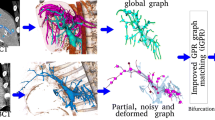
Abstract
The topologies of vascular trees embedded inside soft tissues carry important information which can be successfully exploited in the context of the computer-assisted planning and navigation. For example, topological matching of complete and/or partial hepatic trees provides important source of correspondences that can be employed straightforwardly by image registration algorithms. Therefore, robust and reliable extraction of vascular topologies from both pre- and intra-operative medical images is an important task performed in the context of surgical planning and navigation. In this paper, we propose an extension of an existing graph-based method where the vascular topology is constructed by computation of shortest paths in a minimum-cost spanning tree obtained from binary mask of the vascularization. We suppose that the binary mask is extracted from a 3D CT image using automatic segmentation and thus suffers from important artefacts and noise. When compared to the original algorithm, the proposed method (i) employs a new weighting measure which results in smoothing of extracted topology and (ii) introduces a set of tests based on various geometric criteria which are executed in order to detect and remove spurious branches. The method is evaluated on vascular trees extracted from abdominal contrast-enhanced CT scans and MR images. The method is quantitatively compared to the original version of the algorithm showing the importance of proposed modifications. Since the branch testing depends on parameters, the parametric study of the proposed method is presented in order to identify the optimal parametrization.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Liver cancer is the 7th most common cause of cancer death in Europe with around 62,200 deaths in 2012 [1]. In order to improve the success rate of interventional liver cancer treatment, several computer-aided approaches have been proposed for both intervention planning and navigation.
The common factor of methods proposed to facilitate hepatic interventions remains the necessity of reliable and robust extraction and analysis of the hepatic vascular structures. The liver vascularization plays an important role in many aspects: the volume that is removed during the hepatectomy depends directly on the topology of the vascular network [2]. Similarly, the registration methods often rely on vascular landmarks [3]. Furthermore, accurate biomechanical models that are to be used in the augmented-reality applications must take into account the mechanical properties of the vascular trees [4, 5].
While extremely important, the automatic robust analysis of liver vascularization remains a challenging problem. First, the liver displays a high inter-patient anatomic variability. Further, the acquisitions following the injection of a contrast agent, necessary for the pre-operative identification of vascular trees, often suffer from noise due to the respiratory motion which in turn deteriorates the quality of segmentation that is necessary for the extraction of the topological structure in the process known as skeletonization.
Several methods of skeletonization of vascular structures have been proposed in the literature. In [6], the thinning is applied to the binary mask to obtain the skeleton. Since the thinned structure remains a binary image, additional analysis is needed to extract the topological information. A skeletonization method is also proposed in [7], however, since the method works with surface meshes, it requires the extraction of such a mesh from the binary mask. In [8], another method based on Dijkstra minimum-cost spanning tree and which takes as input a binary mask is presented. In their paper, the authors test the method on phantom and clinical data acquired in neurosurgery. The same method is further extended and tested for airways tree skeletonization in [9].
In this paper, we focus on the skeletonization of vascular trees extracted from medical patient-specific data. We suppose that the skeletonization is performed on binary maps obtained by the automatic segmentation [10]. In this case, the resulting binary map typically suffers from artefacts, such as important surface bumps (see Fig. 1), making the skeletonization process very challenging. Therefore, we propose an extension of a graph-based method presented in [8] and [9]: first, we modify the algorithm to make is less sensitive w.r.t. the quality of the input data. Second, we propose a set of tests allowing for automatic removal of spurious or false branches relying on geometric criteria. The proposed skeletonization method is evaluated on 3 porcine and 32 human vascular trees and the results are compared to the original version of the algorithm. Moreover, a parametrization study is presented, showing the influence of method parameters on the quality of constructed skeleton.
2 Methods
2.1 Minimum-Cost Spanning Tree
The method based on the minimum-cost spanning tree [8] is applied directly to the binary map of the vascular structure which is converted into a weighted 3D graph where each voxel belonging to the segmented volume is represented by a weighted node. The method constructs the skeleton in two steps: First, the spanning tree is constructed iteratively starting from the root voxel. In the actual version of the algorithm, the root voxel is selected interactively in the area close to the root of the vascular tree. This is the only step of the method which currently requires manual intervention. Then, the edges between voxels are constructed recursively using a sorted heap: in each step, the head of the heap having the minimal weight is marked and all its unmarked neighbors are inserted into the heap. In [8], the sorting weight w(v) of a voxel v is defined as \(\frac{1}{r_b(v)}\), where \(r_b(v)\) is the shortest distance of the voxel v from the boundary of the segmented vessel. We employ a modified definition the sorting weight \(w(v) = w(u)+\frac{d(u,v)}{r_b(v)^k}\) where u is the voxel preceding v in the minimum-cost spanning tree, and d(u, v) is the Euclidean distance between the voxels u and v. The metric \(r_b(v)\) is pre-computed for each voxel of the input binary map. The modified definition of the heap-sorting weight makes the skeletonization process less sensitive to the bumps of the segmented vascular tree illustrated in Fig. 1. For each visited voxel, its distance from the tree root \(d_r\) is stored.
In the second phase of the method, the centerlines are extracted recursively from the spanning tree as in [8]:
-
The 0-order path \(P_0\) is constructed as the shortest path connecting the root and the voxel with the highest \(d_r\) in the graph, that is the voxel which is at the farest outlet from the root.
-
Any n-order path \(P_n\) for \(n>0\) is extracted in three steps. First, an expansion step is performed so that for each voxel r of the path \(P_{n-1}\), a set \(V_r\) of all voxels accessible throught the minimum-cost spanning tree from r is constructed. \(V_r\) is the set of all voxels that are linked to r and not belonging to \(P_{n-1}\). Second, a voxel \(t\in V_v\) having the maximum value \(d_r\) is found. Finally, the new path is constructed as the shortest path from r to t, denoted as (r, t).
Different scenarios occurring during skeletonization of automatically segmented vessels. T-shape scenarios: (a) false path due to the surface bump, (b) correct branch. V-shape scenarios: (c) false path due to the vessel thickening, (d) correct branch: the background voxels shown in red. (Color figure online)
2.2 Typical Artefacts of Graph-Based Skeletonization
The algorithm of the centerline extraction performs well on the synthetic data as well as real data where the vascular structures are represented by smooth and well-formed pipes. However, when applied to real data obtained using automatic segmentation of hepatic veins, artefacts appear resulting in false branches, i. e., redundant bifurcations.
First, directly during the construction of a path (r, t), it is verified that none of the voxels except r is already included in a path constructed before, i. e. there is no collision between the new path and any previously constructed path. While such situation should not happen due to the properties of the spanning tree, exceptions might occur, i. e. when holes or plate-shaped structures are present in the input binary mask.
The second type of artefacts is introduced by scenarios depicted in Fig. 2: for the sake of reference, we divide the scenarios in \(T-shapes \) (Fig. 2a, b) and \(V-shapes \) (Fig. 2c, d). In the case of T-shapes, the bumps (a) created by the automatic segmentation are difficult to distinguish from real branches (b). In the case of V-shapes, the problematic scenarios occur when a redundant centerline appears in the graph due to the vessel thickening (c). While from the anatomical point of view, such phenomenon is improbable, it is not rare in binary masks produced by the automatic segmentation.
2.3 Filtering of Spurious Branches
In order to prevent the extraction of false branches, we propose to filter the branches directly during the extraction of paths based on several criteria. First, let us denote R the geometrical position in Cartesian coordinates of the branch candidate root voxel r, T the position of its tip voxel t and finally C the position of voxel c on the parent branch which has the lowest Euclidean distance from T. In the following description of the algorithm, we always refer to Fig. 2.
-
1.
If \(|TR| < L_{\text {thr}}d_b(r)\), the branch candidate is rejected. Here, the \(d_b(r)\) is distance from boundary of the root voxel r and \(L_{\text {thr}}\) is a parameter, length threshold. This parameter is a unitless quantity which determines the relative ratio between a child branch having an acceptable length and the thickness of the parent branch. The goal of this step is to eliminate the short false branches produced either inside the tube of the parent branch or due to the bumps (a).
-
2.
In the next step, the correct V-shapes are identified (d). In this case, there exist voxels on the line CT (connecting the voxels c and t) located outside of the binary mask. Such background voxels guarantee that path is located in a child branch which is correctly detached from the parent branch. Therefore, in this step, the path candidate for which such voxels exist is directly accepted as a new path and the algorithm proceeds with extraction of another path.
-
3.
The branch candidate that made it to this step is sufficiently long but has no background voxel. Therefore, it corresponds either to valid T-shape (b) or invalid V-Shape (c). In order to distinguish the two cases, we introduce the second parameter, angle threshold \(A_{\text {thr}}\). The path candidate is accepted if the angle \(\angle CRT > A_\text {thr}\), otherwise, it is rejected.
The algorithm stops when no new branches are added in the actual order.
3 Results
In all the tests, the skeletonization was always stopped after the extraction of paths of order 5, as the segmentations used for validation contained no reliable branches of a higher order.
3.1 Data
The porcine data sets were acquired from abdominal CT volumes of a female pig scanned in flank and supine positions with voxel resolution of \(0.5\times 0.5 \times \)0.5 mm\(^3\). While both portal and venous phases were obtained for the flank position, only the venous phase was available for the supine configuration, thus resulting in three data sets. The human data sets were acquired from abdominal CT and MR volume with voxel resolution of \(0.68\times 0.68 \times 2.0\) mm\(^3\) and \(1.0\times 1.0\times \)2.5 mm\(^3\), respectively. A retrospective cohort of fifteen patients (4 women, 11 men) that received a bi-phasic magnetic resonance angiography (MRA) and computed tomography angiography (CTA) on the same day were included in this study. The MRA images were acquired on the Achieva 3T scanner (Philips Healthcare, Best, The Netherlands) using a 3D mDIXON sequence and on the Discovery MR450 scanner (GE medical Systems, Milwaukee,WI,USA) using the 3D LAVA sequence. CTA images were acquired using the helical mode on the Brillance 64 scanner (Philips Healthcare, Best, The Netherlands) and the BrightSpeed system (GE Medical Systems, Milwaukee, WI, USA). Vascular structures were segmented in each data set using an automatic method [10].
3.2 Evaluation of the Skeletonization
The graph-based skeletonization is evaluated in Table 1 for each vascular tree. First, a large number of path candidates is extracted from the spanning tree. The most significant reduction of candidates happens due to the insufficient length of the candidate branch: more than 85% of candidates are false child branches, i. e. too short when compared to the thickness of the parent branch. Less than 10% of candidates are removed because of crossing with other branches (potential cycles). Finally, less then 5% of candidates are accepted as new paths: majority of them are accepted as V-shape vessels.
3.3 Method Parametrization
As introduced in Sect. 2.3, the graph-based skeletonization method requires two parameters: the length threshold \(L_\text {thr}\) and the parent–child angle threshold \(A_\text {thr}\). As for the latter, very low sensitivity was observed w.r.t. this parameter: only from 1 to 2 false V-shape candidates appeared during the extraction and were eliminated by setting the threshold to 20\(^\circ \). This observation confirms that the construction of the spanning tree already prevents the formation false V-shape paths.
In contrast, the value of \(L_\text {thr}\) influences the filtering significantly. Intuitively, a logical choice for this parameter is 1; it should be sufficient to filter all the candidates having the length shorter that the thickness of the parent branch. However, this assumption is not valid when bumps are present in the input data (Fig. 2). The size of the bumps can make it difficult to distinguish them from short real branches; see Fig. 1 showing a sample of the real data used for testing.
In order to study the influence of \(L_\text {thr}\) on the skeletonization of the vascular trees, we have performed a scaling of the parameter starting from 1.0 to 2.0; the result is presented in Fig. 3. The plot shows that the number of accepted branches decreases rapidly from 1.0 to 1.4; then, the decreasing slope becomes less steep. Although it is not possible to specify the cut-off value which would allow for reliable filtering of the false branches while keeping the real ones, we propose using the value 1.4 as the threshold. The values in Table 1 have been obtained with this parameter.
Comparison of the number of accepted branches; cN stands for the case N. The results given by our method are displayed in blue and green. The color gradient corresponds to increased values of k from \(k=1\) (blue) to \(k=7\) (green). The results of Verscheure’s algorithm are displayed in yellow. The red horizontal line depicts the number of real branches for each case. (Color figure online)
3.4 Original vs. Modified Skeletonization Method
In order to assess the method, we have compared the proposed approach to the method presented in [8]. We used 32 segmentations performed using the ITK-Snap semi-automatic segmentation algorithm based on active contour (Snakes, [11]) on computed tomography and magnetic resonance images. The segmentations corresponds either to human hepatic and splenic arteries or to the human hepatic portal vascular system.
The best accuracy computed as the number of accepted branches, i. e. the value for which the error in the branch number is minimal, is obtained for both algorithms with \(L_\text {thr}=2.5\) regardless the values of the other parameters. Thus, this value was used for the comparison of the two methods. With higher values true branches are rejected as outliers. Figure 4 shows the number of accepted branches by the two algorithms versus the real number of branches determined manually by an expert. The consistency of the branches were checked manually for all datasets. The evaluation shows that the number of branches extracted with the original algorithm proposed by Verscheure is slightly superior when compared to the values obtained by our algorithm. Most of the time, a false branch detected by Verscheure’s algorithm runs in parallel to another branch (see Fig. 5). Interestingly, even without an angle threshold, our modified method produces on average 3 times less parallel branches when compared to the original method. The failing cases are encountered for very noisy segmentation maps for which the bumps are too large for the false branches to be filtered out by the length threshold. Another important source of differences between the two algorithms is given by false loops which occur when distance between distinct branches remains under the resolution of the segmented image, leading to topological errors.
The shape of the extracted centerlines depend also on the exponent k employed in the definition of the heap-sorting weight \(w(v) =w(u)+\frac{d(u,v)}{r_b(v)^k}\): For \(k \le 3\), the centerlines tend to be rather straight and do not follow the shape of the vessel properly. This is particularly obvious for tortuous vessels such as the splenic arteries. Therefore, we recommend to use \(k>3\).
4 Conclusions
In this paper, we have presented a method of automatic skeletonization of vascular trees. The algorithm was assessed on binary masks automatically segmented from abdominal CT scans of a porcine liver. It was shown that it is possible to select parametrization to suppress the false branches introduced by the noise in the input data. In the actual case, we have further smoothed the extracted skeleton using cubic splines. The illustration of the complete tree is shown in Fig. 6.
In the future work, we plan to integrate the algorithm to an automatic pipeline employed in an augmented-reality framework for the intra-operative navigation in hepatic surgery.
References
World health organization. http://www.who.int. Accessed 12 Jun 2015
Mise, Y., Tani, K., Aoki, T., Sakamoto, Y., Hasegawa, K., Sugawara, Y., Kokudo, N.: Virtual liver resection: computer-assisted operation planning using a three-dimensional liver representation. J. Hepato-Biliary-Pancreat. Sci. 20(2), 157–164 (2013)
Ambrosini, P., Ruijters, D., Niessen, W.J., Moelker, A., van Walsum, T.: Continuous roadmapping in liver tace procedures using 2D-3D catheter-based registration. Int. J. Comput. Assist. Radiol. Surg. 10(9), 1357–1370 (2015)
Peterlík, I., Duriez, C., Cotin, S.: Modeling and real-time simulation of a vascularized liver tissue. In: Medical Image Computing and Computer-Assisted Intervention-MICCAI 2012. Springer 50–57 (2012)
Plantefève, R., Peterlik, I., Haouchine, N., Cotin, S.: Patient-specific biomechanical modeling for guidance during minimally-invasive hepatic surgery. Ann. Biomed. Eng. 44(1), 139–153 (2016)
Lee, T.C., Kashyap, R.L., Chu, C.N.: Building skeleton models via 3-D medial surface axis thinning algorithms. CVGIP. Graph. Models Image Process. 56(6), 462–478 (1994)
Piccinelli, M., Veneziani, A., Steinman, D.A., Remuzzi, A., Antiga, L.: A framework for geometric analysis of vascular structures: application to cerebral aneurysms. IEEE Trans. Med. Imaging 28(8), 1141–1155 (2009)
Verscheure, L., Peyrodie, L., Dewalle, A.S., Reyns, N., Betrouni, N., Mordon, S., Vermandel, M.: Three-dimensional skeletonization and symbolic description in vascular imaging: preliminary results. Int. J. Comput. Assist. Radiol. Surg. 8(2), 233–246 (2013)
Valencia, L.F., Pinzón, A.M., Richard, J.C., Hoyos, M.H., Orkisz, M.: Simultaneous skeletonization and graph description of airway trees in 3D CT images. In: XXVème Colloque GRETSI (2015)
Yushkevich, P.A., et al.: User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31(3), 1116–1128 (2006)
Kass, M., Witkin, A., Terzopoulos, D.: Snakes: active contour models. Int. J. Comput. Vision 1(4), 321–331 (1988)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Plantefève, R., Kadoury, S., Tang, A., Peterlik, I. (2017). Robust Automatic Graph-Based Skeletonization of Hepatic Vascular Trees. In: Cardoso, M., et al. Intravascular Imaging and Computer Assisted Stenting, and Large-Scale Annotation of Biomedical Data and Expert Label Synthesis. LABELS STENT CVII 2017 2017 2017. Lecture Notes in Computer Science(), vol 10552. Springer, Cham. https://doi.org/10.1007/978-3-319-67534-3_3
Download citation
DOI: https://doi.org/10.1007/978-3-319-67534-3_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-67533-6
Online ISBN: 978-3-319-67534-3
eBook Packages: Computer ScienceComputer Science (R0)