arachidonic acid, vitamin B2, and vitamin B12 are produced by fermentation exclusively. Since biotechnical vitamin B2 production has almost completely replaced chemical synthesis, other processes might follow. The replacement of chemical by microbiological and/or enzymatic processes, nowadays often traded under the name “white biotechnology”, is encouraged by the increasing costs for disposing of wastes and a shift towards renewable educts to fulfil the requirements of sustainability. This chapter focuses on vitamin B2 and long-chain polyunsaturated fatty acids because these processes are perfect examples for the application of fungi on an industrial scale. Only the most important physiological features and biotechnological strategies for their improvement are discussed.
2 II. Vitamin B2
Riboflavin is the most frequently used name of 7,8-dimethyl-10-(d-1’-ribityl)isoalloxazine because it indicates its yellow (latin flavus) color as a pure solid. Synonyms are lactoflavin, lactochrome and ovoflavin, referring to the sources it derives from. Riboflavin or its derivatives have key functions in energy metabolism, maintenance of a healthy skin and muscles, support of the immune and nervous systems, and promotion of cell growth and division. It is the precursor for the coenzymes flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD), important electron carriers in redox reactions. Additionally, both flavo-coenzymes participate in bioluminescence, light sensing, phototropism, DNA protection against UV light damage, and reset of the circadian clock (Cashmore et al. 1999). Since higher eukaryotes cannot produce riboflavin, it has the status of a vitamin. Because of light sensitivity and poor resorption, riboflavin deficiency is often a result of surveys on the nutritional status. Therefore supplementation of food (Fletcher et al. 2004) and feed is performed. Overdosing due to dietary supplementation is impeded by the direct excretion of riboflavin with the urine. In industrialized countries processed food is often fortified by the use of riboflavin as a colorant (E 101). About 70% of industrially produced riboflavin is applied in animal feed, since productive livestock, especially poultry and pigs, show growth retardation and diarrhea in the case of riboflavin deficiency. According to a SRI Consulting Report from 2005, the need for industrially produced riboflavin was estimated to be 7000 t year–1. Deficiency symptoms like dermatitis can be avoided by a nutritional intake of 0.3–1.8 mg day–1 for humans and 1–4 mg kg–1 for animals (Eggersdorfer and Adam 1996). One cup of milk per day is sufficient to fulfil this requirement.
For about half of a century riboflavin production was in the domain of the chemical industry and followed a multi-step process starting with 3,4-xylidine and d-ribose. The latter was provided by chemical conversion of glucose or more recently by fermentation from glucose using transketolase-deficient Bacillus spp. strains. The first commercial single-step riboflavin fermentations came up in the 1940s, employing the anaerobic Gram-positive bacterium Clostridium acetobutylicum or the more efficient hemiascomycetes Eremothecium ashbyi or Ashbya gossypii (Wickerham et al. 1946; Perlman 1979). All companies manufacturing riboflavin by fermentation shut down their production plants in the 1960s, being competitively disadvantaged by the chemical processes (Lago and Kaplan 1981).
Today, riboflavin production by modern microbiology has become a paradigm of environmentally superior white biotechnology replacing classic chemistry. After Merck (USA) resumed fermentation by A. gossypii in 1974, the success story started with an A. gossypii production plant of BASF (Germany) in 1990. The market share of riboflavin produced by the microbial processes increased from 5% in 1990 to 75% in 2002. Initially the two naturally vitamin B2-overproducing fungi A. gossypii and Candida famata, and later the metabolically engineered Gram-positive bacterium Bacillus subtilis, were used. Today C. famata is no longer used and chemical riboflavin production came to an end, leaving only A. gossypii- and B. subtilis-based production methods. Both processes are the outcome of classical strain improvement, genetic engineering, and process optimization. The B. subtilis process was recently compared with the A. gossypii process by Hohmann and Stahmann (2010). Because A. gossypii seems to be much better in productivity, the major part of this chapter deals with it and a minor part discusses relevant data of C. famata.
2.1 A. Ashbya gossypii
2.1.1 1. The Riboflavin Biosynthesis Pathway
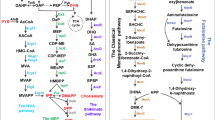
Riboflavin biosynthesis was found in plants and most microorganisms but not in higher eukaryotes. Two precusors, guanosine triphosphate, also needed for RNA, and ribulose-5-phosphate, provided by pentose phosphate pathway, are the roots of this anabolism (Fig. 11.1; Bacher 1991). Despite the dephosphorylation of the ribityl residue, which is catalyzed by an unspecific phosphatase, all enzymes are known and all genes are cloned for A. gossypii (Revuelta et al. 1995). While reduction of the ribosyl residue comes prior to deamination of the diamino pyrimidinone in fungi, the sequential order of these reactions is inverted in bacteria (Burrows and Brown 1978). Because the last step, formation of riboflavin, is a dismutation of two molecules of 6,7-dimethyl-8-ribityllumazine, in total two molecules of ribulose-5-phosphate and one molecule of guanosine triphosphate are needed. Therefore the activity of 3,4-dihydroxy-2-butanone-4-phosphate synthase must be double in comparison to the other enzymes. Detailed insight into riboflavin biosynthesis and the catalytic mechanisms of the involved enzymes is described in comprehensive reviews (Fischer and Bacher 2005).
Riboflavin biosynthesis pathway in Ashbya gossypii or bacteria where desamination takes place before reduction. An enzyme catalysing a dephosphorylation of ArPP is not characterized. Structures are adapted from Bacher (1991). 1: guanosine triphosphate; 2: 2,5-diamino-6-ribosylamino-4 (3H)-pyrimidinone-5’-phosphate; 3: 5-amino-6-ribosylamino-2,4 (1H,3H)-pyrimidinedione-5’-phosphate; 4: 2,5-diamino-6-ribitylamino-4 (3H)-pyrimidinone-5’-phosphate; 5: 5-amino-6-ribitylamino-2,4 (1H,3H)-pyrimidinedione-5’-phosphate; 6: 5-amino-6-ribitylamino-2,4 (1H,3H)-pyrimidinedione; 7: ribulose-5’-phosphate; 8: 3,4-dihydroxy-2-butanone 4-phosphate; 9: 6,7-dimethyl-8-ribityllumazine; 10: riboflavin. I: Rib1p, GTP cyclohydrolase II; II: Rib7p, DARPP reductase; III: Rib2p, DArPP deaminase; IV: Rib3p, DHBP synthase; V: Rib4p, DRL synthase; VI: Rib5p, riboflavin synthase;
2.1.2 2. Isolation of RIB Genes
The yeast Saccharomyces cerevisiae played an important role in the genetic analysis of riboflavin biosynthesis. Mating between riboflavin-auxotrophic mutants obtained by random mutagenesis and the screening of haploid strains revealed six complementation groups (Oltmanns and Bacher 1972). The S. cerevisiae genes RIB1 to RIB5 and RIB7 were then cloned by functional complementation of the respective auxotrophic mutants (Revuelta et al. 1994). The RIB genes of A. gossypii could not be identified based on the S. cerevisiae RIB sequence information because neither the genome of S. cerevisiae nor that of A. gossypii was available at that time. Functional expression of A. gossypii cDNA or genomic DNA libraries in the S. cerevisiae riboflavin auxotrophs allowed cloning of the A. gossypii RIB genes (Revuelta et al. 1995).
2.1.3 3. Regulation of Riboflavin Overproduction
Overproduction of riboflavin by A. gossypii cultivated on glucose as carbon and energy source was found to be linked to the formation of ascospores (Fig. 11.2). Supplementation of the medium with cAMP or non-metabolizable derivatives of cAMP arrested the fungus in the state of a vegetative mycelium without riboflavin overproduction. This phenomenon is an argument for a cAMP-dependent signaling cascade controlling riboflavin overproduction (Schlösser et al. 2001). An independent approach to study regulation was performed by chemostatic cultivation. A. gossypii did not overproduce riboflavin at constant dilution rates. Dilution rate down-shifts causing nutritional stress resulted in sporulation of the culture and riboflavin overproduction. Increased RIB3, RIB4, and RIB5, but not RIB2 and RIB7, mRNA levels were detected after the nutritional down-shift (Schlösser et al. 2007). In batch culture, the DHBP synthase activity increased 50-fold when growth rate declined due to an increased transcription initiation, as revealed by promoter–reporter fusion experiments (Schlösser et al. 2001). Whereas the molecular mechanism of rib gene regulation in A. gossypii has still to be elucidated, glucose repression could be excluded (Schlösser et al. 2001).
Since the initiation of RIB gene expression in A. gossypii correlates with sporulation a linked regulatory mechanism for asci development and the induction of riboflavin overproduction can be suggested. A. gossypii, originally described as severe but today a negligible plant pathogen, was isolated from plant tissues like cotton balls (Gossypium hirsutum). They were reported to become yellowish (Batra 1973) probably due to pigmentation with riboflavin when infected by the fungus. Riboflavin can protect the hyaline A. gossypii spores against UV-induced damage (Stahmann et al. 2001). A. gossypii is unable to penetrate the epidermis of plant tissue and therefore depends on insects with piercing–sucking mouthparts like Dysdercus nigrofasciatus. They take up and distribute the spores while they feed on the plants. A riboflavin pigmentation of A. gossypii-infected tissue, probably also taking place in citrus fruits (Dammer and Ravello 1990), might preferentially attract the insects to infected plants.
Time course of growth and sporulation in a culture of the A. gossypii wild-type strain. Riboflavin production starts when the growth rate declines (A). A medium was used where other nutrients but not glucose limited growth. Some cells differentiate into asci (B). They liberate spindle-shaped ascospores (C)
2.1.4 4. Classical and Molecular Tools for Strain Improvement
The wild type of A. gossypii isolated by Ashby and Novel (1926) is a natural overproducer of riboflavin, accumulating 100 mg g–1 biomass under growth-limiting cultivation conditions (Schlosser et al. 2001). The selection of improved production strains is possible by visual inspection of colonies obtained by the cultivation of a randomly mutagenized A. gossypii population on agar plates. Submerged production of haploid monokaryotic spores, which can be easily harvested by extraction with lipophilic solvents, allows the generation of classical mutants at a high throughput.
Nowadays, a broad spectrum of molecular tools is available including plasmid transformation, replacement mutagenesis with dominant marker genes, restriction enzyme-mediated chromosomal integration, and transposon mutagenesis (Wright and Philippsen 1991). Replacement mutagenesis with a high site-specificity is possible with less than 50 bp homology regions flanking the DNA fragment, which has to be integrated into the genome (Wendland et al. 2000). Pop-out of dominant markers, e.g. the kan gene which provides geneticin resistance, is possible via short repetitive DNA sequences. This option allows a repeated use of the same marker for consecutive chromosomal DNA manipulations.
Today the A. gossypii genome sequence is publicly available (Gattiker et al. 2007). Previously, functional complementation of Saccharomyces cerevisiae mutants with A. gossypii genomic libraries was the method of choice to isolate genes. This was possible because introns are rare in the A. gossypii genome.
2.1.5 5. Engineering of Pathways
2.1.5.1 a) Riboflavin Pathway
Additional copies of RIB3, RIB4, and RIB5 were integrated into the genome of A. gossypii (Althöfer et al. 1999) by use of a geneticin resistance cassette controlled by the A. gossypii TEF promoter (Steiner and Philippsen 1994). The original RIB promoters caused more than a doubling in productivity when the itaconate-resistant mutant Ita-GS-01 (Schmidt et al. 1996b) was used as the parental strain (Althöfer et al. 1999). Integration of additional copies of RIB1, RIB2, RIB 4, and RIB7 into the genome of A. gossypii LU21 also resulted in a significant increase in riboflavin production (Althöfer et al. 2003). Flanking of the geneticin resistance gene by two TEF promoter elements facilitated a pop-out of the resistance marker. The marker-free strains fulfill the legal requirements for self-cloning strains according to EC Council Directive 98/81. The above-mentioned two examples are applications of restriction enzyme-mediated integration. Although successful, the method has the disadvantage of risking a disruption of the loci involved in product formation (Casas-Flores et al. 2004). Site-specific insertion of linear DNA by double cross-over into the A. gossypii genome is possible in A. gossypii (Steiner et al. 1995). The low frequency of homologous recombination needs larger screening efforts but provides strains with a defined molecular structure.
2.1.5.2 b) Riboflavin Precursor Supply
2.1.5.2.1 Purine and Glycine Metabolism
An important step in the purine biosythesis pathway, phospho-ribosylamine synthesis catalyzed by phosphoribosyl-pyrophospate amidotransferase, is negatively regulated at the transcriptional level by adenine in A. gossypii. For a deregulated expression of this amidotransferase in A. gossypii, the encoding AgADE4 gene was functionally linked to the strong constitutive promoter of the glyceraldehyde 3-phosphate dehydrogenase gene. Furthermore a feed-back resistant amidotransferase mutein designed by three amino acid exchanges was constructed by site-directed mutagenesis. Overexpression of the mutein gene in the wild type of A. gossypii led to an increased riboflavin production in shaking flask cultivation (Jimenez et al. 2005).
Glycine supplementation is known to stimulate riboflavin overproduction by A. gossypii (Demain 1972), presumably by overcoming a metabolic limitation in de novo biosynthesis of the riboflavin precursor GTP (Fig. 11.3).
This idea is supported by the effect of the glycine analog aminomethyl phosphonic acid which did not increase, but competitively reduced riboflavin production (Monschau et al. 1999), suggesting that glycine did not induce riboflavin production via a regulatory function, but alleviated as a substrate a possible glycine shortage. Overexpression of GLY1 encoding a threonine aldolase increased riboflavin production only if the medium was supplemented with l-threonine. Since A. gossypii prefers uptake of l-threonine and excretion of glycine, the boosting effect exceeds simple supplementation of the medium with the precursor of purine biosynthesis (Fig. 11.4; Monschau et al. 1998).
Glycine pathways in Ashbya gossypii. Glycine, a flux limiting precursor of riboflavin, can be produced by at least three pathways. For the threonine aldolase pathway, enzyme activity and the corresponding gene (GLY1) were investigated. Evidence for the existance of a serine hydroxymethyltransferase and a glyoxylate aminotransferase using glutamate as amino donor was given by enzyme activity assays with crude extracts. Increased riboflavin production of a mutant disrupted in SHM2, encoding the cytosolic serine hydroxymethyltransferase, was explained by a reduced flux from glycine to l-serine
Elevated glycine supply also explains the enhanced riboflavin overproduction of SHM2-disrupted A. gossypii mutants. SHM2 encodes the cytosolic serine hydroxymethyltransferase catalyzing the conversion of glycine into serine by consumption of methylene tetrahydrofolate (Schlüpen et al. 2003). In vivo 13C-labeling experiments confirmed that this cytosolic serine hydroxymethyltransferase was a C-1 compound producer during growth but a glycine-consuming enzyme during the riboflavin production phase (Fig. 11.4).
2.1.5.2.2 Isocitrate Lyase and Isocitrate Dehydrogenase
Since technical production uses plant triglycerids as carbon and energy source, the glyoxylate pathway came under consideration as a flux-limiting bottle-neck. Indeed, A. gossypii mutants resistant to inhibitors of isocitrate lyase, e.g. itaconate, showed increased riboflavin production (Schmidt et al. 1996a, b). Isocitrate lyase and malate synthase constitute the anaplerotic glyoxylate shunt providing malate from acetyl-CoA, which is derived e.g. from β-oxidation of fatty acids.
The genes of both enzymes are induced upon glucose depletion (Maeting et al. 1999) and assimilation of the intracellular reserve lipids (Fig. 11.5; Stahmann et al. 1994) or upon growth on plant lipids. The mutation leading to itaconate resistance increased isocitrate lyase-specific activity (Schmidt et al. 1996b), probably favoring anaplerosis but also 3,4-dihydroxy-2-butanone 4-phosphate synthase specific activity, was significantly increased (Schlösser et al. 2001). The itaconate resistance might be the result of a pleiotropic mutation, affecting up-regulation of both the glyoxylate shunt and the riboflavin pathway. A first transcription factor identified to have an impact on riboflavin production was AgBAS1. A truncation of amino acids 632–743 led to an enhanced productivity but avoided an auxotrophy for adenine, which was seen when the gene was replaced with a dominant marker (Mateos et al. 2006).
Isocitrate lyase and malate synthase are transported into the peroxisomes of A. gossypii (Maeting et al. 1999) an organelle in which β-oxidation takes place (Maeting et al. 2000). Interestingly, a knock-out of the gene encoding a peroxisomal isocitrate dehydrogenase led to a reduced production of riboflavin. Overexpression of the gene using the constitutive TEF promoter led to an increase in riboflavin production. A higher activity of the peroxisomal NADP-dependent isocitrate dehydrogenase may lift the intracellular levels of NADPH, which in turn might be limiting in A. gossypii cells assimilating unsaturated fatty acids liberated from plant oils (Althöfer et al. 2001).
2.1.6 6. Riboflavin Excretion and Vacuolar Accumulation
Kinetic studies of riboflavin influx and efflux with riboflavin auxotrophic or overproducing A. gossypii mutants indicated the presence of active riboflavin transport systems (Förster et al. 2001). Inhibition of riboflavin uptake by extracellular FMN or FAD led to an increase in the apparent riboflavin efflux during the early production phase, indicating the presence of a separate excretion carrier.
A. gossypii transports the yellow pigment not only into the cultivation medium. A significant amount is transported into the vacuole and is accumulated there so that crystals can be formed. Disruption of VMA1 encoding a subunit of the vacuolar ATPase did not interfere with the viability of A. gossypii, but no vacuolar riboflavin was detectable and instead all was excreted into the culture medium (Förster et al. 1999).
2.1.7 7. Production and Downstream Processing
The industrial riboflavin production process utilizes soybean oil and soybean meal as carbon and energy source. Since these are present in the fermentation broth as osmotically inactive emulsified lipid droplets or solid particles a high nutrient load is possible. A. gossypii secretes a 35-kDa lipase (Stahmann et al. 1997) hydrolyzing the triglycerides in the culture medium into free fatty acids, which are then ingested by the fungus. In stirred or shaken systems the lipase is inactivated within minutes due to aggregation at the gas/water and lipid/water interfaces. Furthermore, elevated concentrations of free fatty acids interfere with lipase production (Stahmann et al. 1997).
Riboflavin-overproducing cells accumulate significant amounts of the product as intracellular crystals. A heating step after completion of the fermentation run induces self-lysis and liberates riboflavin from the biomass (Kurth 1992). Heating of the fermentation broth and slow cooling over several hours promote the formation of riboflavin crystals in the broth, facilitating separation of the crystals from the supernatant by decantation (Faust et al. 1991). A partial purification is carried out by resuspension of the riboflavin-containing precipitate in diluted acid, heating, and decantation (Grimmer et al. 1992). Since A. gossypii production strains with increased copy numbers of homologous genes are classified as self-clones, remains of the production organism in the feed application product are tolerated.
2.2 B. Candida famata
Ten years ago Candida famata, considered synonymous to Candida flareri by the ATCC, was one of two fungi applied for riboflavin production (Stahmann et al. 2000).
Production strains, improved by COORS (USA) and afterwards used by ADM (USA), were described to produce more than 20 g l–1 riboflavin (Heefner 1988). C. famata was one of the organisms used for early studies of riboflavin biosynthesis (Audley and Goodwin 1962). A special feature is that it can grow on n-alkanes, allowing the composition of a defined medium with biotin for riboflavin production (Olczyk 1978).
Like A. gossypii this fungus is also a natural riboflavin overproducer. It does not grow with hyphae causing high viscosity but grows as a yeast, and therefore it has advantages in stirred vessel cultivation concerning gas exchange. A. gossypii and C. famata have in common that their overproduction of riboflavin, as found in wild-type strains, can be enhanced by supplementation of the medium with glycine (Heefner 1988).
Probably the regulation of the RIB genes is negatively affected by iron ions in C. famata. Improvement of production was observed with mutants resistant to increased concentrations of iron (Heefner et al. 1988). But keeping the iron concentration in the culture medium below 15 μM to prevent its negative effect needs an extra effort. Unfortunately, the iron effect has not been studied on a physiological or molecular level yet.
Mutants of C. famata resistant to 2-deoxyglucose (Heefner et al. 1993) are probably mutated in a transcription factor that has an impact on riboflavin biosynthesis genes. The very first step in metabolism, i.e. uptake of the carbon source, as well as glucose repression mechanisms are the targets of this antimetabolite.
Antimetabolites were reported to be useful in the selection of mutants with improved riboflavin production. Tubercidin, an inhibitor of purine biosynthesis, is an example found in patent literature (Heefner et al. 1992). A transformation system was established for C. famata using a leu2 mutant as recipient and the LEU2 gene of Saccharomyces cerevisiae as marker (Voronovsky et al. 2002). The C. famata RIB genes were identified by functional complementation of C. famata RIB mutants. The genes were also found to be functional in Pichia guilliermondii auxotrophs (Dmytruk et al. 2004). An interesting product for the future might be FMN, which was found to be overproduced when the flavin kinase gene was overexpressed (Yatsyshyn et al. 2009).
3 III. Polyunsaturated Fatty Acids
Polyunsaturated fatty acids (PUFAs) are vitamin-like compounds with structural function in cell membranes and lipid tissue. They serve as precursors for the biosynthesis of hormones like leucotriens, prostaglandins, and thromboxanes. PUFAs are aliphatic monocarboxylic acids with two to six double bonds in a biologically active cis-configuration within their C18- to C24-backbone. They are categorized as ω-3 or ω-6, depending on the position of the first double bond, counting from the terminal carbon atom of the fatty acid. Their biosynthesis starts from oleic acid (18:1) and is continued by a sequence of desaturase and elongase reactions (Fig. 11.6; Marszalek and Lodish 2005). Since humans cannot introduce double bonds at the ω-3 or ω-6 position, linoleic acid (18:2, ω-6) and α-linolenic acid (18:3, ω-3) are essential. They belong to the vitamin F group.
Arachidonic acid (20:4, ω-6; ARA) and docosahexaenoic acid (22:6, ω-3; DHA) are not essential but of commercial interest. They are needed for the developing brain, neuronal tissue, and retina of the human fetus and are therefore contained in human breast milk. Studies on supplemented infant formula suggested a correlation between PUFA content in the cerebral cortex of breast-fed infants and better cognitive and visual functions of infants fed with infant formula supplemented with long-chain PUFAs (Heird and Lapillonne 2005).
Eukaryotic biosynthesis of fatty acids starts with fatty acid synthetase (FAS) in the cytosol (Ratledge 2004). Parallel elongation and desaturation steps take place in the membranes of the endoplasmatic reticulum
For DHA and ARA, commercial-scale processes based on natural isolates or classically derived microbial strains are available. Attempts to employ molecular engineering techniques in PUFA production were successful. When Saccharomyces cerevisiae, not a natural PUFA producer, was provided with the appropriate desaturase and elongase genes one or several biosynthetic steps towards eicosapentanoic acid (20:5, ω-3; EPA) or DHA starting from the corresponding precursor fatty acids were possible (Parker-Barnes et al. 2000). A series of patent applications from DuPont published in 2004 and 2005 disclosed EPA production in the genetically modified oleogenic yeast Yarrowia lipolytica.
DHA is found in phospholipids of different cell types, e.g. in the photosensitive part of the retina DHA, accounts for more than 60% of the total fatty acid content. Functioning as signal molecule precursor, beneficial effects of DHA on the prevention of cardiovascular diseases, probably by down-regulation of the intracellular mechanism leading to the expression of pro-atherogenic genes, are well documented. Fish is the most important source of DHA in human nutrition. It cannot synthesize DHA by itself but marine microorganisms at the lower end of the food chain are the genuine DHA producers. In view of the over-exploitation of marine fish resulting in ever-reducing catch size, fish oil is not considered a sustainable source for DHA so that microbial alternatives are needed.
Crypthecodinium cohnii, a marine microalga, consists of triglycerides with 30% DHA. In flask cultivations with ethanol as the carbon source 83 g l–1 biomass accumulated with a lipid triglyceride content between 30% and 50% (Swaaf et al. 2003). The genera Schizochytrium, Thraustochytrium, and Ulkenia, marine heterotroph protists that are classified as Labyrinthulida, are preferred sources of DHA. In laboratory-scale fermentations with a Schizochytrium sp. strain high cell densities of 63 g l–1 dry biomass were obtained with a fatty acid content of 70%, of which 37% were DHA (Ganuza et al. 2008). DHA-containing products acquired from protist fermentations are offered by Martek Biosciences and Advanced BioNutrition (both USA) focusing on the human nutrition and aquaculture market, respectively.
ARA, also a natural component of breast milk, supports neonatal eye and brain development (Alessandri et al. 2004). Breast-fed infants have higher ARA blood levels than formula-fed infants. Infant formulas fortified with ARA and DHA have been sold in Europe for more than 10 years. After permission by the FDA, they were introduced in the United States in 2002.
In contrast to DHA, which can be obtained from fish oil or microbial processes, industrial ARA production solely relies on fermentation. First publications about the mycelial fungus Mortierella alpina as ARA producer appeared in the late 1980s.
3.1 A. Mortierella alpina
Mortierella alpina, a filamentous zygomycete, produces lipids rich in ARA when grown at 18 °C (Lindberg and Molin 1993). Its presense in the spores and prefered degradation during their germination suggests a role as a store for carbon and energy (Lounds et al. 2007). The pattern of the fatty acids produced could be influenced by the addition of a specific delta 5-fatty acid desaturase inhibitor, i.e. dioxabicyclo [3.3.0] octane derivatives like sesamin, or a selection of mutants with absent or reduced delta 5-fatty acid desaturase activity (Kawashima et al. 1992).
A number of M. alipina genes encoding enzymes, which are involved in the synthesis of polyunsaturated fatty acids, were cloned, e.g. delta 5-fatty acid (Knutzon et al. 1998) delta 6-fatty acid (Sakuradani et al. 1999a), delta 9-fatty acid (Sakuradani et al. 1999b), and delta 12-fatty acid (Huang et al. 1999) desaturases. Convincingly, verification of the genes functionality by expression, e.g. in Aspergillus oryzae, resulted in a shift to more than 70% of linoleic acid (18:2, ω-6) in the cell’s fatty acid composition (Sakuradani et al. 1999c). Two transformation systems were established, one working with uracil auxotrophs (Takeno et al. 2004) and a second using zeocin as dominant marker (Takeno et al. 2005). By use of the latter, ARA production was increased after transformation with a gene encoding a rate-limiting elongase (Takeno et al. 2005). More details concerning breeding of M. alpina is presented by a recent mini-review (Sakuradani et al. 2009).
The accumulation of lipid starts in M. alpina cells when growth becomes limited, e.g. after consumption of the nitrogen source, and depends on the high activity of malic enzyme needed for NADPH supply in fatty acid biosynthesis (Zhang and Ratledge 2008). The highest titers with glucose as carbon source reported so far are 20 g l–1 of PUFAs, with up to 70% ARA produced by M. alpina mutants (Sakuradani et al. 2009).
Since the production of biodiesel today makes glycerol available at low costs, strains were screened to use this carbon source for ARA production (Hou 2008). The morphology of the fungal mycelium in the reactor is of specific concern as only small pellets with a diameter of 1–2 mm allow sufficient mass transfer and keep the broth viscosity at an acceptable level. After fermentation the mycelium is harvested by filtration, dried, and solvent-extracted. Alternatively, the mycelium is homogenized as an aqueous suspension and extracted with a water-immiscible organic solvent. Safety of M. alpina concerning its products in food was investigated more than a decade ago (Streekstra 1997) and recently ARA-containing biomass in rats (Nisha et al. 2009).
Fungal ARA is produced and marketed as ARASCO by Martek Biosciences (USA). In 2004 Cargill (USA) entered into a joint venture with Wuhan Alking Bioengineering Co. Ltd (China), an ARA producer serving the Chinese infant formula business. Suntory, a leading Japanese beverage manufacturer and a pioneer in microbial PUFA production, provides a fungal ARA oil under the trade name SUNTGA.
4 IV. Conclusions
Mineral oil-based chemical riboflavin production, successfully used for half a century, has been completely replaced by two microbial processes for almost a decade. These save half of the costs, reduce waste and energy requirements, and use renewable resources like sugars or plant oils. A major point is that more than 100 biochemical conversions take place in a single cell and these work in a single vessel. A complex multi-step plant was replaced by a single-step process. The key to this remarkable success were decades of strain development and improvement. A combination of selection after random mutagenesis and genetic engineering after rational design resulted in strains not only improved by “removal of a bottle-neck” but by generation of a “network of anabolic highways” and a deletion of unwished reaction. A sensible use of “high-tech” like NMR or genome sequencing has allowed phenomena to be explained, but simple techniques are sometimes more powerful, like a comparison of the yellow color of different strains on agar plates allowing the prediction of which one will be better in the 100 m3 vessel.
The coexistance of a fast bacterial process using Bacillus subtilis and a much slower fungal process applying A. gossypii can mean that both are much more efficient than chemistry and not under pressure now because the market is growing and the selling price is high. This is probably true for both riboflavin and ARA, because they are needed in small dosages. The design of both A. gossipii and M. alpina strains is not at an end. Each step has improving potential and some steps are not known at all.
An open point is the transport of riboflavin and its precursors across membranes. Neither the transporter accumulating the pigment in the vacuole nor the carrier excreting it against a concentration gradient are known. Also little is known about the facilitation of shuttle processes in the cell, e.g. between peroxisomes and mitochondria.
A problem in studying fatty acid biosynthesis is the lack of in vitro assays for desaturases and elongases which are probably working on membranes. Overexpression and in vivo data do help indirectly but determination of specific activities would shift the research on the most relevant level for their direct comparison.
References
Alessandri JM, Guesnet P, Vancassel S, Astorg P, Denis I, Langelier B, Aïd S, Poumès-Ballihaut C, Champeil-Potokar G, Lavialle M (2004) Polyunsaturated fatty acids in the central nervous system: evolution of concepts and nutritional implications throughout life. Reprod Nutr Dev 44:509–538
Althöfer H, Seulberger H, et al (1999) Genetic method for producing riboflavin. Patent WO 9961623
Althöfer H, Zelder O, et al (2001) Monocellular or multicellular organisms for production of riboflavin. Patent WO 0111052
Aranceta J, Serra-Majem L, Perez-Rodrigo C, Llopis J, Mataix J, Ribas L, Tojo R, Tur JA (2001) Vitamins in Spanish food patterns: the eVe study. Public Health Nutr 4:1317–1323
Ashby SF, Nowell N (1926) The fungi of stigmatomycosis. Ann Bot 40:69–83
Audley A, Goodwin N (1962) Studies on the biosynthesis of riboflavin. 7. The incorporation of adenine and guanine into riboflavin and into nucleic acid purines in Eremothecium ashbyii and Candida flareri. Biochem J 84:587–592
Bacher A (1991) Biosynthesis of flavins. In: Müller F (ed) Chemistry and biochemistry of flavoenzymes, vol 1. CRC, Boca Raton, pp 215–259
Batra LR (1973) Nematosporaceae (Hemiascomycetidae): taxonomy, pathogenicity, distribution and vector relations. USDA Technol Bull 1469:1–71
Burrows RB, Brown GM (1978) Presence in Escherichia coli of a deaminase and a reductase involved in biosynthesis of riboflavin. J Bacteriol 136:657
Casas-Flores S, Rosales-Saavedra T, et al (2004) Three decades of fungal transformation: novel technologies. Methods Mol Biol 267:315–325
Cashmore AR, Jarillo JA, Wu Y, Liu D (1999) Cryptochromes: blue light receptors for plants and animals. Science 284:760–765
Dammer KH, Ravelo HG (1990) Verseuchung von Leptoglossus gonagra (Fabr.) mit Nematospora coryli Peglion und Ashbya gossypii (Ashby et Novell) Guilliermond in einer Zitrusanlage der Republik Kuba. Arch Phytopathol Pflanzensch 26:71–78
de Swaaf ME, Pronk JT, Sijtsma L (2003) Fed-batch cultivation of the docosahexaenoic-acid-producing marine alga Crypthecodinium cohnii on ethanol. Appl Microbiol Biotechnol 61:40–43
Demain AL (1972) Riboflavin oversynthesis. Annu Rev Microbiol 26:369–388
Eggersdorfer M, Adam G (1996) Vitamins, chapter 1, A27. In: Ullmann (ed) Ullmann’s encyclopedia of industrial chemistry. VCH, Weinheim, pp 521–530
Faust T, Meyer J, et al (1991) Process for the separation of riboflavin from a fermentation suspension. Patent EP 0438767
Fischer M, Bacher A (2005) Biosynthesis of flavocoenzymes. Nat Prod Rep 22:324–350
Fletcher RJ, Bell IP, Lambert JP (2004) Public health aspects of food fortification: a question of balance. Proc Nutr Soc 63:605–614
Förster C, Santos MA, Ruffert S, Krämer R, Revuelta JL (1999) Physiological consequence of the disruption of the VMA1 gene in the riboflavin overproducer Ashbya gossypii. J Biol Chem 274:9442–9448
Förster C, Revuelta JL, et al (2001) Carrier-mediated transport of riboflavin in Ashbya gossypii. Appl Microbiol Biotechnol 55:85–89
Ganuza E, Anderson AJ, Ratledge C (2008) High-cell-density cultivation of Schizochytrium sp. in an ammonium/pH-auxostat fed-batch system. Biotechnol Lett 30:1559–1564
Gattiker A, Rischatsch R, et al (2007) Ashbya genome database 3.0: a cross-species genome and transcriptome browser for yeast biologists. BMC Genomics 8:9
Grimmer J, Kiefer H, et al (1992) Process for the purification of riboflavin obtained by fermentation. Patent EP 0464582
Heefner DL, Yarus MJ, Boyts A, Burdzinski LA (1987) Riboflavin production with yeast. Patent EP 0231605A2
Heefner D, Weaver, CA, Yarus MJ, Burdzinski LA, Gyure DC, Foster EW (1988) Riboflavin producing strains of microorganisms, method for selecting, and method for fermentation. Patent WO 88/09822
Heefner DL, Weaver CA, Yarus MJ, Burdzinski LA (1992) Method for producing riboflavin with Candida famata. Patent US 005164303A
Heefner D, Boyts A, Burdzinski L et al (1993) Efficient riboflavin production with yeast. Patent US 005231007 A
Heird and Lapillonne (2005) The role of essential fatty acids in development. Annu Rev Nutr 25:23.1–23.23
Hohmann HP, Stahmann KP (2010) Biotechnology of riboflavin production. In: Lew M, Hung WL (eds) Comprehensive natural products chemistry II. Elsevier, Oxford, pp 115–139
Hou CT (2008) Production of arachidonic acid and dihomo-gamma-linolenic acid from glycerol by oil-producing filamentous fungi, Mortierella in the ARS culture collection. J Ind Microbiol Biotechnol 35:501–506
Huang YS, Chaudhary S, Thurmond JM, Bobik EG Jr, Yuan L, Chan GM, Kirchner SJ, Mukerji P, Knutzon DS (1999) Cloning of delta12- and delta6-desaturases from Mortierella alpina and recombinant production of gamma-linolenic acid in Saccharomyces cerevisiae. Lipids 34:649–659
Jimenez A, Santos MA, et al (2005) Metabolic engineering of the purine pathway for riboflavin production in Ashbya gossypii. Appl Environ Microbiol 71:5743–5751
Kawashima H, Akimoto K, Yamada H, et al (1992) Process for production of 8,11-eicosadienoic acid. Patent EPA 0535941 A1
Knutzon DS, Thurmond JM, Huang YS, Chaudhary S, Bobik EG Jr, Chan GM, Kirchner SJ, Mukerji P (1998) Identification of delta5-desaturase from Mortierella alpina by heterologous expression in bakers’ yeast and canola. J Biol Chem 273:29360–29366
Kurth R (1992) Process for the enhancement of riboflavin levels in spray-dried riboflavin fermentation extracts. Patent EP 0487985
Lago BD, Kaplan L (1981) Vitamin fermentations: B2 and B12. Adv Biotechnol 3:241–246
Lindberg AM, Molin G (1993) Effect of temperature and glucose supply on the production of polyunsaturated fatty acids by the fungus Mortierella alpina in fermenter cultures. Appl Microbiol Biotechnol 39:450–455
Lounds C, Eagles J, Carter AT, MacKenzie DA, Archer DB (2007) Spore germination in Mortierella alpina is associated with a transient depletion of arachidonic acid and induction of fatty acid desaturase gene expression. Arch Microbiol 188:299–305
Maeting I, Schmidt G, Sahm H, Revuelta JL, Stierhof YD, Stahmann KP (1999) Isocitrate lyase of Ashbya gossypii - transcriptional regulation and peroxisomal localization. FEBS Lett 444:15–21
Maeting I, Schmidt G, Sahm H, Stahmann KP (2000) Role of a peroxisomal NADP-specific isocitrate dehydrogenase in the metabolism of the riboflavin overproducer Ashbya gossypii. J Mol Catal 10:335–343
Marszalek JR, Lodish HF (2005) Docosahexaenoic acid, fatty acid-interacting proteins, and neuronal function: breastmilk and fish are good for you. Annu Rev Cell Dev Biol 21:633–657
Mateos L, Jiménez A, Revuelta JL, Santos MA (2006) Purine biosynthesis, riboflavin production, and trophic-phase span are controlled by a Myb-related transcription factor in the fungus Ashbya gossypii. Appl Environ Microbiol 72:5052–5060
Monschau N, Sahm H, Stahmann KP (1998) Threonine aldolase overexpression plus threonine supplementation enhanced riboflavin production in Ashbya gossypii. Appl Environ Microbiol 64:4283–4290
Monschau N, Stahmann KP, Sahm H, Zelder O (1999) Uni- or multicellular microorganisms for the production of riboflavin. Patent EP 0930367 A2
Nisha A, Muthukumar SP, Venkateswaran G (2009) Safety evaluation of arachidonic acid rich Mortierella alpina biomass in albino rats – a subchronic study. Regul Toxicol Pharmacol 53:186–194
Olczyk C (1978) A n-Alkanes as a substratum for riboflavin production. I. Investigations of the dynamics of the flavinogenesis in chosen yeasts of the genus Candida. Pol J Pharmacol Pharm 30:83–88
Oltmanns O, Bacher A (1972) Biosynthesis of riboflavin in Saccharomyces cerevisiae: the role of genes rib1 and rib7. J Bacteriol 110:818–822
Parker-Barnes JM, Das T, Bobik E, Leonard AE, Thurmond JM, Chaung LT, Huang YS, Mukerji P (2000) Identification and characterization of an enzyme involved in the elongation of n-6 and n-3 polyunsaturated fatty acids. Proc Natl Acad Sci USA 97:8284–8289
Perlman D (1979) Microbial process for riboflavin production. In: Peppler H, Perlman D (eds) Microbial technology, microbial processes, vol 1, 2nd edn. Academic, New York, pp 521–527
Plaut GWE (1954) Biosynthesis of riboflavin. II. Incorporation of C14-labelled compounds into ring A. J Biol Chem 211:111–116
Ratledge C (2004) Fatty acid biosynthesis in microorganisms being used for single cell oil production. Biochimie 86:807–815
Revuelta DJL, Sanios Garcia MA, Garcia Ramirez JJ, Gonzales-Hernandez GA, Buitrago Sema MJ (1994) Saccharomyces cerevisiae recombinant riboflavin production. Patent WPI 94-177113
Revuelta DJL, Buitrago SMJ, Santos GMA (1995) Riboflavin synthesis in fungi. Patent WO 9526406-A
Sakuradani E, Kobayashi M, Shimizu S (1999a) Delta6-fatty acid desaturase from an arachidonic acid-producing Mortierella fungus. Gene cloning and its heterologous expression in a fungus, Aspergillus. Gene 238:445–453
Sakuradani E, Kobayashi M, Shimizu S (1999b) Delta9-fatty acid desaturase from arachidonic acid-producing fungus. Unique gene sequence and its heterologous expression in a fungus, Aspergillus. Eur J Biochem 260:208–216
Sakuradani E, Kobayashi M, Ashikari T, Shimizu S (1999c) Identification of Delta12-fatty acid desaturase from arachidonic acid-producing mortierella fungus by heterologous expression in the yeast Saccharomyces cerevisiae and the fungus Aspergillus oryzae. Eur J Biochem 261:812–820
Sakuradani E, Ando A, Ogawa J, Shimizu S (2009) Improved production of various polyunsaturated fatty acids through filamentous fungus Mortierella alpina breeding. Appl Microbiol Biotechnol 84:1–10
Schlösser T, Schmidt G, et al (2001) Transcriptional regulation of 3,4-dihydroxy-2-butanone 4-phosphate synthase. Microbiology 147:3377–3386
Schlösser T, Wiesenburg A, et al (2007) Growth stress triggers riboflavin overproduction in Ashbya gossypii. Appl Microbiol Biotechnol 76:569–578
Schmidt G, Stahmann KP, Sahm H (1996a) Isolation and characterization of isocitrate lyase from the riboflavin-producing fungus Ashbya gossypii. Microbiology 142:411–417
Schmidt G, Stahmann KP, Kaesler B, Sahm H (1996b) Correlation of isocitrate lyase activity and riboflavin formation in the riboflavin overproducer Ashbya gossypii. Microbiology 142:419–426
Stahmann KP, Kupp C, Feldmann SD, et al (1994) Formation and degradation of lipid bodies found in the riboflavin-producing fungus Ashbya gossypii. Appl Microbiol Biotechnol 42:121–127
Stahmann KP, Böddecker T, Sahm H (1997) Regulation and properties of a fungal lipase showing interfacial inactivation by gas bubbles, or droplets of lipid or fatty acid. Eur J Biochem 244:220–225
Stahmann KP, Revuelta JL, Seulberger H (2000) Three biotechnical processes using Ashbya gossypii, Candida famata, or Bacillus subtilis compete with chemical riboflavin production. Appl Microbiol Biotechnol 53:509–516
Stahmann KP, Arst HN, et al (2001) Riboflavin, overproduced during sporulation of Ashbya gossypii, protects its hyaline spores against ultraviolet light. Environ Microbiol 3:545–550
Steiner S, Philippsen P (1994) Sequence and promoter analysis of the highly expressed TEF gene of the filamentous fungus Ashbya gossypii. Mol Gen Genet 242:263–271
Steiner S, Wendland J, Wright MC, et al (1995) Homologous recombination as the main mechanism for DNA integration and cause of rearrangements in the filamentous ascomycete Ashbya gossypii. Genetics 140:973–987
Streekstra H (1997) On the saftey of Mortierella alpina for the production of food ingredients, such as arachidonic acid. J Biotechnol 56:153–165
Takeno S, Sakuradani E, Murata S, Inohara-Ochiai M, Kawashima H, Ashikari T, Shimizu S (2004) Establishment of an overall transformation system for an oil-producing filamentous fungus, Mortierella alpina 1S-4. Appl Microbiol Biotechnol 65(4):419–425
Takeno S, Sakuradani E, Tomi A, Inohara-Ochiai M, Kawashima H, Shimizu S (2005) Transformation of oil-producing fungus, Mortierella alpina 1S-4, using Zeocin, and application to arachidonic acid production. J Biosci Bioeng 100:617–622
Voronovsky AA, Abbas CA, Fayura LR, Kshanovska BV, Dmytruk KV, Sybirna KA, Sibirny AA (2002) Development of a transformation system for the flavinogenic yeast Candida famata. FEMS Yeast Res 2:381–388
Wendland J, Ayad-Durieux Y, et al (2000) PCR-based gene targeting in the filamentous fungus Ashbya gossypii. Gene 242:381–391
Wickerham LJ, Flickinger MH, Johnsten RM (1946) The production of riboflavin by Ashbya gossypii. Arch Biochem 9:95–98
Wright P, Philippsen P (1991) Replicative transformation of the filamentous fungus Ashbya gossypii with plasmids containing Saccharomyces cerevisiae ARS elements. Gene 109:99–105
Yatsyshyn VY, Ishchuk OP, Voronovsky AY, Fedorovych DV, Sibirny AA (2009) Production of flavin mononucleotide by metabolically engineered yeast Candida famata. Metab Eng 11:163–167
Zhang Y, Ratledge C (2008) Multiple isoforms of malic enzyme in the oleaginous fungus Mortierella alpina. Mycol Res 112:725–730
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2011 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Stahmann, KP. (2011). Production of Vitamin B2 and a Polyunsaturated Fatty Acid by Fungi. In: Hofrichter, M. (eds) Industrial Applications. The Mycota, vol 10. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-11458-8_11
Download citation
DOI: https://doi.org/10.1007/978-3-642-11458-8_11
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-11457-1
Online ISBN: 978-3-642-11458-8
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)