Abstract
During our combined 45 plus years of operating confocal microscopes and managing core microscopy facilities, and through teaching our Basic Confocal Microscopy Workshop at several venues, we have found that students and technicians who are novice users of confocal microscopes are often instructed by their mentors to go to the confocal system and collect some images. Often the implied message is that it should be easy and quick since it is only a microscope. Unfortunately, all too often the advisor of the student or supervisor of the technician does not have a full understanding of the complexity of a confocal microscope. Unless these novice users are in a situation where others have the time and knowledge to properly train them, their initial efforts often amount to an exercise in futility because key parameters are not properly considered. This leads to specimens that are not prepared properly and a lack of understanding of how to operate the confocal microscope in a way that maintains the fidelity of the specimen information. In too many instances, this lack of user training is exacerbated further because there is little or no daily oversight of the setup and maintenance of the microscope. In this combined scenario, neither the experimental preparation nor the microscopes are capable of producing the highest-quality information. As with the first edition, the goal of this book is to provide beginning and intermediate users of confocal microscopes of all types the basic knowledge required to effectively utilize the full capability of confocal imaging to provide high-quality image data. Each chapter has been thoroughly revised and updated to reflect advances in the field. We have also added information on new detectors and emerging techniques such as super-resolution microscopy and specimen clearing, updated the software and operation chapter, and added a chapter on how to do co-localization fluorescence microscopy studies correctly.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
1.1 Introduction to the Second Edition of Basic Confocal Microscopy
Since publication of the first edition of Basic Confocal Microscopy in 2011, a number of advances have occurred influencing several aspects of confocal microscopy technology, including a number of super- and enhanced resolution techniques, specimen preparation methods, lasers, detectors, and operating and image analysis software. For the purpose of discussions throughout this text, we will define enhanced resolution techniques as those that improve resolution from the historical Abbe defined limit of 200 nm down to approximately 140 nm and super-resolution techniques to those that provide resolution well below 100 nm. While techniques and technology have improved, the basics that must be understood to generate high-quality confocal images remain constant. In this second edition, we will address how these recent changes have improved the performance and expanded the research applications of confocal imaging but will also maintain the introductory concept to confocal imaging that made the first edition a success.
Because of advances that make possible imaging deep into thick specimens, we have added information on tissue clearing techniques that complements the deep imaging capability of some modern confocal systems. Tissue clearing, including techniques such as X-CLARITY@, benzyl alcohol/benzyl benzoate (BABB), 3Disco and iDisco (dichloromethane/dibenzylether), and several others, are mechanisms to increase the depth from which useful image information can be extracted. This provides exciting possibilities for improving our understanding of three-dimensional relationships between structures in large regions of tissue. The protocols and advantages and disadvantages of several tissue clearing techniques will be discussed. Also in the area of sample preparation, we will provide information on antigen retrieval protocols and extend the discussion of available fluorescent probes available for confocal microscopy.
There have been rapid and significant advances in the hardware configurations of confocal microscopes in recent years, and now, unlike 7–8 years ago, systems are sold with a complete configuration of diode lasers rather than gas lasers such as argon or helium neon. This range of new diode lasers, along with high-sensitivity detectors such as the gallium arsenide phosphide (GaAsP) detectors, has made it possible to detect very low signals in point scanning instruments. Likewise, significant advances in cooled charged coupled devices (cCCD) and scientific complementary metal oxide semiconductors (sCMOS) now allow faster and more sensitive image capture in microscopes where a full field is captured in a single process. These detector advances have greatly expanded our capability to examine samples with low levels of fluorescence and to attenuate probe intensity to minimize specimen damage in live cell imaging or samples that rapidly photobleach. High-sensitivity detectors have also contributed to the development of new technologies such as the enhanced resolution Zeiss Airyscan and Leica HyVolution instruments that can exceed the 200 nm resolution level obtainable with standard confocal configurations and can approach 140 nm resolution. While not the sub-100 nm resolution level of super-resolution techniques such as STORM (stochastic optical reconstruction microscopy) , PALM (photoactivated localization microscopy ), STED (stimulated emission depletion microscopy ), and SIM (structured illumination microscopy ), these enhanced resolution instruments provide a cost-effective mechanism to beat the diffraction limitations of light present in standard confocal configurations.
As often discussed, confocal microscopes enhance many research projects. However, it is not always the best instrument for some imaging situations. For example, imaging a thin monolayer of cells may be better served with an epifluorescence wide-field microscope than optical sectioning with a confocal microscope. Likewise, while super-resolution instruments have greatly enhanced our understanding of some biological principles, they typically serve a specific research purpose and may have limited depth of imaging capabilities (Wentao et al. 2016), require special fluorochromes (Dempsey et al. 2011), or have other difficult-to-meet requirements for optimal image quality (Ashdown et al. 2014). As a consequence, super-resolution instruments are often great at addressing specific research questions, but they typically are not suitable for other research applications better suited for standard confocal imaging. A discussion of these techniques, their applications, and comparisons to more traditional methods has been added to Chap. 8 where different types of confocal systems are presented.
A number of advances have also taken place in operating software and analysis of two-dimensional and three-dimensional images. Chapter 9 in the first edition described in detail the Zeiss AIM software used to operate the LSM 510 line of instruments. About the time the first edition of Basic Confocal Microscopy was published, Zeiss introduced a new operating system (ZEN) that had many additions to improve the user interface. Operating software for other confocal systems have also advanced and now include similar functions such as online libraries for protocol development. Chapter 9 from the first edition has been updated to include information from these new operating systems (the Leica LAS software will be described in Chap. 9 of this edition) while still covering the basic setup of the microscope for optimal image quality. For analysis of co-localization and correlation of two molecules in two-dimensional (2-D) space, we have also added a new chapter by Dr. Teng-Leong Chew that includes the implementation and interpretation of various co-localization and correlation coefficients.
Even though confocal technology has advanced, the basics of optical imaging and the principles for collection and analysis of publication quality images and data remain essentially the same. It is our hope that by presenting information on these updates in confocal imaging that we will be able to maintain our goal of providing the requisite basic information for confocal imaging in a well-organized manner that will assist novice users in understanding the basics of confocal imaging.
1.2 Why an Introductory Text on Confocal Microscopy?
The premise of the first edition was that during our combined 35 plus years of operating confocal microscopes and managing core microscopy facilities, and through teaching our Basic Confocal Microscopy Workshop at several venues, we found that students and technicians that are novice users of confocal microscopes are often instructed by their mentors to go to the confocal system and collect some images. Often the implied message is that it should be easy and quick since it is only a microscope. Unfortunately, all too often the advisor of the student or supervisor of the technician does not have a full understanding of the complexity of a confocal microscope. Unless these novice users are in a situation where others have the time and knowledge to properly train them, their initial efforts often amount to an exercise in futility because key parameters are not properly considered. This leads to specimens that are not prepared properly and a lack of understanding of how to operate the confocal microscope in a way that maintains the fidelity of the specimen information. In too many instances, this lack of user training is exacerbated further because there is little or no daily oversight of the setup and maintenance of the microscope. In this combined scenario, neither the experimental preparation nor microscopes are capable of producing the highest-quality information. Now with well over 45 years of combined experience in managing core microscopy facilities, we have unfortunately found that the premise of the first edition of Basic Confocal Microscopy (that many faculty, technicians, and students operating confocal microscopes do not adequately understand the technology) is still valid.
Good confocal microscopy is obviously dependent upon proper specimen preparation and the correct setup of various microscope parameters. However, even if an excellent confocal image is collected, there is often a poor understanding of how to properly display the full richness of the information contained in the image and how best to analyze two-dimensional (2-D) and 3-D confocal images. There is an abundance of good image processing and analysis software available to the user. However, these robust programs also provide the capability of inappropriately manipulating the data or inadvertently degrading the image information. A lack of understanding of basic digital imaging and image processing theory frequently results in improper image processing in 2-D programs such as Image J and FIJI (NIH freeware), Photoshop (Adobe Systems, Inc., San Jose, CA), MetaMorph (Molecular Devices, Sunnyvale, CA), or others and in more advanced 3-D volumetric programs such as AMIRA (Thermo Fisher Scientific, Hillsboro, OR), IMARIS (Bitplane, Concord, MA), or VoxBlast (VayTek, Inc., Fairfield, IA).
The goal of this book is to provide beginning and intermediate users of confocal microscopes a resource that can be used to address many of the frequently asked questions concerning confocal imaging and to provide a strong foundation for maximizing the data obtained from experiments involving confocal microscopy. While most of the information is directly relevant to single-photon scanning laser systems, much of the information also applies to spinning disk, multiphoton, and enhanced and super-resolution confocal systems. In several chapters specific comparisons of the technology that differentiates these systems will be made and advantages and disadvantages of each presented. The information presented will also provide the background information necessary when moving forward to complex imaging protocols such as Forster (or fluorescence) resonance energy transfer (FRET), fluorescence recovery after photobleaching (FRAP), fluorescence lifetime imaging (FLIM), and other advanced techniques.
1.3 Historical Perspective
It has long been recognized by microscopists that as the thickness of the specimen increases, light emerging from scattering objects above and below the focal plane of the microscope degrade the quality of the image. This occurs primarily because of reduced image contrast. The loss of contrast is caused by impinging light produced from the out-of-focus planes. Like turning on the lights in a movie theater, this stray light reduces the signal-to-noise ratio (SNR) and obscures important image details. The various factors affecting the axial (Z) resolution (ability to distinguish two small objects as separate and distinct along the axial axis) were explored by Berek in 1927 (Berek 1927). In Berek’s analysis , the three key elements affecting image quality were (1) spreading of the light beam emerging from objects in the specimen, (2) the magnification of the image, and (3) the sensitivity of the detection system. For Berek, the detection system was the observer’s eye. However, in the modern age of microscopy, the eye has been replaced with more sensitive detectors. With regard to Berek’s item 2, microscopists have always worked with the highest magnification required for maintaining image data fidelity. This leaves the spread of out-of-focus light into the image plane as the last of Berek’s parameters that needs to be minimized to obtain good axial resolution . Obviously, if one could limit the projection of out-of-focus light onto the image, then a significant gain in resolution should be achieved. The removal of the obscuring out-of-focus light is precisely what the confocal microscope is designed to do, and the subsequent gain in axial resolution remains the biggest advantage of confocal microscopy. However, as will be described in subsequent chapters, several other advantages accrue from the confocal design, including increases in lateral resolution.
The first confocal microscope is generally credited to Marvin Minsky (Minsky 1988). In his 1957 patent application, Minsky described a microscope in which the typical wide-field illumination arrangement was replaced with one in which a point source is focused to a small spot within the specimen. Light arising from the illuminated spot is focused by the objective lens to a small spot at the image plane. Thus, a point source of light is in conjugate focus (confocal) at the specimen and at the image plane (Fig. 1.1a). Placing a small pinhole aperture made of an opaque material at the image plane permits only the light coming from the focal point of the specimen to pass to the detector. In contrast, light coming from above and below the plane of focus will not be in focus at the image plane and will be rejected by the opaque material surrounding the pinhole. This confocal setup can also be achieved in an epi-illumination setup (Fig. 1.1b). The confocal arrangement dramatically improves contrast by removing the out-of-focus light originating above and below the focal plane. The arrangements diagramed in Fig. 1.1 are not the only possible designs. Since its inception, various other designs have been introduced for creating the required confocality of focus at the specimen and image planes.
Optical train for confocal microscope in conventional (a) and epi-illumination setups (b). The light path of the confocal beam is represented by the gray lines. In the conventional arrangement, light from the photon source is focused onto the entrance pinhole (a). This pinhole provides a bright focused point source. Light from this point source is collected by the condenser lens and focused to a spot (b) within the sample. The light emerging from the focused spot within the specimen is collected by the objective lens and focused at a second (exit) pinhole (c). Points a, b, and c are in conjugate focus (confocal). The path of light emerging outside of the focal point B is represented by the dotted black lines and arrives at the exit pinhole out of focus. Thus, most of this light is rejected and not transmitted to the detector
In an epi-illumination setup (b), the objective lens acts as both the condenser and objective lens. Light returning from the specimen is diverted by the dichroic (dichromatic beam splitter), and this diverted light (dark gray lines) is focused on the exit pinhole (dark gray lines). As with the conventional arrangement, light from above or below the focal point in the specimen arrives at the pinhole out of focus (not depicted) and so is rejected. Conventional wide-field fluorescence systems lack the pinhole so all out-of-focus light becomes a component of the final image as shown in Fig. 1.3
Of course, a single point within a specimen does not provide much information about the specimen. In order to acquire full details across the lateral (X-Y) focal plane of the specimen, the spot must be scanned across the image and the image information collected sequentially. In Minsky ’s original design, the scanning was produced by translating the specimen laterally. This method was slow and prone to vibration, both of which presented problems for biological work. A notable advance for the use of point scanning instruments in biology was made in the 1980s with the development of the ability to raster the illumination across the specimen rather than translating the stage. This allowed for faster scan rates without the introduction of vibration. The publication of images of biological samples using the beam-scanning instrument (White et al. 1987) spurred an extreme interest in confocal microscopy for biological research.
Arguably, the development of beam scanning along with concurrent advancements in laser technology, fluorescent labels, lens design, and computer processing really set the stage for the rapid deployment of laser scanning confocal microscopy as a key tool for cell biological research. However, laser scanning instruments are not the only mechanism for implementing confocal microscopy. A parallel development occurred based on Paul Nipkow ’s invention of a method for converting an optical image into an electrical signal that could be transmitted over a cable (Nipkow 1884). Nipkow’s technique converted the 2-D image information into a 1-D serial signal by scanning the image using a spinning wheel with precisely placed rectangular holes. The holes were arranged in a spiral pattern around the wheel such that when the wheel was spun, the small areas being sampled changed. The moving holes filled in the gaps between the initially sampled regions. In 1967 Eggar and Petráň (Eggar and Petráň 1967; Petráň et al., 1968) modified the design of the Nipkow disk by including multiple spirals in a single wheel. They then used the spinning disk to provide both illuminating and imaging pinholes for a confocal microscope.
As with point scanning microscopes, over the years several different arrangements have been designed for spinning disk confocal microscopes. Figure 1.2 illustrates one such arrangement for an epi-illumination system. In this design, light is passed through the pinholes, directed onto the specimen, and the image light passes back through conjugate pinholes in the disk as it spins. By including sufficient numbers of pinholes and spinning the disk at a suitable speed, a real-time confocal image of the specimen can be obtained that can be viewed by eye or collected directly by a detector. One of the key benefits of this type of confocal microscope compared to laser scanning instruments is that spinning disks allow much faster image acquisition times. Further information on the design and use of spinning disk confocal systems is given in Chap. 8.
Design of an epi-illumination spinning disk confocal microscope. Although multiple areas of the specimen will be illuminated at once, to simplify the diagram only light from one pinhole is depicted. As in Fig. 1.1 only focused light reaches the detector. Since light emitted from all pinholes reaches the detector simultaneously, image collection is rapid, but resolution and often overall signal is compromised in spinning disk systems as discussed in Chap. 8
The Minsky and Petráň microscopes define the two principal implementations of confocal microscopy: the sequential scan (point scan) and spinning disk (multipoint scan, area scan) microscopes, respectively. As one might imagine, however, variations on these two schemes have been designed to overcome specific limitations of each for specific applications. A nice review of some of these implementations is provided by Shinya Inuoué (Inuoué 2006). Of course, the full power of imaging a thin plane within a specimen is best exploited by scanning multiple thin planes in succession and reconstructing a high-resolution 3-D map of the specimen by stacking the 2-D images. As described in Chaps. 6, 7, 8, 9 and 10, key advances in digital imaging , detectors, and improved computer power over the last two decades now provide a convenient method of capturing, storing, and displaying sequentially acquired image information in both 2-D and 3-D formats.
While the above approaches to confocal imaging are still prominent, robust, and very important in today’s research environment, more recently the development of enhanced and super-resolution confocal microscopes has significantly expanded the field of confocal and fluorescence microscopy. The importance of these techniques in furthering our understanding of many biological principles was evidenced by awarding of the 2014 Nobel Prize in Chemistry to Drs. Eric Betzig, Stefan Hell, and William Moerner. While the enhanced and super-resolution techniques may use different approaches for improving resolution, most use deconvolution, or mathematically reassigning signal generated by photons back to its point of origin, to improve resolution. A detailed discussion of deconvolution will be presented in Chap. 8, and examples of how some of these techniques are used to beat the 200 nm lateral resolution limits due to diffraction of light will be discussed in several chapters.
1.4 Is the Confocal Hype Legitimate?
Why has confocal microscopy revolutionized the way many laboratories image their samples? The simple answer is that the use of specific wavelengths of light, typically emitted from a laser, and the use of pinholes or some other mechanism to eliminate or reassign out-of-focus light as briefly mentioned above (and described in more detail in Chap. 9), has significantly increased our ability to resolve and co-localize small structures and molecules in high contrast images. An example of this is shown in Fig. 1.3. Wide-field images (Fig. 1.3a) contain large amounts of out-of-focus light that significantly deteriorates image resolution and contrast making it difficult to observe specific structures and detail. A confocal image (Fig. 1.3b) from the same region of the same sample clearly shows increased resolution and contrast making it much easier to discern the structures present in the section of heart muscle shown.
Wide-field fluorescent (top) and single-photon confocal scanning laser microscope (CSLM ) (bottom) images taken from a 100 m thick vibratome section of mouse heart stained for f-actin (green) and connexin 43 (red). In the wide-field image, out-of-focus light that contributes to the formation of the image significantly decreases the resolution and contrast of the image. Use of the pinhole in the confocal image to remove the out-of-focus light results in an image of much higher contrast and resolution as shown by the striated pattern of the myocyte sarcomeres and distinct cell: cell junctions labeled by the connexin 43 antibody
The improvement in image quality in Fig. 1.3 is obvious, but the confocal image in Fig. 1.3b remains limited by the diffraction of light and instrument configuration. As an example of further improvement in image quality available with enhanced resolution techniques, Fig. 1.4 shows a comparison of images from the same optical field of the hippocampus in a brain slice collected in confocal and enhanced (Airyscan) resolution mode with a Zeiss LSM 800 Airyscan confocal microscope . The improved resolution, as shown by the punctate staining in the Airyscan mode, is apparent, while the loss of resolution in the normal confocal mode is evidenced by the diffuse distribution of the green fluorescence in the tissue.
Comparison of images collected on the Zeiss LSM 800 in normal confocal and Airyscan modes from the hippocampal region of a brain slice. Resolution of the Airyscan images is significantly improved over that seen in the image collected in confocal mode showing the punctate presynaptic terminals labeled with M2 mAChRs (red) and the GluN1 subunit of the NMDA receptor important for synaptic plasticity and learning
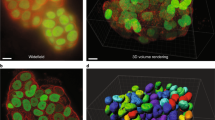
With the development of sensitive detectors, fast computing capabilities, and high-density media for storage, confocal imaging technology has grown rapidly. These advancements have made it possible to collect a large number of optical sections through a sample and to rapidly reconstruct them into a high-resolution high contrast projection of the sample where all detail is in focus (Fig. 1.5). Further advances in imaging software have made the use of 3-D data sets an important element in studying most biological systems. Many of these advances will be discussed in subsequent chapters of this book. However, both confocal imaging hardware and digital imaging software technologies are advancing at a very rapid pace making it essential that researchers stay vigilant in determining how confocal imaging may benefit their individual research programs.
Confocal optical sections (Z-series) through a section of intestine stained with multiple fluorescent dyes. Images were collected at 1 m intervals through a 50 m thick section of tissue, and every other section (2 m intervals) is shown in (a). All sections were then projected into a single composite image as shown in (b). The procedures for collection and projection of data sets are discussed in later chapters. Blue, DAPI stain for nuclei; red, f-actin stain; green, green fluorescent protein; yellow, mRNA-stabilizing protein
The answer to the above question about confocal hype is obviously a resounding yes. Even though commercially available systems have only been available for about 30 years, and well-equipped confocal systems often cost $500 K or more and can be expensive to maintain, the thousands of publications that utilize confocal imaging and the large range of applications from biological to material samples imaged clearly indicate that confocal microscopy has revolutionized the way many laboratories perform their research. Recent advances including spectral imaging, new fluorochromes and lasers, and increased imaging speed and resolution all indicate that confocal imaging will continue to be an important component of the imaging sciences in many fields of investigation.
1.5 The Ten Commandments of Confocal Imaging
As part of our Basic Confocal Microscopy Workshop, we often have students create a list of Confocal Commandments, which are comprised of statements we make that might be considered unequivocal in nature. The following is a list of some of these commandments that we have collected over the years that need to be considered by all undertaking the task of learning and using confocal microscopy as a research tool. These commandments establish some general guidelines to consider when using a confocal microscope, preparing a specimen, and handling digital images, which are all integral and equal parts of operating a confocal microscope. In fact, how we process and present the images we collect is every bit as important as how we do the initial data collection. The various chapters in this book will expand on the basic principles that lead to these commandments.
Our Ten Commandments of confocal imaging are as follows.
1.5.1 The Perfect Microscope and the Perfect Microscopist Do Not Exist
As we will discuss in great detail, physical factors inherent when using photons to produce microscopic images and the characteristics intrinsic to the design of many microscopes result in limitations in the amount of light that can be collected and restricts the obtainable resolution. While super-resolution systems have circumvented some of these defects, the confocal systems commonly available in core facilities and laboratories still must contend with these limitations. This makes it even more critical that the operator understands and adheres to proper preparation of specimens and knows how to appropriately set up the microscope before capturing images. Although some limitations can be minimized by selection of optimal microscope components, they cannot be totally eliminated. Even with the best microscope optics available, the physical nature of light and refractive index mismatch as the light passes through the several interfaces in the optical path of the microscope and specimen will result in image defects. These defects result in the loss of signal and resolution.
Moreover, even with optimal image quality, the human element of understanding image collection and data interpretation is often a limiting factor in getting the most out of a microscope. North (2006), in a feature article for the Journal of Cell Biology, noted that all data are subject to interpretation and that in microscopy a great number of errors are introduced in complete innocence. A common example is the frequent interpretation that the appearance of the color yellow in a sample stained with green and red emitting fluorophores indicates co-localization. However, many factors may affect this interpretation. Without a thorough understanding of sample preparation, optics, imaging parameters, and data analysis, an incorrect conclusion of co-localization may be reached in complete innocence. Several reasons why yellow in an image generated from a sample stained with green and red fluorophores may not represent true co-localization will be discussed in subsequent chapters.
1.5.2 Confocal Microscopy Is More Than a Confocal Microscope
To effectively use a confocal microscope, investigators must have an understanding of specimen fixation and processing, antigen-antibody interactions, fluorescence theory, microscope optics and hardware components, and the handling of digital images for both image enhancement and analysis protocols. Each of these topics will be addressed in subsequent commandments and discussed in detail throughout the text.
The fact that performing confocal microscopy is much more than operating a microscope is illustrated by the sequence of the following chapters. It is essential that information on specimen preparation, fluorescence theory, and the basics of digital imaging be provided prior to material on confocal instrumentation if users are to understand the operation of a confocal microscope and be able to get the optimum amount of information from their samples.
1.5.3 During Specimen Processing the Integrity of the Specimen Must Be Maintained as Much as Possible
The integrity of the specimen includes the 3-D architecture. A major advantage of confocal imaging when compared to wide-field epifluorescence imaging is the acquisition of high-resolution, high contrast images which can be obtained through the Z-axis of a sample and the capability of software programs to reconstruct the 3-D nature of cells and tissues (Fig. 1.5).
Biological confocal microscopy often involves antigen staining to localize specific molecules and structures. It is essential that specimen fixation and subsequent processing maintain, as much as possible, the antigenicity of a specimen and the in vivo localization of cell and tissue antigens, analytes, structural components, etc. This may require extensive adjustment of protocols involving time, temperature, pH, and concentrations of fixatives and primary and secondary antibody solutions. Many of these issues, such as antigenicity and antibody penetration, have become more relevant as tissue clearing techniques have increased in popularity. Chapter 4 will address advantages and disadvantages of tissue clearing protocols, and Chap. 5 will address antigen-antibody interactions, labeling strategies, and potential problems that may arise during staining of samples with various fluorochromes.
Once successful processing protocols are developed, it is also essential that specimens be mounted properly to maintain the 3-D architecture of the sample. Chapter 4 also presents information on various aspects of specimen preparation including the use of various fixatives, buffers, mounting media, and strategies for mounting specimens to insure maintenance of the 3-D architecture of the specimen .
1.5.4 Photons Are Your Friends and Signal-to-Noise Ratio (SNR) Is King
Many factors including microscope optics and fluorochrome characteristics tend to reduce the number of photons available for formation of an image. At the same time that we are trying to maximize the number of photons (signal) collected, microscope hardware such as detectors and electronics introduce electronic noise that may result in a poor SNR. As a result, the operator must always be aware of the SNR in an image in an effort to establish operating parameters that maximize image quality while minimizing specimen damage. The SNR problem is being addressed by the development of new, high-sensitivity detectors such as the GaAsP and hybrid detectors discussed in Chap. 7. However, although these new detectors provide a better set of tools, their sensitivity is not infinite, and so sufficient signal-to-noise ratio remains a problem in many imaging protocols. In particular, a high SNR is critical for some forms of enhanced resolution microscopy. Thus, several chapters in this book discuss various aspects of fluorochrome and system properties that affect the SNR and provide suggestions on how to maximize the signal for optimal image quality .
1.5.5 Quantification of Fluorescence in a Confocal Micrograph Is a Challenge and at Best Is Only Semiquantitative
This is perhaps one of the most important commandments when dealing with today’s competitive research environment and the need for quantitative data that is essential for funding opportunities and high-impact publications. Even though a large percentage of researchers using confocal microscopes report quantitative results from their studies, one must use caution when inferring numerical data from images collected with a confocal microscope. Pawley (2000) posed the question “does a fluorescent micrograph reveal the actual location and number of labeled molecules in a cell or tissue” to members of his well-known 3D Microscopy of Living Cells course. Based on responses collected in the course, he published “The 39 Steps: A Cautionary Tale of Quantitative 3-D Fluorescence Microscopy” in BioTechniques. Table 1.1 is an abbreviated list of some of the factors that microscopists using confocal systems must be aware of during every imaging session. The conclusion of Pawley’s paper is that “all you can really be sure of measuring with most laser-scanning confocal microscopes in the fluorescence mode is some feature of the number of photons collected at a particular time.” Throughout the following chapters, we will discuss many of the issues that limit the effectiveness of confocal microscopes as a quantitative research tool and provide tips and suggestions for specimen preparation, imaging parameters, and handling digital images so that as much data as possible can be collected from each image data set .
1.5.6 Scientific Digital Imaging and Normal Digital Imaging (Family Photography) Are Not the Same
The greatest power of digital imaging is that exact copies of data can easily be made. This is excellent when archiving data and reverting to the original files when image processing does not result in the desired effect. However, while it may seem obvious that much of the processing we do on images collected with over-the-shelf digital cameras should not be done with scientific images, the innocence of the investigator again may be a problem. For example, when adjusting the contrast and brightness of a confocal image in programs such as Photoshop, the gamma function should always be used rather than the contrast and brightness functions. Gamma corrections should also be performed only after the histogram stretch functions are completed. While rules such as this are not important in family photography applications, not applying them correctly to digital images collected for scientific applications has the potential to alter the appearance of the data.
As discussed extensively in Chaps. 6 and 12, it is essential that an original, unaltered file of the data is archived for reference. All changes in the image should be made only on a copy of the original file. There are specific guidelines that have been published by several groups, including the Microscopy Society of America (http://www.microscopy.org), that specifically state how scientific digital images should be handled. More information concerning these guidelines and the ethics of handling digital images generated for scientific studies will be provided in Chaps. 6, 9, 10, 11 and 12 on processing of confocal images and the ethics associated with the presentation of the images.
Most hardware used for the collection and display of digital images utilizes software that includes some form of image processing prior to rendering the image. Frequently, manufacturers do not make this information available resulting in images that are collected without a full understanding of how they have been processed by the hardware used in image capture. While this is typically not a problem in recreational photography, processing of scientific data by collection devices prior to saving the information should always be a concern. Whenever possible, when working with images collected as scientific data, a thorough understanding of how the images are collected and processed by the system hardware is desirable. Unfortunately, this information is sometimes difficult to obtain from the manufacturer of the equipment or even worse, considered proprietary and so never revealed. We strongly feel that equipment and software manufacturers owe it to the scientific community to make critical information that can affect image fidelity readily available .
1.5.7 Your Image Is Your Data: Garbage in Will Result in Garbage Out
One should always be detail oriented in sample preparation , image collection, and handling digital images. The factors listed in Table 1.1 and by Pawley (2000) that affect quantitative confocal microscopy imaging are equally important in the acquisition of images for qualitative studies in which “pretty” pictures to demonstrate a scientific point are required. Without heeding each of the factors, it is unlikely that publication quality confocal images will be generated or that data collection from images will be maximized .
1.5.8 The Resolution and Bit Depth Present in a Digital Image Are a One-Way Street
After image capture the resolution of an image is set, and image processing protocols will not improve or increase the resolution of an image. While it may be possible through gamma, contrast, and brightness functions, and other types of algorithms such as sharpening filters to improve the aesthetic appearance of an image, as will be seen in Chaps. 6, 7, 10, and 11, once an image is collected with hardware and software available on a system, any structures that can be resolved in the image will be present. Using software to increase the number of pixels in a digital image will not improve the resolution, but only result in the creation of pixels by interpolation. These pixels are created by an algorithm such as averaging neighboring pixel values and appear as the computer “believes” they should look.
One may argue that image processing through deconvolution improves the resolution of a data set, but the limits of resolution have already been determined by the hardware present on the microscope and the physical properties of the light used to collect it. Deconvolution uses the point spread function (PSF) to mathematically reassign photons from the blur to their point of origin to reduce blur and improve resolution of an image. However, the ultimate resolution was set during collection of the image by factors such as the wavelength of light used and numerical aperture of the objective as discussed in detail in Chap. 7. Deconvolution may enable one to better define the data present, but the limits of resolution were set during collection of the image .
1.5.9 The JPEG (Joint Photographic Experts Group) Image File Format Is EVIL but Useful
This statement applies to any file format that compresses the data and does not allow full recovery of all of the information present in the original file. The JPEG format is the one encountered most often in imaging and so the one we chose to single out. As noted above, resolution is a one-way street, and the original data should be stored as collected. Chapter 6 will show that saving files in the JPEG format results in significant loss of information, and especially damaging to scientific images, this loss is greatest along the edges. All original data should be stored in a lossless format such as a TIF (Tagged Image File) or a proprietary version of a TIF format as provided by the instrument manufacturer. JPEG and other compression formats may be used in situations where images either need to be shared electronically or inserted into formats for lectures, seminars, and posters. In these situations resolution may be sacrificed in favor of smaller file sizes to make handling of images more reasonable. However, these compressed images should never be used as the primary source of data . File format options will be discussed in detail in subsequent chapters.
1.5.10 Storage Media Is Essentially Free and Infinite
The message of this commandment from the first edition of Basic Confocal Microscopy is that it is essential that the original data sets be archived appropriately and that any image manipulation is performed on copies of the data that must also be archived. When the first edition was published, this essentially required several CDs or DVDs to accumulate and store data sets that were typically in the range of a few gigabytes at most. While essentially still true, this commandment from the first edition does need to be qualified to some degree. When compared to data sets from a few years ago, today’s data sets have grown exponentially due to the speed of image collection, the types of imaging present such as light sheet microscopy, and resolution available. This has resulted in a single data set that may approach or even exceed several terabytes. It is also possible to collect several data sets in a fairly short period of time, and cumulative data sets are now approaching a petabyte in size. Thus the mechanism and potential cost of storage has changed significantly in the few short years since publication of the first edition and the term Big Data, and the cost and processing of Big Data, is becoming a concern for many laboratories.
Even though it may now be necessary to have servers with several nodes to store data, and these may be expensive, as noted in the first edition, the cost of data storage is still minimal compared to generating new experiments if the data is questioned and the original files are no longer available. In addition, archiving of data is now a requirement of many funding agencies such as NIH and NSF that have specific policies on data storage and accessibility for the scientific community (https://grants.nih.gov/grants/policy/data_sharing/ and https://www.nsf.gov/sbe/ses/common/archive.jsp), as do many other foundations and funding agencies. For example, NSF guidelines indicate that “for appropriate data sets, researchers should be prepared to place their data in fully cleaned and documented form in a data archive or library within one year after the expiration of an award. Before an award is made, investigators will be asked to specify in writing where they plan to deposit their data set(s).” Additional archiving guidelines will be discussed further in Chap. 12 on Ethics and Resources .
Thus it is no longer acceptable to simply archive data on personal storage devices such as external hard drives, thumb drives, etc., and there may be significant cost involved with storage on Cloud devices or servers supported by universities.
1.6 Summary
These Ten Commandments for confocal imaging provide a set of principles to guide users in a confocal microscopy laboratory. Other commandments have occasionally been added to the list during our workshops, but if close attention is paid to each of the above, and a detailed understanding of the importance of each is developed, users will have a strong understanding of confocal technology for use in their research.
In Chaps. 2, 3, 4, 5, 6, and 7, we present information on the topics of fluorescence, specimen preparation, and digital imaging which are essential for understanding confocal imaging. In subsequent chapters we present information on various types of confocal instruments, the proper setup of operating parameters for confocal imaging, and appropriate techniques for enhancing and analyzing confocal images. Topics pertinent to the various commandments as well as some frequently asked questions such as:
-
1.
Are these fluorescent markers co-localized?
-
2.
Can I quantify the amount of labeled material present based on the fluorescence intensity which is present?
-
3.
Can I measure the size or area of these structures based on a confocal data set?
-
4.
How deep can I image into my sample?
are addressed. Hopefully by learning the basic principles of confocal imaging, the quality of the confocal imaging experience of many beginning and intermediate users of the technology will be improved.
Literature Cited
Ashdown GW, Cope A, Wiseman PW, Owen DM (2014) Molecular Flow Quantified beyond the Diffraction Limit by Spatiotemporal Image Correlation of Structured Illumination Microscopy Data. Biophys J 107:L21–L23
Berek M (1927) Grundlagen der tiefenwahrnehmung im mikroskop. Marburg Sitzungs Ber 62:189–223
Dempsey GT, Vaughan JC, Chen KH, Bates M, Zhuang X (2011) Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat Methods 8:1027–1036. https://doi.org/10.1038/nmeth.1768
Eggar MD, Petráň M (1967) New reflected light microscope for viewing unstained brain and ganglion cells. Science 157(786):305–307
Inoue S (2006) Foundations of Confocal Scanned Imaging in Light Microscopy. In: Pawley JP (ed) Handbook of Biological Confocal Microscopy, 3rd edn. Springer, New York 985 pp
Minsky, M. (1957) U.S. Patent #3013467. Microscopy Apparatus
Minsky M (1988) Memoir on inventing the confocal scanning microscope. Scanning 10:128–138
Nipkow, P. (1884) German Patent no. 30, 105. Germany
North AJ (2006) Seeing is believing? A beginners’ guide to practical pitfalls in image acquisition. J Cell Biol 172:9–18
Pawley J (2000) The 39 steps: A cautionary tale of quantitative 3-D fluorescence microscopy. BioTechniques 28:884–888
Petráň M, Hadravsky M, Egger MD, Galambos R (1968) Tandem scanning reflectgd light microscope. J Opt Soc Am 58:661–664
Wentao Y, Ziheng J, Dashan D, Xusan Y, Yunfeng X, Qihuang G, Peng X, Kebin S (2016) Super-resolution deep imaging with hollow Bessel beam STED microscopy. Laser Photonics Rev 10(1):147–152. https://doi.org/10.1002/lpor.201500151
White JG, Amos WB, Fordham M (1987) An evaluation of confocal versus conventional imaging of biological structures by fluorescence light microscopy. J Cell Biol 105:41–48
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Price, R.L., Jerome, W.G. (2018). Introduction and Historical Perspective. In: Jerome, W., Price, R. (eds) Basic Confocal Microscopy. Springer, Cham. https://doi.org/10.1007/978-3-319-97454-5_1
Download citation
DOI: https://doi.org/10.1007/978-3-319-97454-5_1
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-97453-8
Online ISBN: 978-3-319-97454-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)