Abstract
Oncogenesis is driven by the accumulation of genetic and epigenetic alterations that result in dysregulation of key oncogenes, tumor suppressor genes, and DNA repair/housekeeping genes. One of the major clinical needs is the discovery and clinical validation of new molecular biomarkers using non-or minimally invasive procedures to assist early diagnosis, prognosis and prediction of response to treatment. Histone methylation has profound effects on nuclear functions such as transcriptional regulation, maintenance of genome integrity and epigenetic inheritance. On the other hand, aberrant DNA methylation can be detected in several biological fluids of patients and could be served as a potential tumor biomarker. In the present chapter we describe latest developments on histone and DNA methylation based biomarkers in Lung cancer.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
- DNA methylation
- Epigenetic silencing
- Lung cancer
- miRNAs methylation
- DNA methylation biomarkers
- Liquid biopsy
- ctDNA
1 Introduction
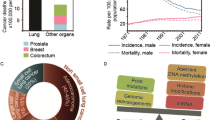
Lung cancer remains the second leading cause of death worldwide, after heart disease with more than 200,000 new cases and 160,000 deaths each year. The high incidence of lung cancer in combination with the very low 5-year survival rate of 17% is the main cause of high mortality rate in this type of cancer [188]. The main subtypes of lung cancer are small cell lung cancer carcinoma (SCLC) and non-small cell lung carcinoma (NSCLC), which includes squamous cell carcinoma, adenocarcinoma, and large cell carcinoma subtypes [38]. NSCLC is the most common type, accounting for approximately 85% of all lung cancer cases. Although smoking remains the major risk factor for all histologies (especially small cell and squamous cell carcinoma), it is important to note that only around 10% of smokers will ultimately develop lung cancer [134]. Globally, an estimated 15% of men and 53% of women with lung cancer are never-smokers. This fact indicates additional risk factors for the disease. Adenocarcinoma, for example, is the most common form among nonsmokers. Other risk factors include exposure to radon, asbestos, and environmental/occupational exposure to polycyclic aromatic hydrocarbons and other pollutants [177]. However, as with smoking, not all exposed to these environmental factors develop lung cancer.
The carcinogenic process is driven by the accumulation of genetic and epigenetic alterations that result in dysregulation of key oncogenes, tumor suppressor genes, and DNA repair/housekeeping genes. The probability that these pathologically important events will occur is not only dependent on the individual’s exposure but also on interpersonal phenotypic variability. Although genetic heterogeneity accounts for some of the variable risk, it does not totally explain this phenomenon [107]. Epigenetic variability, including DNA methylation, histone modifications, and noncoding RNA expression, also contribute to the phenotype of an individual and, accordingly, to the risk of malignancy [108].
Early detection of lung carcinoma could change the disease outcome; in fact, the survival rate can increase dramatically. In the effort to improve early detection, many imaging and cytology-based strategies have been employed; however, none has yet been highly effective, mainly because of limited sensitivity and the huge cost they bear to public health systems [7]. It is now widely accepted that epidemiological risk modeling is required for stratification of individuals for CT screening for early detection of lung cancer [161]. In addition to CT, one of the major clinical needs is now the inclusion of new molecular biomarkers detected in clinical samples using non-or minimally invasive procedures to assist early diagnosis, prognosis and prediction of response to treatment. Understanding the molecular pathways within lung cancer, and focusing on their molecular heterogeneity, is the most effective way towards the development of novel diagnostic and therapeutic tools. In the last decade, a plethora of molecular factors all involved in lung carcinogenesis have been evaluated as prognostic biomarkers [8].
2 Histone Methylation
Histone post-translational modifications include methylation, acetylation, phosphorylation and ubiquitination; through the modulation of chromatin structure, histones play a significant role in creating gene transcriptional activation or repression [224]. Their role is crucial for precise coordination and organization of the open and closed chromatin structure during many dynamic processes such as DNA replication, repair, recombination, and transcription. Changes in local or global chromatin structure have been found to be the key features of many if not all tumors, indicating that such epigenetic changes may make a potential contribution to carcinogenesis [184, 207].
Histone methylation has profound effects on nuclear functions such as transcriptional regulation, maintenance of genome integrity and epigenetic inheritance [132]. For example, histone methylation on arginine or lysine residues can either activate or repress gene transcription, depending on which particular arginine or lysine residue become modified [103]. Methylation and demethylation on arginine or lysine residues in histone tails are reversible modifications that are tightly controlled by histone methyltransferases and histone demethylases. Such dynamic balance of methylation and demethylation is frequently altered in tumorigenesis and pathogenesis of other disorders as well [31, 45, 87, 220].
There are three histone methylation states: monomethyl (me1), dimethyl (me2) or trimethyl (me3) [162]. In general, methylation of H3K4, H3K36 and H3K79 is generally considered to activate genes while methylation of H3K9, H3K27, H3K56, H4K20 and H1.4K26 causes transcriptional repression [105]. H3K4me1 is related with enhancer functions and participates in gene repression in metazoans [30, 66], nucleosome dynamics and chromatin regulation of yeast stress-responsive genes [141]. H3K4me2 is connected to gene repression and transcription in yeast [130, 151], whereas H3K4me3 is linked to active transcription and is present around transcriptional start sites [14, 219].
Lysine-specific demethylation is facilitated by two families of enzymes, of which the JmjC (JumonjiC) domain-containing family of histone demethylases (JHDMs) is the major one. KDM proteins are divided in two subgroups; KDM1 and KDM 2-7 [201]. Unfortunately, there are few studies on KDM demethylases in lung tumors. KDM5B (lysine-specific demethylase 5B), also known as JARID1B (jumonji AT-rich interactive domain 1B) or PLU-1, is one member of the JHDMs subfamily which has recently attracted much attention [64]. Famous oncogenes such as E2F1 and E2F2 are downstream genes in the KDM5B pathway [65, 114]. Recently, KDM5B was found to stimulate NSCLC cell proliferation and invasion by affecting p53 expression [183]. KDM4A and KDM4B remove the tri- and dimethylated marks from H3K9 and H3K36 thus leading to gene repression while KDM4D can only move a methyl group from a trimethylated mark of H3K36. In non-neoplastic tissues, expression of KDM- 4C is especially high in the testes and expression in the lung is very low. KDM4A and KDM4B have a generally higher expression in non-neoplastic tissues the highest levels being found in ovary and spleen, but they are moderately expressed also in the lung [105]. Another recent study was undertaken to investigate the immunohistochemical expression of KDM4A, KDM4B and KDM4D in a set of 188 lung carcinomas. The results were associated with tumor histology, parameters describing the spread of the tumors, and survival of the patients. As an additional marker, the antibody to H3 trimethylated state was used. KDM4A and KDM4D play a role in spread of the lung carcinomas. Further, cytoplasmic KDM4A positivity associates with patient survival. These results are in line with the supposed role of KDMs in epigenetic regulation of cancer cells, affecting proliferation, apoptosis and DNA repair mechanisms [192].
In lung cancer, several global histone modifications have been associated with survival; in particular, decreased levels of H3K4diMe have been associated with poor outcome [178]. Furthermore, the combination of several histone modifications have been reported to predict survival (H3K4me2, H3K9ac, and H2AK5ac) [13], and H4K20me3 downregulation has been associated with poor prognosis in patients with stage I lung adenocarcinoma [210].
Over the last decade, many studies have revealed epigenetic aberrations involving histone modifications in lung cancer. Miyanaga et al. [139] treated 16 NSCLC cell lines with HDAC inhibitors and both displayed antitumor activities in 50% of the cell lines tested. They also conducted gene expression profiling and created a nine-gene classifier which predicts HDAC inhibitor drug sensitivities. Another group compared lung cancer cells with normal lung cells, and they found that lung cancer cells displayed aberrant histone H4 modification patterns with hyperacetylation of H4K5/H4K8, hypoacetylation of H4K12/H4K16, and loss of H4K20 trimethylation [210]. These findings indicate an important role for histone H4 modifications and highlight H4K20me3 as a potential diagnostic biomarker and therapeutic target for lung cancer. Another study has shown that lower global levels of histone modifications are predictive of a more aggressive cancer phenotype in lung adenocarcinoma [178].
Additionally, the differential expression pattern of HATs and HDACs in the tumor samples, as compared to normals, may have important implications for the management of the patients [147]. HDAC1 gene expression appears to correlate with lung cancer progression; overexpression of HDAC1 and HDAC3 correlates with poor prognosis in pulmonary adebocarcinoma patients [137, 138, 171]. HDAC3 was also found in elevated levels in 92% cases of SCC tumors using antibody microarrays for detection of target proteins [15].
Histone deacetylase inhibitors (HDIs) might beneficially contribute to tumor treatment, by reducing the responsiveness of tumor cells to the TNF mediated activation of the NF-B pathway. This is shown in NSCLC cells treated with HDIs which down-regulated TNF-receptor-1 mRNA, protein levels, and surface protein expression, and consequently responded to TNF-treatment with attenuated NF-B nuclear translocation and DNA binding [78]. Treatment with trichostatin A (TSA) resulted in a dose dependent reduction of H157 lung cancer cells by apoptosis with nuclear fragmentation and an increase in the sub-G0/ G1 fraction. TSA initiated apoptosis by activation of the intrinsic mitochondrial and extrinsic/Fas/FasL system death pathways [94,95,95]. TSA is also a powerful NSCLC cell radiosensitizer, enhancing G2/M cell cycle arrest, promoting apoptosis, interfering with DNA damage repair and synergistically causing cell death when combined with other HDAC inhibitors, such as vorinostat [180, 231]. It has been shown that vorinostat inhibits telomerase activity by reducing hTERT expression [113] and decreases bcl-2 expression [100].
The first compound clinically used as an LSD1 inhibitor is tranylcypromine, a monoamine oxidase inhibitor approved more than 50 years ago for treatment-refractory depression. More potent and specific LSD1 inhibitors are presently under preclinical and early clinical development. The methylation of lysine 27 of histone H3, H3K27, is regulated by the enhancer of zeste homolog 2 (EZH2), the catalytic domain of the polycomb repressive complex 2 (PRC2). Trimethylation of H3K27 by EZH2 leads to silencing of PRC2 target genes that are involved in stem cell differentiation and embryonic development. EZH2 is overexpressed in a variety of cancers, including NSCLC. 3-Deazaneplanocin A (DZNep) is an EZH2 inhibitor that leads to reduced trimethylated H3K27 levels in breast cancer cells and the de-repression of aberrantly silenced genes [172].
Aberrant histone methylation is a relatively recently discovered feature in NSCLC, which is reflected in the scarceness of studies using agents affecting histone methylation. It was recently shown that EZH2 knockdown as well as indirect EZH2 inhibition using 3-deazaneplanocin A (DZNep) could prime NSCLC cell lines to the effect of the topoisomerase inhibitor etoposide [53]. Aberrant histone demethylation in NSCLC however is not extensively studied so far in NSCLC. LSD1 knockdown as well as LSD1 inhibition using pargyline suppressed invasion, migration, and proliferation in lung cancer specimens [211]. To our knowledge, there are as yet no studies investigating combination therapies for lung cancer using LSD1 inhibitors [172].
Changes in the number of methyl residues in lysine residues of H3K9, H3K27 and H3K36 through lysine methylation/demethylation is very important since it affects the expression of genes by loosening or tightening the attachment of DNA to the nucleosome [98].
3 DNA Methylation in Lung Cancer
DNA methylation is the most studied epigenetic regulatory mechanism. CpG island methylation is mediated by different DNA methyltransferases (DNMTs) that can lead to gene silencing. Three active DNMTs (DNMT1, DNMT3a, and DNMT3b) are in charge to transfer a methyl group from S-adenosyl-L-methionine to the CpG islands 5′-cytosine carbon [32, 55, 150] DNMT1 is primarily involved in the maintenance methylation after DNA replication, while DNMT3a and b are responsible of de novo DNA methylation [47, 118, 119, 235]. During the last years DNA methylation is gaining ground as a potential biomarker for diagnosis, staging, prognosis, and monitoring of response to therapy. The field of DNA methylation based markers for prognosis and diagnosis is still emerging and its widespread use in clinical practice needs to be implemented [83]. As DNA methylation is often considered an early event in carcinogenesis, tumor-specific methylation has a great potential to be used as a screening and/or diagnostic tool in a non-invasive and cost-effective way.
Hypermethylation includes tumor suppressor gene inactivation through promoter methylation, is a hallmark of lung cancer and tends to occur as an early event in carcinogenesis [21, 236]. Tumor suppressor genes can be inactivated through a combined ation of promoter methylation in one allele and the presence of mutation or deletion in the other; in dominantly acting suppressor gene loci inactivation of one allele is generally insufficient to lead to clonal selection, since the protein can still be produced from the other normal allele. However, there is also evidence that in some cases partial inactivation of one allele by promoter methylation can contribute to carcinogenesis and be sufficient for clonal selection [24].
Lung cancer involves an accumulation of genetic and epigenetic events in the respiratory epithelium. Mutations and copy number alterations play a well-known role in oncogenesis, though epigenetic alterations are, in fact, more frequent than somatic aberrations in lung cancer [28]. During the neoplastic progression from hyperplasia to adenocarcinoma, promoter methylation of specific tumor suppressor genes, along with the overall number of hypermethylated genes seems to be increased [115].
3.1 Tumor Suppressor Gene Inactivation Through Gene Promoter Methylation
Many of the tumor suppressor genes that are hypermethylated in lung cancer are found to be hypermethylated in other types of solid tumors as well. Moreover, some are specific, although many are not. In premalignant and malignant states, promoter methylation is commonly observed in genes involved with crucial functions, including cell cycle control, proliferation, apoptosis, cellular adhesion, motility, and DNA repair [108]. Up to now, there is some evidence for a CpG island methylator phenotype (CIMP), a tumor phenotype characterized by widespread hypermethylation of a panel of genes, in lung cancer [125,126,126, 131, 186]. This is not wholly surprising, since the group of enzymes that catalyze the covalent attachment of the methyl group to the cytosine base (DNMTs), are upregulated in NSCLC [94,95,95, 116].
3.2 lncRNAs and miRNAs Methylation in Lung Cancer
It has been recently shown that abnormal promoter methylation does not affect only protein coding genes but can also affect various noncoding RNAs that may play a role in malignant growth [128]. To identify which long non-coding RNAs (lncRNAs) are involved in non-small cell lung cancer (NSCLC), Feng et al. analyzed microarray data on gene expression and methylation and identified 8500 lncRNAs that are expressed differentially between tumor and non-malignant tissues; 1504 of these were correlated with mRNA expression. Two of the lncRNAs, LOC146880 and ENST00000439577, were positively correlated with expression of two cancer-related genes, KPNA2 and RCC2, respectively. High expression of these two lncRNAs was also associated with poor survival. Analysis of lncRNA expression in relation to DNA methylation has shown that LOC146880 expression was down-regulated by DNA methylation in its promoter [51].
MicroRNAs (miRNAs) also play an important role in cancer development and progression, altering several biological functions by affecting targets through either their degradation or suppression of protein encoded. It has been recently shown that miR-1247, is downregulated in various cancers, but its biological role in non-small-cell lung cancer (NSCLC) is unknown. Furthermore, Stathmin 1 (STMN1) was found to be an immediate and functional target of miR-1247. The expression of STMN1 was significantly increased in NSCLC cell lines but was decreased by 5-Aza treatment. In addition, miR-1247 upregulation partially inhibited STMN1-induced promotion of migration and invasion of A549 and H1299 cells. These results indicate that miR-1247 was silenced by DNA methylation. Therefore, miR-1247 and its downstream target gene STMN1 may be a future target for the treatment of NSCLC [234].
3.3 Genomic Hypomethylation
DNA hypomethylation at CpG dinucleotides was the first epigenetic abnormality to be identified in cancer cells, over three decades ago. The degree of hypomethylation of genomic DNA was shown to correlate with the severity of the cancer; genome-wide DNA methylation decreased as the tumor progressed from a benign proliferating mass to metastatic invasive cancer [217].
A possible explanation for the mechanism of reduced DNA methylation contribution to carcinogenesis is that hypomethylation of genomic DNA favors mitotic recombination between repetitive sequences resulting in chromosomal instability. Mitotic recombination normally occurs at a high frequency in human cells [60, 69]. Since recombination depends on the homology between nucleotide sequences, repetitive sequences are especially permissive to recombination events, resulting in gross chromosomal anomalies, including chromosomal rearrangements, deletions, and/or translocations [217].
Another mechanism through which DNA hypomethylation contributes to carcinogenesis is reactivation of transposable elements. It was shown already many years ago that SINEs and LINEs together make up approximately 45% of the human genome [106] and are usually methylated in normal tissues. LINEs belong to the class of transposable elements that lack LTRs at their ends. LINEs, which are part of the LINE-1 (or L1) family, constitute approximately 17% of the human genome and are the only transposable elements capable of autonomous transposition [17].
In lung cancer, genomic hypomethylation may be a late event in tumorigenesis in contrast to gene-specific hypermethylation, which can occur early during cancer development. However, currently there is not a clear consensus on the timing, as Anisowicz et al. [4] found that hypomethylation was associated with NSCLC progression from normal to lung cancer. DNA hypomethylation in lung cancers, as this was shown by high-resolution CpG methylation mapping, occur specifically at repetitive sequences [166], including heterochromatin repeats (e.g., satellite DNA), SINEs (short interspersed nuclear elements), LINEs (long interspersed nuclear elements), LTR (long terminal repeat) elements, and segmental duplications in subtelomeric regions. However cancer-specific hypomethylation at repeat regions was not conserved between the individual tumors indicating randomness for targeting repeat sequences for demethylation in cancer [57]. In NSCLC widespread hypomethylation has been associated with genomic instability [35] that could result in oncogene activation [49] and loss of imprinting [101]. In lung cancer, hypomethylation tends to occur at nuclear elements, long terminal repeat (LTR) elements, segmental duplicates, and subtelomeric regions. On the contrary, loss of methylation is much less common at non-repetitive sequences [166].
In addition to the genomic loss of methyl content, gene-specific hypomethylation has been reported for several loci, including MAGEA [58, 97], TKTL1 [86], BORIS [70, 167], DDR131 14-3-3s [160, 185], and TMSB10 [60]. MAGE overexpression with an associated loss of methylation is a common event in lung cancer, as it has been observed in 75–80% of NSCLC [81].
3.4 EMT and DNA Methylation
EMT is a fundamental and conserved process characterized by loss of cell adhesion and increased cell motility. EMT is essential for numerous developmental processes including mesoderm formation and neural tube formation and wound healing. However, initiation of metastasis involves invasion, which has many phenotypic similarities to EMT, including a loss of cell-cell adhesion and an increase in cell mobility [200].
EMT is regulated by a variety of growth factors including epidermal growth factor (EGF), platelet derived growth factors (PDGFs), fibroblast growth factor-2 (FGF-2), and transforming growth factor-beta (TGF-β) [85] and is characterized by the loss of CDH1 (E-cadherin), a trans-membrane protein that is required for adherent junctions [109]. Following the loss of epithelial markers, there is a corresponding increase in mesenchymal markers, for example VIM (vimentin), CDH12 (N-cadherin) FN1 (fibronectin), ACTA2 (alpha-smooth muscle actin), and increased activity of MMP (matrix metalloproteinases) [168, 221]. Recent studies have shown that a multilayer regulatory network of transcription factors controls EMT. The most studied network is the regulation through SNAIL (SNAI1 and SNAI2), ZEB (ZEB1 and ZEB2), and TWIST (TWIST1) family members, which are referred as EMT transcription factors (EMT-TF) [190].
In NSCLC, DNA methylation of a subset of genes related to EMT leads to their transcriptional inactivation [120]. One of the master regulators of EMT, TWIST, binds to the CDH1 promoter and recruits the CHD4/nucleosome remodeling and deacetylase complex (CHD4/NuRD complex, also known as Mi2/NuRD complex) by direct interaction to several of its components as MTA2, CHD4, and RBBP7 [56]. In addition, MTA2 directly recruits the histone deacetylase HDAC2. The TWIST/CHD4/NuRD complex represses CHD1 expression by nucleosome remodeling as well as deacetylation of histones. The biological relevance of this mechanism of transcription regulation was demonstrated within the context of metastasis of two types of cancers, lung and breast cancer, since depletion of the components of the TWIST/CHD4/ NuRD complex suppressed cell migration and invasion in cell culture and murine models of cancer metastasis. This work [56] shows that not only DNA methylation but also other chromatin modifications, as nucleosome remodeling and histone modifications, play a role during cancer metastasis.
4 Smoking and DNA Methylation
Some epigenetic alterations reported for lung cancer may be smoking-specific, since they occur at greater frequency in smokers and increase with increasing smoking duration and intensity [90, 123, 202]. Genes reported to undergo smoking specific promoter hypermethylation include APC, FHIT, RASSF1A, and CCND2 [50, 203]. Also, the frequency of promoter hypermethylation of p16INK4a, MGMT, RASSF1A, MTHFR, and FHIT is greater in the NSCLC tumors of smokers relative to nonsmokers [91, 123, 209]. Moreover, RARb, p16INK4a, FHIT, and RASSF1A promoter hypermethylation increases with increasing smoking intensity [3, 71, 228].
DNMT1 expression is elevated in smokers with lung cancer, likely due to tobacco-specific nitrosamines that reduce DNMT1 ubiquitination and degradation [118, 119]. Additionally, it is widely accepted tha smoking-induced chronic inflammation and increased reactive oxygen species generation lead to increased DNA methylation [144].
Damiani et al. [34] developed an in vitro model that mimics the field cancerization observed in chronic smokers and identified several epigenetic changes and their kinetics. More specifically, immortalized normal human bronchial epithelial cells (HBECs) were exposed for 12 weeks to two cigarette carcinogens; methylnitrosurea (MNU) and benzo(a)pyrenediolepoxide 1 (BPDE). Stable knockdown of DNMT1, but not DNMT3 prevented cell transformation after exposure to these carcinogens. HBECs transform to a fibroblast like mesenchymal form after 4 weeks of carcinogen exposure. Significant reductions in miR-200b and miR-200c, were observed at 4 weeks exposure and was sustained upon cell transformation at 12 weeks. Interestingly, these two microRNAs are involved in regulating and inhibiting the EMT. Further studies revealed that expression of these EMT-regulating microRNAs are initially reduced by transcriptionally inactive chromatin at 4 weeks, followed by cytosine methylation-mediated repression at their promoters [19].
Interestingly long-term exposure to carcinogenic stimuli would imply a later selection of existing clones, thus genes that are silenced due to the duration or amount of tobacco smoking, are likely later stage contributors to this disease. Experimentally, wide genomic hypomethylation and promoter hypermethylation of RASSF1A and RARb were observed when normal small-airway epithelial cells and immortalized bronchial epithelial cells were exposed to cigarette smoke condensate [125, 126]. There is also experimental evidence indicating that cigarette condensate decreases nuclear levels of H4K16ac and H4K2me3 in respiratory epithelial cells [133].
Conversely, RASSF2, TNFRSF10, BHLHB5, and BOLL have been reported to be hypermethylated more frequently in NSCLC of patients who never smoked [108]. Moreover, chronic inflammation, which occurs in response to cigarette smoking, also plays an important role in lung cancer development, stimulating cellular turnover and proliferation. Inflammation has long been associated with DNA methylation in lung cancer [11, 136]. There is evidence that reactive oxygen species, generated during chronic inflammation, target transcriptional repressors and lead to increased levels of DNA methylation [144].
Cigarette smoke also inhibits the metabolism and storage of folate [143]. It has been shown on studies based in experimental models, that nitrates, nitrous oxide, cyanates, and isocyanates found in tobacco smoke transform folate, a major source of methyl groups for 1-carbon metabolism, into a biologically inactive compound [1, 89]. In additional support of this, reduced serum folate levels have been observed in smokers relative to nonsmokers [145, 152]. One-carbon metabolism is a critical pathway in the DNA methylation process, and depletion of folate can impact negatively the availability of s-adenosylmethionine, the primary methyl donor in the cytosine methylation reaction. Consequently, folate deficiency can result in chromosomal damage through impaired nucleotide synthesis and aberrant DNA methylation [25, 48].
5 Hypermethylated Genes in Lung Cancer
DNA 5′-cytosine hypermethylation is an early event in lung carcinogenesis [28, 83]. Many genes are hypermethylated in lung cancer including p16, PAK3, NISCH, KIF1A, OGDHL, BRMS1, FHIT, CTSZ, CCNA1, NRCAM, LOX, MGMT, DOK1, SOX15, TCF21, DAPK, RAR, RASSF1, CYGB, MSX1, BNC1, CTSZ, and CDKN2A [6, 44, 46, 72, 80, 82, 140, 149, 176, 182, 191, 205, 215].
The percent of hypermethylation for each gene varies, for example p16 and MGMT are hypermethylated in 100% of patients with pulmonary SqCC up the 3 years before cancer diagnosis. p16 inhibits cyclin-dependent kinases 4 and 6, which after binding cyclin D1, phosphorylate and inactivate the retinoblastoma (Rb) tumor suppressor gene, blocking cell cycle progression [218]. p16 is lost in ~70% of lung cancer cases, often by promoter methylation, promoting the G1 to S phase transition [181]. Interestingly, p16 methylation occurs in normal-appearing epithelium from smokers and precursors lesions, and increases as the disease progresses [20]. The specific mechanisms by which each gene hypermethylation event promotes cancer vary, but most of them include repression of tumor suppressor genes with subsequent activation of genes promoting cell growth and cell cycle progression [6, 44, 46, 72, 80, 82, 140, 149, 176, 191, 205, 215].
Some of the most often studied hypermethylated genes in lung cancer include p16INK4a, RASSF1A, APC, RARb, CDH1, CDH13, DAPK, FHIT, and MGMT. Although p16INK4a is hypermethylated, mutated, or deleted frequently in NSCLC, with estimates for the prevalence of alteration of this gene around 60%, p14arf, which is also encoded on the CDKN2A gene, is inactivated much less commonly (8–30% of NSCLC) [54, 204]. On the other hand, p16INK4a is disrupted in less than 10% of SCLC patients. In addition, RASSF1A is deleted or hypermethylated in 30–40% of NSCLC and 70–100% of SCLC, FHIT is deleted or hypermethylated in 40–70% of NSCLC and 50–80% of SCLC and finally TSLC1 is hypermethylated in an estimated 85% of NSCLC [204].
Hypermethylation of CDKN2A has been identified in premalignant lesions, thus may occur early in the tumorigenesis of some lung cancers types [18]. Promoter methylation of RASSF1A, APC, ESR1, ABCB1, MT1G, and HOXC9 have been associated with stage I NSCLC [117] suggesting they also are an early event in lung cancer. CpG island methylation of homeobox-associated genes is also common in stage I lung cancer, appearing in nearly all early-stage tumors [165]. Conversely, other commonly hypermethylated genes, such as hDAB2IP, H-Cadherin, DAL-1, and FBN2, have been associated with advanced-stage NSCLC [29, 229], suggesting these changes may occur at a later point during cancer progression. However, it is important to note that later involvement does not preclude the importance of the modification in the development of the disease, as these modifications may play key roles in the ability of the cancer to continue to develop in its advanced state, to slide over host immunity or exogenous cancer treatments, or to metastasize locally. Furthermore, due to the heterogeneity and the unique molecular signature of lung cancer, it is critical that these generalized “temporal” observations are kept in perspective; an early event in 1 tumor may not occur until later on in another [108].
Promoter methylation of CDKN2A and PTPRN2 has been shown to be one of the earliest events in cellular hyperplasia. Subsequently, studies have shown aberrant promoter hypermethylation of RASSF1A, CDH13, MGMT, and APC in lung cancer [92, 117, 158, 226]. Methylation of SHOX2, in bronchial aspirates as a biomarker, was identified in a 250-patient case-control study with 78% sensitivity and 96% specificity [99]. Hypermethylation of each CDKN2A, CDX2, HOXA1, and OPCML individually distinguished lung adenocarcinoma from healthy donors with a sensitivity of 67–86% and a specificity of 74–82% and showed significant DNA methylation even in stage I tumor samples [206]. Moreover, hypermethylation of the DAPK promoter was found in 34% of lung cancer samples. Taking into consideration the different histological subtypes of NSCLC, DAPK promoter methylation was more frequently observed in squamous cell carcinoma than in adenocarcinoma and large cell carcinoma; however, these differences were not statistically significant [142].
In sputum, tumor cells can be identified by atypical cell morphology. Sputum collection is a procedure that can be done easily and non-invasively by the patient. However, sampling may be inadequate because of the presence of epithelial cells resulting in underestimation of the methylation level in cancer cells. Sputum cytology is still implemented as standard diagnostic tool for lung cancer diagnosis, although in developed countries, it was replaced by tumor biopsies/tumor cytology. Over the last decade, research on sputum cytology for risk assessment and recurrence of early lung cancer brought new insights and advanced highly sensitive molecular techniques [135].
Analysis of the RASSF1A and 3OST2 promoters methylation in sputum specimen demonstrated a combined sensitivity of 85% with a specificity of 74% [77]. Promoter methylation of 31 genes was also analyzed in sputum of lung cancer patients in two independent cohorts to define a gene unique methylation signature for lung cancer risk assessment [111]. Accurate diagnosis was made for 71–77% of the patients using the promoter methylation signature of seven of these genes (PAX5β, PAX5α, Dal-1, GATA5, SULF2, and CXCL14). Whang et al. observed 55% MLH1 promoter hypermethylation of the tumor samples obtained from stage I and II patients. Further evaluation demonstrated a similar promoter hypermethylation in 38% of the sputum samples. Finally, they reported a 72% concordance of sputum samples matched to tissue biopsies [213]. A different study found that CDKN2A was methylated in 80.2% of tumor tissues and showed a frequency of 74.7% in sputum specimens. Several studies have evaluated the correlation between tissue and sputum samples. Hypermethylation of the best studied gene, CDKN2A, seems to be higher in tumor samples than in sputum with an interquartile range of 84–37% to 74–32%, respectively [33, 40, 122].
In serum and plasma of cancer patients cell free DNA from necrotic and apoptotic cancer cells have been detected [12]. A lot of genes have been evaluated in lung cancer patients to identify specific and sensitive targets for early lung cancer detection in clinical trials. In NSCLC, 75–87% of serum samples corresponding to their matched tissue samples for promoter hypermethylation of RASSF1A, CDKN2A, RARb, CDH13, FHIT, and BLU. In a study evaluating lung cancer risk using this panel of six genes, a sensitivity of 73% and a specificity of 82% were reported, with a concordance between tumor tissues and corresponding matched plasma samples, of 75% [74]. Promoter methylation of CDKN2A, DAPK, PAX5b, and GATA5 was analyzed in blood but it was 0.2–0.6-fold lower than in tissue biopsy samples [23]. Subsequent studies have shown CDKN2A methylation in blood, but the results given are very different in different studies, varying from 22.2% to 75.7% [16, 195]. Hypermethylation for DAPK was found in 35% of the bronchial epithelium and in 41% of blood samples from smokers whereas the remaining samples from nonsmokers were unaffected, showing smoking−/lung cancer-associated methylation changes [169].
In a very recent study, Daugaard I et al. compared the genome-wide methylation pattern in tumor and tumor adjacent normal lung tissues from four lung adenocarcinoma patients using DNA methylation microarrays and identified 74 differentially methylated regions (DMRs), 15 of which were validated and can be targeted as biomarkers in LAC [36]. Another study demonstrated that SPAG6 and L1TD1 are tumor-specifically methylated in NSCLC DNA methylation is involved in the transcriptional regulation of these genes and tumor-cell growth suppressing properties of L1TD1 in NSCLC cells [2].
In the past, abberant estrogen receptor (ER) regulation has been associated with various lung pathologies, but so far its involvement in lung cancer initiation and/or progression has remained unclear. Tekpli et al., aimed to assess in vivo and in vitro ER expression and its possible epigenetic regulation in non-small cell lung cancer (NSCLC) samples and their corresponding normal tissues and cells, and they reported significantly lower ERα and ERβ expression levels in the NSCLC tissue samples compared to their normal adjacent tissue samples. They also found that in tumor and normal lung tissues, smoking was associated with decreased ER expression and that normal lung tissues with a low ERβ expression level exhibited increased smoking-related DNA adducts. Taken together, these results indicate that decreased ER expression mediated by DNA methylation may play a role in NSCLC development [199].
6 DNA Methylation Based Biomarkers
The virtually universal presence of DNA hypermethylation in all types of cancer makes it an ideal candidate tumor biomarker. Compared with other molecular marker classes such as mRNA and proteins, DNA methylation has many advantages. First, DNA methylation is a covalent modification of DNA, so it is chemically stable and can survive harsh conditions for long periods of time. Second, through simple procedures it can be readily amplifiable and easily detectable. In addition, contrary to cancer-specific mutations, which are relatively rare and present in different gene positions, the incidence of aberrant methylation of specific CGIs is much higher, and moreover such methylation can be discovered by genome-wide screening procedures. Finally, DNA methylation has been detected in a number of body fluids of patients with cancer. In lung cancer, aberrant DNA methylation can be detected in the ctDNA, in sputum, in bronchoalveolar lavage and saliva of patients [8].
DNA hypermethylation in lung cancer patients can be detected in a plethora of biological samples, including bronchoscopic washings/brushings, sputum samples, and blood (plasma and serum), all of which are less invasive and easier on the patient than a tumor biopsy [5]. The clinical significance of detecting methylation biomarkers in blood could facilitate the evaluation of tumor progression next to routine screening. Nevertheless, it could be an indication of invasiveness, reflecting an advanced tumor stage [135].
6.1 Early Detection
Lung cancer mortality could be reduced significantly with the early detection of the disease. However, only about 15% of lung tumors are localized in the time of diagnosis, with the majority presenting at an advanced stage [38]. Five-year survival for lung cancer is markedly better for early-stage patients, with a less than 10% 5-year survival for advanced-stage patients vs greater than 70% for early-stage patients [68]. Cytology is by far the gold standard method for lung cancer diagnosis in minimally-invasive respiratory samples, despite its low sensitivity. Spiral computed tomography has shown promise for the early detection of lung cancer, but it has a high false positive rate [52], with as many as 30% of indeterminate nodules identified by computed tomography found ultimately to be benign [79], indicating that there is a need for development of additional markers to increase specificity. As discussed previously, promoter hypermethylation can be an early event in lung carcinogenesis and, as such, may have utility in early detection of the disease.
Promoter hypermethylation of p16INK4a has been observed in NSCLC precursor lesions [115], and PTPRN2 promoter methylation is reported to be an early event in pulmonary adenocarcinoma, with detectable changes in the premalignant atypical adenomatous hyperplasia [177].
More important, some of these early epigenetic events can be detected by non- or minimally invasive sample collection techniques, the most important characteristic for cancer-screening applications. For example, aberrant DNA methylation can be detected in sputum [109, 127], bronchoalveolar aspirate/lavage [37, 43, 175] and saliva [75, 189] in patients with lung cancer. For example, CDKN2A and MGMT promoter methylation was detected in sputum as long as 3-years before lung cancer diagnosis [148] and promoter methylation of p16INK4a, MGMT, PAX5b, DAPK, GATA5, and in another study RASSF1A was detected in sputum 18 months before lung cancer diagnosis [22].
Diaz et al. aimed to identify epigenetic biomarkers with clinical utility for cancer diagnosis in minimally or non-invasive specimens to improve the accuracy of current technologies. They identified nine cancer-specific hypermethylated genes in early-stage lung primary tumors, four of which (BCAT1, CDO1, TRIM58 and ZNF177) presented consistent CpG island-hypermethylation compared to non-malignant tissue and were associated with transcriptional silencing. It was shown that this epigenetic signature achieved higher diagnostic efficacy in bronchial fluids as compared with conventional cytology for lung cancer diagnosis, indicating that minimally-invasive epigenetic biomarkers have emerged as promising tools for cancer diagnosis [41].
However, specificity can be an issue for some of these early markers; they can also be detected in individuals who never developed the disease, something that underscores the importance of multimarker panels. Interestingly, promoter methylation of p16INK4a has been detected in sputum from former and current smokers [21]. However, not all single markers are nonspecific, as exemplified by SHOX2 promoter methylation, which has demonstrated good sensitivity (68–78%) and specificity (95–96%) for NSCLC in bronchial aspirates (AUC, 86–94%) samples [43].
Breath capture methods are also evaluated for early detection of lung cancer. Breath capture methods can be based on direct breathing into an analysis platform or on the collection of exhaled breath through cooling devices (exhaled breath condensates, EBCs) [164]. EBC-based lung cancer diagnosis has recently become more relevant, especially since studies have reported that EBCs can also be used to detect DNA mutations and DNA methylation patterns in lung cancer patients [39]. A recent study demonstrated promoter hypermethylation of CDKN2A in EBC of 40% of the NSCLC patients that were analyzed using fluorescent quantitative methylation-specific PCR (F-MSP) [222]. However, DNA methylation of DAPK, PAX5beta, and RASSF1A has been also assayed in EBCs of lung cancer patients showing high variability between each individual [62]. The discrepancies between different reports might be explained through the fact that EBC is a highly diluted mixtures of compounds. Thus, EBC-based diagnosis of lung cancer requires appropriate stringent standardization protocols in order to reduce variability and increase sensitivity of the technique. Nevertheless, collecting EBCs is a promising new strategy of diagnosis of lung diseases, including lung cancer [135].
Three studies reported methylation of p16 and RARbata; two studies showed methylation of APC, RASSF1A, DAPK, SHP1P2, DLEC1, KLK10, and SFRP1. The other genes were reported to be methylated only once. The genes found to be hypermethylated in over 30% of NSCLC (based on at least two independent studies) were APC and RASSF1A. The methylation frequency of DAPK between different studies varied from 26.1% to 68.4%. Except for DAPK, the methylation frequencies of other genes had little differences across studies. Most of the studies involved controls; therefore, comparison of the data across cases and controls was possible. Methylation-specific PCR techniques have been employed by most of the studies to quantify the methylation statues of genes [104].
Many studies have demonstrated that hypermethylation in promoter region of RARb gene could be found with high prevalence in tumor tissue and autologous controls such as corresponding non-tumor lung tissue, sputum and plasma of the NSCLC patients, but due to the small number of subjects included in the individual study, the statistical power is limited. Hua et al. performed a meta-analysis using a systematic search strategy in PubMed, EMBASE and CNKI databases and calculated the pooled odds ratio (OR) of RARb promoter methylation in lung cancer tissue versus autologous controls. The results show a strong and significant correlation between tumor tissue and autologous controls of RARb gene promoter hypermethylation prevalence across studies, indicating that RARb promoter methylation may play an important role in carcinogenesis of the NSCLC [76].
Another team performed a meta-analysis to review the diagnostic ability of CDH13 methylation in NSCLC as well as in its subsets. Thirteen studies, including 1850 samples were included in this meta-analysis. In a validation stage, 126 paired samples from TCGA were analyzed and 5 out of the 6 CpG sites in the CpG island of CDH13 were significantly hypermethylated in lung adenocarcinoma tissues but none of the 6 CpG sites was hypermethylated in squamous cell carcinoma tissues. These pooled data showed that the methylation status of the CDH13 promoter is strongly associated with lung adenocarcinoma. The CDH13 methylation status could be a promising diagnostic biomarker for diagnosis of lung adenocarcinoma [157].
Han et al. investigated the correlation of hMLH1 promoter hypermethylation and NSCLC using 13 studies by comprising 1056 lung cancer patients via a meta-analysis. Initially, they observed that loss of hMLH1 protein expression was significantly associated with its promoter hypermethylation, hMLH1 gene inactivation through hypermethylation contributed to the tumorigenesis of NSCLC, and that there is a correlation between histologic subtypes/disease stages (TNM I + II vs III + IV) and hypermethylation status of hMLH1 gene.Finally, they found that NSCLC patients with hMLH1 hypermethylation and subsequent low expression levels of hMLH1 have a short overall survival period than those patients with normal expression of hMLH1 gene. Thus, they concluded that hMLH1 hypermethylation should be an early diagnostic marker for NSCLC and also a prognostic index for NSCLC. hMLH1 is an interesting therapeutic target in human lung cancers [63].
Abnormal miRNA expression and promoter methylation of genes detected in sputum may provide biomarkers for non-small lung cancer (NSCLC). In a recent study, they evaluated the individual and combined analysis of the two classes of sputum molecular biomarkers for NSCLC detection and they found that integrated analysis of 2 miRNAs (miR-31 and miR-210) and 2 genes (RASSF1A and 3OST2) yields higher sensitivity (87.3%) and specificity (90.3%) compared with the individual panels of the biomarkers (P < 0.05) [194].
6.2 Prognostic Biomarkers
Promoter methylation of RASSF1A [214, 227], PTEN, DAPK[198], p16INK4a [59, 94,95,95, 146], Wif-1, CXCL12 [197], DLEC1, MLH1 [179], CDH1, CDH [96], APC [26, 208], RUNX3 [227], SPARC and DAL1 have all been associated with NSCLC outcome [27, 196, 230]. In addition, DNMT1 overexpression in NSCLC is associated with decreased survival [94,95,95, 116] and DNMT3b, only in patients younger than 65 years [223]. Along similar lines, the CpG island methylator phenotype has also been correlated with prognosis in NSCLC. Relative to advanced-stage lung cancers, chemotherapeutic recommendations are not as clear for earlystage disease, with no true consensus regarding the optimal approach [193]. Early-stage lung cancer can be controlled locally, but exhibits a high recurrence rate. Completely resected stage IB and II tumors have a near-50% recurrence rate, with a median time to recurrence of 1 year [88]. Patients with stage IA tumors are less likely to experience a recurrence, although certain IA subsets have high recurrence rates [187]. Methylation of p16INK4a, RASSF1A, CDH13, and APC has been associated with early recurrence in surgically treated stage I NSCLCs [27]. The combination of FHIT and p16INK4a promoter methylation has also been associated with recurrence in stage I NSCLC [94,95,95].
The results of a metaanalysis suggest that FHIT hypermethylation is associated with an increased risk and worse survival in NSCLC patients. FHIT hypermethylation, which induces the inactivation of FHIT gene, plays an important role in the carcinogenesis and clinical outcome and may serve as a potential drug target of NSCLC [225]. Another recent study was the first to investigate SFRP3 expression and its potential clinical impact on non-small cell lung carcinoma (NSCLC). WNT signaling components present on NSCLC subtypes were preliminary elucidated by expression data of The Cancer Genome Atlas (TCGA). They identified a distinct expression signature of relevant WNT signaling components that differ between adenocarcinoma (LUAD) and squamous cell carcinoma (LUSC). Of interest, canonical WNT signaling is predominant in LUAD samples and non-canonical WNT signaling is predominant in LUSC. In line, high SFRP3 expression resulted in beneficial clinical outcome for LUAD but not for LUSC patients. Moreover, DNA hypermethylation of SFRP3 was evaluated in the TCGA methylation dataset resulting in epigenetic inactivation of SFRP3 expression in LUAD, but not in LUSC, and was validated by pyrosequencing of our NSCLC tissue cohort and in vitro demethylation experiments. Immunohistochemistry confirmed SFRP3 protein downregulation in primary NSCLC and indicated abundant expression in normal lung tissue. Thus, the above results indicate that SFRP3 acts as a novel putative tumor suppressor gene in adenocarcinoma of the lung possibly regulating canonical WNT signaling [173]. Functional analysis revealed that overexpressed STXBP6 in A549 and H1299 cells significantly decreased cell proliferation, colony formation, and migration, and increased apoptosis. Finally, significantly lower survival rates (P < 0.05) were observed when expression levels of STXBP6 were low, providing a basis for the genetic etiology of lung adenocarcinoma [112]. Moreover, recently Zhang et al. found that PAX6 gene was specifically methylated in NSCLC, and demonstrated the effect of promoter methylation of PAX6 gene on clinical outcome in NSCLC, indicating the methylated PAX6 may be useful biomarkers for prognostic evaluation in NSCLC [233]. Interestingly, it is found for the first time that TMEM196 acts as a novel functional tumour suppressor inactivated by DNA methylation and is an independent prognostic factor of lung cancer. Multivariate analysis showed that patients with TMEM196 expression had a better overall survival [127].
Targeted therapies can be successfully used in a subset of patients with lung adenocarcinomas (ADC), but they are not appropriate for patients with squamous cell carcinomas (SCC). In addition, there is a need for the identification of prognostic biomarkers that can select patients at risk of relapse in early stages. It has been shown that a high prometastatic serine protease TMPRSS4 expression is an independent prognostic factor in SCC. Similarly, aberrant hypomethylation in tumors correlates with high TMPRSS4 expression and could be used as an independent prognostic predictor in SCC. The inverse correlation between expression and methylation status was also observed in cell lines. In vitro studies showed that treatment of cells lacking TMPRSS4 expression with a demethylating agent significantly increased TMPRSS4 levels. In conclusion, TMPRSS4 is a novel independent prognostic biomarker regulated by epigenetic changes in SCC and a potential therapeutic target in this tumor type, where targeted therapy is still underdeveloped [212].
6.3 Methylated ctDNA as a Biomarker in Liquid Biopsy
Several studies have reported the potential of investigating tumor-specific methylation in blood for the screening and diagnosis of lung cancer. Determination of the methylation patterns of multiple genes to obtain complex ctDNA methylation signatures can contribute importantly to cancer development and/or progression. In recent years, methylation specific PCR has been successfully applied in the area of evaluating gene hypermethylation in the ctDNA, leading to highly sensitive and specific methodologies for NSCLC diagnosis.
6.3.1 Methylated ctDNA as a Marker for Early Diagnosis
Various gene promoters were found to be differentially methylated in ctDNA of lung cancer patients and healthy controls. These differences have been evaluated towards early detection of lung cancer and are summarized in Table 1. Epigenetically regulated genes have been evaluated for this purpose, such as Short stature homebox 2 (SHOX2) [100, 103], doublecortin like kinase 1 (DCLK1) [156], septin9 (SEPT9) [155], ras association domain family 1 isoform A (RASSF1A) and retinoic acid receptor B2 (RARB2) [154]. It is important to note that a large proportion of cases in these studies are late-stage cancers. Therefore, it has to be an inclusion of patients amenable to therapy in order to validate a biomarker useful for the screening and diagnosis of lung cancer [121].
Zhang Y et al. evaluated the methylation status of 20 tumor-suppressor genes in serum of NSCLC patients using methylation-specific PCR [232]. They report that nine genes (APC, CDH13, KLK10, DLEC1, RASSF1A, EFEMP1, SFRP1, RARbeta, and p16 (INK4A) were hypermethylated in NSCLC patients. The methylation frequencies in the plasma were consistent with those in the paired tumor tissues. The above results indicated that methylated alteration of multiple genes played important roles in NSCLC pathogenesis and the methylated genes in ctDNA might be potential candidate epigenetic biomarkers for NSCLC detection [54]. As the human 8-oxoguanine DNA glycosylase (hOGG1) gene promoter is frequently methylated in NSCLC, Qin et al. evaluated whether genetic or epigenetic alterations of hOGG1 are associated with increased risk of non-small cell lung cancer. The methylation profiles of peripheral blood mononuclear cell specimens from 121 NSCLC patients and 121 controls were determined through methylation-specific PCR of hOGG1. hOGG1 methylation-positive carriers had a 2.25-fold greater risk of developing NSCLC than methylation-free subjects. Furthermore, the demethylating agent 5 aza-2′-deoxycytidine restored hOGG1 expression in NSCLC cell lines. These data provide strong evidence of an association between peripheral blood mononuclear cell hOGG1 methylation and the risk of NSCLC in a Chinese population [159].
6.3.2 Methylated ctDNA as a Prognostic Marker
DNA methylation can be indicative of tumor aggressiveness and risk of cancer recurrence due to residual disease after surgical resection and/or chemotherapy. ctDNA has a short half-life (~2 h), and its persistence in the blood following surgery has been linked to poor prognosis [42]. In the context of early stage malignancies, prognostic biomarkers are urgently needed to distinguish patients who are cured with surgery alone, from those at high risk of disease recurrence who may benefit from adjuvant chemotherapy. The prognostic significance of gene promoter ctDNA methylation has been described in several studies, although most of them evaluate late-stage cancers, as summarized in Table 2.
Detection of methylated breast cancer metastasis suppressor-1 (BRMS1) and (sex determining region Y)-box 17 (SOX17) in operable and advanced NSCLC, was shown to have a negative impact on survival [9, 10]. In contrast, SFN (14–3-3 Sigma) promoter methylation was correlated with a reduced risk of death [163].
In SCLC evaluation of doublecortin-like kinase 1 (DCLK1) promoter region methylation may be useful in both early diagnosis and prediction of the course of lung cancer [156].
6.3.3 Methylated ctDNA in the Prediction and Monitoring of Response to Therapy
Several studies have reported the detection of tumor-specific methylation in plasma for tracking a patient’s response to therapy as summarized in Table 3. The value of methylated ctDNA in plasma to predict response to therapy has also been investigated, although it is important to distinguish cfDNA from leukocytic DNA, because DNA methylation marks are coupled tightly to cellular differentiation and vary by cell type [73].
Wang and colleagues observed that there is an elevated level of adenomatous polyposis coli (APC) and RASSF1A promoter methylation in ctDNA within 24 h after cisplatin-based therapy, consistent with chemotherapy-induced cell death [216]. Moreover, methylation status of SHOX2, RASSF1A and RARB2 has shown potential to monitor disease recurrence after surgery and chemotherapy [174]. A recent manuscript addresses the role of O6-methylguanine-DNA methyltransferase (MGMT) as a biomarker in the oncogenesis of cancer and the opportunity of turning this gene into a drugable target in neuroendocrine tumours of the lung. Studies in brain tumours conclude that MGMT promoter methylation is considered a strong predictive factor for a favourable outcome for treatment with temozolomide, e.g. alkylating agent. In NSCLC MGMT promoter methylation is not a prognostic and predictive factor, hence temozolomide has no place. Temozolomide can be considered a ‘personalized’ treatment if the predictive role of the gene is further confirmed [67]. Another example of the use of DNA methylation as a predictive biomarker, are patients with unmethylated checkpoint with forkhead and ring finger domains (CHFR) promoter who survived longer when receiving EGFR tyrosine kinase inhibitors as second-line treatment, compared to conventional chemotherapy [170]. Furthermore, Ramirez et al. found that 14-3-3 sigma methylation in pretreatment serum may be an important predictor of NSCLC outcome in patients treated with platinum based chemotherapy [163]. Another study profiled DNA methylation in SCLC, patient-derived xenografts (PDX) and cell lines at single-nucleotide resolution. DNA methylation patterns of primary samples are distinct from those of cell lines, whereas PDX maintain a pattern closely consistent with primary samples. Clustering of DNA methylation and gene expression of primary SCLC revealed distinct disease subtypes among primary patient samples with similar genetic alterations which were histologically indistinguishable. SCLC is notable for dense clustering of high-level methylation in discrete promoter CpG islands, in a pattern clearly distinct from other lung cancers and strongly correlated with high expression of the E2F target and histone methyltransferase gene EZH2. Pharmacologic inhibition of EZH2 in a SCLC PDX markedly inhibited tumor growth [153]. Finally, with the demonstration that combined epigenetic therapy has efficacy in lung cancer patients [84], future applications of methylated ctDNA for monitoring the activity of demethylating agents will soon come to the forefront [121]. Thus, without careful study design, blood-based methylation profiles can be confounded by variation in relative circulating proportions of leukocyte types associated with outcome, such as immune response [107].
7 Conclusions
DNA methylation is a very early step in tumorigenesis and analysis of DNA methylation in clinical samples is very informative. DNA methylation markers have potential as prognostic markers and, accordingly, have been studied and reported widely in the literature. It is now known that a variety of hypermethylated tumor suppressor genes is implicated in lung cancer oncogenesis and have been associated with prognosis. Moreover, detection of DNA methylated sequences in plasma samples is a very important liquid biopsy approach that allows continuous monitoring of tumor evolution in real ime, in a non-invasive way.
References
Abu Khaled M, Watkins CL, Krumdieck CL (1986) Inactivation of B12 and folate coenzymes by butyl nitrite as observed by NMR: implications on one-carbon transfer mechanism. Biochem Biophys Res Commun 135:201–207
Altenberger C, Heller C, Ziegler B, Tomasich E, Marhold M, Topakian T et al (2017) SPAG6 and L1TD1 are transcriptionally regulated by DNA methylation in non-small cell lung cancers. Mol Cancer 16:1
Andujar P, Wang J, Descatha A, Galateau-Sallé F, Abd-Alsamad I, Billon-Galland MA et al (2010) p16INK4A inactivation mechanisms in non-small-cell patients with lung cancer occupationally exposed to asbestos. Lung Cancer 67:23–30
Anisowicz A, Huang H, Braunschweiger KI, Liu Z, Giese H, Wang H et al (2008) A highthroughput and sensitive method to measure global DNA methylation: application in lung cancer. BMC Cancer 8:222
Ansari J, Shackelford RE, El-Osta H (2016) Epigenetics in non-small cell lung cancer: from basics to therapeutics. Transl Lung Cancer Res 5(2):155–171
Bailey-Wilson JE, Amos CI, Pinney SM, Petersen GM, de Andrade M, Wiest JS et al (2004) A major lung cancer susceptibility locus maps to chromosome 6q23-25. Am J Hum Genet 75:460–474
Baldwin DR, Duffy SW, Wald NJ, Page R, Hansell DM, Field JK (2011) UK lung screen (UKLS) nodule management protocol: modelling of a single screen randomised controlled trial of low-dose CT screening for lung cancer. Thorax 66(4):308–313
Balgkouranidou I, Liloglou T, Lianidou ES (2013) Lung cancer epigenetics: emerging biomarkers. Biomark Med 7(1):49–58
Balgkouranidou I, Chimonidou M, Milaki G, Tsarouxa EG, Kakolyris S, Welch DR et al (2014) Breast cancer metastasis suppressor-1 promoter methylation in cell-free DNA provides prognostic information in non-small cell lung cancer. Br J Cancer 110:2054–2062
Balgkouranidou I, Chimonidou M, Milaki G, Tsaroucha E, Kakolyris S, Georgoulias V et al (2016) SOX17 promoter methylation in plasma circulating tumor DNA of patients with non-small cell lung cancer. Clin Chem Lab Med 54:1385–1393
Balkwill F, Coussens LM (2004) Cancer: an inflammatory link. Nature 431:405–406
Bardelli A, Pantel K (2017) Liquid biopsies, what we do not know (yet). Cancer Cell 31(2):172–179
Barlesi F, Giaccone G, Gallegos-Ruiz MI, Loundou A, Span SW, Lefesvre P et al (2007) Global histone modifications predict prognosis of resected non small-cell lung cancer. J Clin Oncol 25(28):4358–4364
Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z et al (2007) High-resolution profiling of histone methylations in the human genome. Cell 129:823–837
Bartling B, Hofmann HS, Boettger T, Hansen G, Burdach S, Silber RE et al (2005) Comparative application of antibody and gene array for expression profiling in human squamous cell lung carcinoma. Lung Cancer 49:145–154
Bearzatto A, Conte D, Frattini M, Zaffaroni N, Andriani F, Balestra D et al (2002) p16(INK4A) Hypermethylation detected by fluorescent methylation-specific PCR in plasmas from non-small cell lung cancer. Clin Cancer Res 8(12):3782–3787
Beck CR, Garcia-Perez JL, Badge RM, Moran JV (2011) LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet 12:187–215
Belinsky SA (2005) Silencing of genes by promoter hypermethylation: key event in rodent and human lung cancer. Carcinogenesis 26:1481–1487
Belinsky SA (2015) Unmasking the lung cancer epigenome. Annu Rev Physiol 77:453–474
Belinsky SA, Nikula KJ, Palmisano WA, Michels R, Saccomanno G, Gabrielson E et al (1998) Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci U S A 95:11891–11896
Belinsky SA, Klinge DM, Dekker JD, Smith MW, Bocklage TJ, Gilliland FD et al (2005) Gene promoter methylation in plasma and sputum increases with lung cancer risk. Clin Cancer Res 11:6505–6511
Belinsky SA, Liechty KC, Gentry FD, Wolf HJ, Rogers J, Vu K et al (2006) Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high-risk cohort. Cancer Res 66:3338–3344
Belinsky SA, Grimes MJ, Casas E, Stidley CA, Franklin WA, Bocklage TJ et al (2007) Predicting gene promoter methylation in non-small-cell lung cancer by evaluating sputum and serum. Br J Cancer 96(8):1278–1283
Berger AH, Knudson AG, Pandolfi PP (2011) A continuum model for tumour suppression. Nature 476:163–169
Blount BC, Mack MM, Wehr CM, MacGregor JT, Hiatt RA, Wang G et al (1997) Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proc Natl Acad Sci U S A 94:3290–3295
Brabender J, Usadel H, Danenberg KD, Metzger R, Schneider PM, Lord RV et al (2001) Adenomatous polyposis coli gene promoter hypermethylation in non-small cell lung cancer is associated with survival. Oncogene 20:3528–3532
Brock MV, Hooker CM, Ota-Machida E, Han Y, Guo M, Ames S et al (2008) DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med 358:1118–1128
Brzezianska E, Dutkowska A, Antczak A (2013) The significance of epigenetic alterations in lung carcinogenesis. Mol Biol Rep 40:309–325
Chen H, Suzuki M, Nakamura Y, Ohira M, Ando S, Iida T et al (2005) Aberrant methylation of FBN2 in human non-small cell lung cancer. Lung Cancer 50:43–49
Cheng J, Blum R, Bowman C, Hu D, Shilatifard A, Shen S et al (2014) A role for H3K4 monomethylation in gene repression and partitioning of chromatin readers. Mol Cell 53:979–992
Chi P, Allis CD, Wang GG (2010) Covalent histone modifications – miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer 10:457–469
Christman JK (2002) 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene 21:5483–5495
Cirincione R, Lintas C, Conte D, Mariani L, Roz L, Vignola AM et al (2006) Methylation profile in tumor and sputum samples of lung cancer patients detected by spiral computed tomography: a nested case–control study. Int J Cancer 118(5):1248–1253
Damiani LA, Yingling CM, Leng S, Romo PE, Nakamura J, Belinsky SA (2008) Carcinogeninduced gene promoter hypermethylation is mediated by DNMT1 and causal for transformation of immortalized bronchial epithelial cells. Cancer Res 68:9005–9014
Daskalos A, Logotheti S, Markopoulou S, Xinarianos G, Gosney JR, Kastania AN et al (2011) Global DNA hypomethylation-induced deltaNp73 transcriptional activation in non-small cell lung cancer. Cancer Lett 300:79–86
Daugaard I, Dominguez D, Kjeldsen TE, Kristensen LS, Hager H, Wojdacz TK et al (2016) Identification and validation of candidate epigenetic biomarkers in lung adenocarcinoma. Sci Rep 6:35807
de Fraipont F, Moro-Sibilot D, Michelland S, Brambilla E, Brambilla C, Favrot MC (2005) Promoter methylation of genes in bronchial lavages: a marker for early diagnosis of primary and relapsing non-small cell lung cancer? Lung Cancer 50:199–209
Dela Cruz CS, Tanoue LT, Matthay RA (2011) Lung cancer: epidemiology, etiology, and prevention. Clin Chest Med 32:605–644
Dent AG, Sutedja TG, Zimmerman PV (2013) Exhaled breath analysis for lung cancer. J Thorac Dis 5(Suppl 5):S540–S550
Destro A, Bianchi P, Alloisio M, Laghi L, Di Gioia S, Malesci A et al (2004) K-ras and p16(INK4A)alterations in sputum of NSCLC patients and in heavy asymptomatic chronic smokers. Lung Cancer 44(1):23–32
Diaz-Lagares J, Mendez-Gonzalez D, Hervas M, Saigi MJ, Pajares D, Garcia AB et al (2016) A novel epigenetic signature for early diagnosis in lung cancer. Clin Cancer Res 22(13):3361–3371
Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M et al (2008) Circulating mutant DNA to assess tumor dynamics. Nat Med 14:985–990
Dietrich D, Kneip C, Raji O, Liloglou T, Seegebarth A, Schlegel T et al (2012) Performance evaluation of the DNA methylation biomarker SHOX2 for the aid in diagnosis of lung cancer based on the analysis of bronchial aspirates. Int J Oncol 40:825–832
Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M et al (2006) DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet 38:1378–1385
Ellinger J, Kahl P, von der Gathen J, Rogenhofer S, Heukamp LC, Gutgemann I et al (2010) Global levels of histone modifications predict prostate cancer recurrence. Prostate 70:61–69
Estécio MR, Yan PS, Ibrahim AE, Tellez CS, Shen L, Huang TH et al (2007) High-throughput methylation profiling by MCA coupled to CpG island microarray. Genome Res 17:1529–1536
Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E et al (2007) MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A 104:15805–15810
Fang JY, Xiao SD (2003) Folic acid, polymorphism of methyl-group metabolism genes, and DNA methylation in relation to GI carcinogenesis. J Gastroenterol 38:821–829
Feinberg AP, Vogelstein B (1983) Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 301:89–92
Feng Q, Hawes SE, Stern JE, Wiens L, Lu H, Dong ZM et al (2008) DNA methylation in tumor and matched normal tissues from non-small cell patients with lung cancer. Cancer Epidemiol Biomarkers Prev 17:645–654
Feng N, Ching T, Wang Y, Liu B, Lin H, Shi O et al (2016) Analysis of microarray data on gene expression and methylation to identify long non-coding RNAs in non-small cell lung cancer. Sci Rep 6:37233
Field JK, Baldwin D, Brain K, Devaraj A, Eisen T, Duffy SW et al (2011) CT screening for lung cancer in the UK: position statement by UKLS investigators following the NLST report. Thorax 66:736–737
Fillmore CM, Xu C, Desai PT, Berry JM, Rowbotham SP, Lin Y-J et al (2015) EZH2 inhibition sensitizes BRG1 and EGFR mutant lung tumours to TopoII inhibitors. Nature 520:239–242
Fischer JR, Ohnmacht U, Rieger N, Zemaitis M, Stoffregen C, Manegold C et al (2007) Prognostic significance of RASSF1A promoter methylation on survival of non-small cell patients with lung cancer treated with gemcitabine. Lung Cancer 56:115–123
Forde PM, Brahmer JR, Kelly RJ (2014) New strategies in lung cancer: epigenetic therapy for non-small cell lung cancer. Clin Cancer Res 20:2244–2248
Fu J, Qin L, He T, Qin J, Hong J, Wong J et al (2011) The TWIST/Mi2/NuRD protein complex and its essential role in cancer metastasis. Cell Res 21(2):275–289
Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW et al (2003) Induction of tumors in mice by genomic hypomethylation. Science 300(5618):489–492
Glazer CA, Smith IM, Ochs MF, Begum S, Westra W, Chang SS et al (2009) Integrative discovery of epigenetically derepressed cancer testis antigens in NSCLC. PLoS One 4:e8189
Gu J, Berman D, Lu C, Wistuba II, Roth JA, Frazier M et al (2006) Aberrant promoter methylation profile and association with survival in patients with non-small cell lung cancer. Clin Cancer Res 12:7329–7338
Gu Y, Wang C, Wang Y, Qiu X, Wang E (2009) Expression of thymosin beta10 and its role in non-small cell lung cancer. Hum Pathol 40:117–124
Gupta PK, Sahota A, Boyadjiev SA, Bye S, Shao C, O’Neill JP et al (1997) High frequency in vivo loss of heterozygosity is primarily a consequence of mitotic recombination. Cancer Res 57(6):1188–1193
Han W, Wang T, Reilly AA, Keller SM, Spivack SD (2009) Gene promoter methylation assayed in exhaled breath, with differences in smokers and lung cancer patients. Respir Res 10:86
Han Y, Shi K, Zhou SJ, Yu DP, Liu ZD (2016) The clinicopathological significance of hMLH1 hypermethylation in non-small-cell lung cancer: a meta-analysis and literature review. Onco Targets Ther 9:5081–5090
Han M, Xu W, Cheng P, Jin H, Wang X (2017) Histone demethylase lysine demethylase 5B in development and cancer. Oncotarget 8(5):8980–8991
Hayami S, Yoshimatsu M, Veerakumarasivam A, Unoki M, Iwai Y, Tsunoda T et al (2010) Overexpression of the JmjC histone demethylase KDM5B in human carcinogenesis: involvement in the proliferation of cancer cells through the E2F/RB pathway. Mol Cancer 9:59
Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD et al (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39:311–318
Hiddinga BI, Pauwels P, Janssens A, van Meerbeeck JP (2016) O6-Methylguanine-DNA methyltransferase (MGMT): a drugable target in lung cancer? Lung Cancer. pii:S0169-5002(16)30412-3
Hoffman PC, Mauer AM, Vokes EE (2000) Lung cancer. Lancet 355:479–485
Holt D, Dreimanis M, Pfeiffer M, Firgaira F, Morley A, Turner D (1999) Interindividual variation in mitotic recombination. Am J Hum Genet 65(5):1423–1427
Hong JA, Kang Y, Abdullaev Z et al (2005) Reciprocal binding of CTCF and BORIS to the NY-ESO-1 promoter coincides with derepression of this cancer-testis gene in lung cancer cells. Cancer Res 65:7763–7774
Hong YS, Roh MS, Kim NY et al (2007) Hypermethylation of p16INK4a in Korean non-small cell patients with lung cancer. J Korean Med Sci 22:S32–S37
Hoque MO, Kim MS, Ostrow KL et al (2008) Genomewide promoter analysis uncovers portions of the cancer methylome. Cancer Res 68:2661–2670
Houseman EA, Accomando WP, Koestler DC et al (2012) DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13:86
Hsu HS, Chen TP, Hung CH, Wen CK, Lin RK, Lee HC et al (2007) Characterization of a multiple epigenetic marker panel for lung cancer detection and risk assessment in plasma. Cancer 110(9):2019–2026
Hu YC, Sidransky D, Ahrendt SA (2002) Molecular detection approachesνfor smoking associated tumors. Oncogene 21:ν7289–ν7297
Hua F, Fang N, Li X, Zhu S, Zhang W, Gu J (2014) A meta analysis of the relationship between RARb Gene promoter methylation and non-small cell lung cancer. PLoS One 9(5):e96163
Hubers AJ, Brinkman P, Boksem RJ, Rhodius RJ, Witte BI, Zwinderman AH et al (2014) Combined sputum hypermethylation and eNose analysis for lung cancer diagnosis. J Clin Pathol 67(8):707–711
Imre G, Gekeler V, Leja A et al (2006) Histone deacetylase inhibitors suppress the inducibility of nuclear factor kappaB by tumor necrosis factor-alpha receptor-1 downregulation. Cancer Res 66:5409–5418
Isbell JM, Deppen S, Putnam JB Jr et al (2011) Existing general population models inaccurately predict lung cancer risk in patients referred for surgical evaluation. Ann Thorac Surg 91:227–233
Ito M, Ito G, Kondo M et al (2005) Frequent inactivation of RASSF1A, BLU, and SEMA3B on 3p21.3 by promoter hypermethylation and allele loss in non-small cell lung cancer. Cancer Lett 225:131–139
Jang SJ, Soria JC, Wang L et al (2001) Activation of melanoma antigen tumor antigens occurs early in lung carcinogenesis. Cancer Res 61:7959–7963
Johnstone RW (2002) Histone-deacetylase inhibitors: novel drugs for the treatment of cancer. Nat Rev Drug Discov 1:287–299
Jones PA, Baylin SB (2002) The fundamental role of epigenetic events in cancer. Nat Rev Genet 3:415–428
Juergens RA, Wrangle J, Vendetti FP et al (2011) Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov 1:598–607
Kalluri R, Weinberg RA (2009) The basics of epithelialmesenchymal transition. J Clin Invest 119(6):1420–1428
Kayser G, Sienel W, Kubitz B et al (2011) Poor outcome in primary non-small cell lung cancers is predicted by transketolase TKTL1 expression. Pathology 43:719–724
Ke XS, Qu Y, Rostad K, Li WC, Lin B, Halvorsen OJ et al (2009) Genome-wide profiling of histone h3 lysine 4 and lysine 27 trimethylation reveals an epigenetic signature in prostate carcinogenesis. PLoS One 4:e4687
Kelsey CR, Marks LB, Hollis D et al (2009) Local recurrence after surgery for early stage lung cancer: an 11-year experience with 975 patients. Cancer 115:5218–5227
Khaled MA, Krumdieck CL (1985) Association of folate molecules as determined by proton NMR: implications on enzyme binding. Biochem Biophys Res Commun 130:1273–1280
Kim DH, Nelson HH, Wiencke JK et al (2001) p16(INK4a) and histology-specific methylation of CpG islands by exposure to tobacco smoke in non-small cell lung cancer. Cancer Res 61:3419–3424
Kim H, Kwon YM, Kim JS et al (2004) Tumor-specific methylation in bronchial lavage for the early detection of non-small-cell lung cancer. J Clin Oncol 22:2363–2370
Kim JS, Han J, Shim YM, Park J, Kim DH (2005) Aberrant methylation of H-cadherin (CDH13) promoter is associated with tumor progression in primary nonsmall cell lung carcinoma. Cancer 104(9):1825–1833
Kim H, Kwon YM, Kim JS et al (2006a) Elevated mRNA levels of DNA methyltransferase-1 as an independent prognostic factor in primary nonsmall cell lung cancer. Cancer 107:1042–1049
Kim HR, Kim EJ, Yang SH et al (2006b) Trichostatin A induces apoptosis in lung cancer cells via simultaneous activation of the death receptor-mediated and mitochondrial pathway? Exp Mol Med 38:616–624
Kim JS, Kim JW, Han J, Shim YM, Park J, Kim DH (2006c) Cohypermethylation of p16 and FHIT promoters as a prognostic factor of recurrence in surgically resected stage I non-small cell lung cancer. Cancer Res 66:4049–4054
Kim DS, Kim MJ, Lee JY, Kim YZ, Kim EJ, Park JY (2007) Aberrant methylation of E-cadherin and H-cadherin genes in nonsmall cell lung cancer and its relation to clinicopathologic features. Cancer 110:2785–2792
Kim SH, Lee S, Lee CH et al (2009) Expression of cancer-testis antigens MAGE-A3/6 and NY-ESO-1 in non-small-cell lung carcinomas and their relationship with immune cell infiltration. Lung 187:401–411
Kimura H (2013) Histone modifications for human epigenome analysis. J Hum Genet 58:439–445
Kneip C, Schmidt B, Seegebarth A et al (2011) SHOX2 DNA methylation is a biomarker for the diagnosis of lung cancer in plasma. J Thorac Oncol 6:1632–1638
Komatsu N, Kawamata N, Takeuchi S et al (2006) SAHA, a HDAC inhibitor, has profound anti-growth activity against non-small cell lung cancer cells. Oncol Rep 15:187–191
Kondo M, Suzuki H, Ueda R et al (1995) Frequent loss of imprinting of the H19 gene is often associated with its overexpression in human lung cancers. Oncogene 10:1193–1198
Konecny M, Markus J, Waczulikova I et al (2016) The value of SHOX2 methylation test in peripheral blood samples used for the differential diagnosis of lung cancer and other lung disorders. Neoplasma 63:246–253
Kristensen LH, Nielsen AL, Helgstrand C, Lees M, Cloos P, Kastrup JS et al (2012) Studies of H3K4me3 demethylation by KDM5B/Jarid1B/PLU1 reveals strong substrate recognition in vitro and identifies 2,4-pyridine-dicarboxylic acid as an in vitro and in cell inhibitor. FEBS J 279:1905–1914
Kun N, Yujie J, Xuezhu Z (2015) Cell-free circulating tumor DNA in plasma/serum of non-small cell lung cancer. Tumour Biol 36(1):7–19
Labbé RM, Holowatyj A, Yang ZQ (2013) Histone lysine demethylase (KDM) subfamily 4: structures, functions and therapeutic potential. Am J Transl Res 6:1–15
Lander ES, Linton LM, Birren B et al (2009) Initial sequencing and analysis of the human genome. Nature 409(6822):860–921
Langevin SM, Kelsey KT (2013) The fate is not always written in the genes: epigenomics in epidemiologic studies. Environ Mol Mutagen 54:533–541
Langevin SM, Kratzke RA, Kelsey KT (2015) Epigenetics of lung cancer. Transl Res 165(1):74–90
Le Bras GF, Taubenslag KJ, Andl CD (2012) The regulation of cell-cell adhesion during epithelial-mesenchymal transition, motility and tumor progression. Cell Adh Migr 6(4):365–373
Lee SM, Park JY, Kim DS (2012 Aug) Methylation of TMEFF2 gene in tissue and serum DNA from patients with non-small cell lung cancer. Mol Cells 34(2):171–176
Leng S, Do K, Yingling CM et al (2012) Defining a gene promoter methylation signature in sputum for lung cancer risk assessment. Clin Cancer Res 18:3387–3395
Lenka G, Tsai MH, Lin HC, Hsiao JH, Lee YC, Lu TP et al (2017) Identification of methylation- driven, differentially expressed STXBP6 as a novel biomarker in lung adenocarcinoma. Sci Rep 7:42573
Li CT, Hsiao YM, Wu TC et al (2011) Vorinostat, SAHA, represses telomerase activity via epigenetic regulation of telomerase reverse transcriptase in non-small cell lung cancer cells. J Cell Biochem 112:3044–3053
Li X, Su Y, Pan J, Zhou Z, Song B, Xiong E et al (2013) Connexin 26 is down-regulated by KDM5B in the progression of bladder cancer. Int J Mol Sci 14:7866–7879
Licchesi JD, Westra WH, Hooker CM, Herman JG (2008) Promoter hypermethylation of hallmark cancer genes in atypical adenomatous hyperplasia of the lung. Clin Cancer Res 14:2570–2578
Lin RK, Hsu HS, Chang JW, Chen CY, Chen JT, Wang YC (2007) Alteration of DNA methyltransferases contributes to 5’CpG methylation and poor prognosis in lung cancer. Lung Cancer 55:205–213
Lin Q, Geng J, Ma K, Yu J, Sun J, Shen Z et al (2009) RASSF1A, APC, ESR1, ABCB1 and HOXC9, but not p16INK4A, DAPK1, PTEN and MT1G genes were frequently methylated in the stage I non-small cell lung cancer in China. J Cancer Res Clin Oncol 135(12):1675–1684
Lin RK, Hsieh YS, Lin P et al (2010a) The tobacco-specific carcinogen NNK induces DNA methyltransferase 1 accumulation and tumor suppressor gene hypermethylation in mice and lung cancer patients. J Clin Invest 120:521–532
Lin RK, Wu CY, Chang JW et al (2010b) Dysregulation of p53/Sp1 control leads to DNA methyltransferase-1 overexpression in lung cancer. Cancer Res 70:5807–5817
Lin SH, Wang J, Saintigny P, Wu CC, Giri U, Zhang J et al (2014) Genes suppressed by DNA methylation in non-small cell lung cancer reveal the epigenetics of epithelialmesenchymal transition. BMC Genomics 15:1079
Lissa D, Robles AI (2016) Methylation analyses in liquid biopsy. Transl Lung Cancer Res 5(5):492–504
Liu Y, An Q, Li L, Zhang D, Huang J, Feng X et al (2003) Hypermethylation of p16INK4a in Chinese lung cancer patients: biological and clinical implications. Carcinogenesis 24(12):1897–1901
Liu Y, Lan Q, Siegfried JM, Luketich JD, Keohavong P (2006) Aberrant promoter methylation of p16 and MGMT genes in lung tumors from smoking and never-smoking patients with lung cancer. Neoplasia 8:46–51
Liu Z, Zhao J, Chen XF et al (2008) CpG island methylator phenotype involving tumor suppressor genes located on chromosome 3p in non-small cell lung cancer. Lung Cancer 62:15–22
Liu F, Killian JK, Yang M et al (2010a) Epigenomic alterations and gene expression profiles in respiratory epithelia exposed to cigarette smoke condensate. Oncogene 29:3650–3664
Liu Z, Li W, Lei Z et al (2010b) CpG island methylator phenotype involving chromosome 3p confers an increased risk of nonsmall cell lung cancer. J Thorac Oncol 5:790–797
Liu WB, Han F, Jiang X, Chen HQ, Zhao H, Liu Y et al (2015) TMEM196 acts as a novel functional tumour suppressor inactivated by DNA methylation and is a potential prognostic biomarker in lung cancer. Oncotarget 6(25):21225–21239
Lujambio A, Portela A, Liz J, Melo SA, Rossi S, Spizzo R et al (2010) CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene 29(48):6390–6401
Machida EO, Brock MV, Hooker CM et al (2006) Hypermethylation of ASC/TMS1 is a sputum marker for late-stage lung cancer. Cancer Res 66:6210–6218
Margaritis T, Oreal V, Brabers N, Maestroni L, Vitaliano-Prunier A, Benschop JJ et al (2012) Two distinct repressive mechanisms for histone 3 lysine 4 methylation through promoting 3′-end antisense transcription. PLoS Genet 8:e1002952
Marsit CJ, Houseman EA, Christensen BC et al (2006) Examination of a CpG island methylator phenotype and implications of methylation profiles in solid tumors. Cancer Res 66:10621–10629
Martin C, Zhang Y (2005) The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 6:838–849
Marwick JA, Kirkham PA, Stevenson CS et al (2004) Cigarette smoke alters chromatin remodeling and induces proinflammatory genes in rat lungs. Am J Respir Cell Mol Biol 31:633–642
Massion PP, Carbone DP (2003) The molecular basis of lung cancer: molecular abnormalities and therapeutic implications. Respir Res 4:12
Mehta A, Dobersch S, Romero-Olmedo AJ, Barreto G (2015) Epigenetics in lung cancer diagnosis and therapy. Cancer Metastasis Rev 34:229–241
Meng X, Riordan NH (2006) Cancer is a functional repair tissue. Med Hypotheses 66:486–490
Minamiya Y, Ono T, Saito H et al (2010) Strong expression of HDAC3 correlates with a poor prognosis in patients with adenocarcinoma of the lung. Tumour Biol 31:533–539
Minamiya Y, Ono T, Saito H et al (2011) Expression of histone deacetylase 1 correlates with a poor prognosis in patients with adenocarcinoma of the lung. Lung Cancer 74:300–304
Miyanaga A, Gemma A, Noro R et al (2008) Antitumor activity of histone deacetylase inhibitors in non-small cell lung cancer cells: development of a molecular predictive model. Mol Cancer Ther 7:1923–1930
Mungall AJ, Palmer SA, Sims SK et al (2003) The DNA sequence and analysis of human chromosome 6. Nature 425:805–811
Nadal-Ribelles M, Mas G, Millan-Zambrano G, Sole C, Ammerer G, Chavez S et al (2015) H3K4 monomethylation dictates nucleosome dynamics and chromatin remodeling at stress-responsive genes. Nucleic Acids Res 43:4937–4949
Niklinska W, Naumnik W, Sulewska A, Kozłowski M, Pankiewicz W, Milewski R (2009) Prognostic significance of DAPK and RASSF1A promoter hypermethylation in non-small cell lung cancer (NSCLC). Folia Histochem Cytobiol 47(2):275–280
Northrop-Clewes CA, Thurnham DI (2007) Monitoring micronutrients in cigarette smokers. Clin Chim Acta 377:14–38
O’Hagan HM, Wang W, Sen S et al (2011) Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell 20:606–619
Ortega RM, Lopez-Sobaler AM, Gonzalez-Gross MM et al (1994) Influence of smoking on folate intake and blood folate concentrations in a group of elderly Spanish men. J Am Coll Nutr 13:68–72
Ota N, Kawakami K, Okuda T et al (2006) Prognostic significance of p16(INK4a) hypermethylation in non-small cell lung cancer is evident by quantitative DNA methylation analysis. Anticancer Res 26:3729–3732
Ozdağ H, Teschendorff AE, Ahmed AA et al (2006) Differential expression of selected histone modifier genes in human solid cancers. BMC Genomics 7:90
Palmisano WA, Divine KK, Saccomanno G et al (2000) Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res 60:5954–5958
Palmisano WA, Crume KP, Grimes MJ et al (2003) Aberrant promoter methylation of the transcription factor genes PAX5 alpha and beta in human cancers. Cancer Res 63:4620–4625
Patel K, Dickson J, Din S et al (2010) Targeting of 5-aza-2′- deoxycytidine residues by chromatin-associated DNMT1 induces proteasomal degradation of the free enzyme. Nucleic Acids Res 38:4313–4324
Pinskaya M, Morillon A (2009) Histone H3 lysine 4 di-methylation: a novel mark for transcriptional fidelity? Epigenetics 4:302–306
Piyathilake CJ, Macaluso M, Hine RJ, Richards EW, Krumdieck CL (1994) Local and systemic effects of cigarette smoking on folate and vitamin B-12. Am J Clin Nutr 60:559–566
Poirier JT, Gardner EE, Connis N, Moreira AL, de Stanchina E, Hann CL et al (2015) DNA methylation in small cell lung cancer defines distinct disease subtypes and correlates with high expression of EZH2. Oncogene 34(48):5869–5878
Ponomaryova AA, Rykova EY, Cherdyntseva NV et al (2013) Potentialities of aberrantly methylated circulating DNA for diagnostics and post-treatment follow-up of lung cancer patients. Lung Cancer 81:397–403
Powrózek T, Krawczyk P, Kucharczyk T et al (2014) Septin 9 promoter region methylation in free circulating DNA-potential role in noninvasive diagnosis of lung cancer: preliminary report. Med Oncol 31:917
Powrozek T, Krawczyk P, Nicos M, Kuznar-Kaminska B, Batura-Gabryel H, Milanowski J (2016) Methylation of the DCLK1 promoter region in circulating free DNA and its prognostic value in lung cancer patients. Clin Transl Oncol 18:398–404
Pu W, Geng X, Chen S, Tan L, Tan Y, Wang A et al (2016) Aberrant methylation of CDH13 can be a diagnostic biomarker for lung adenocarcinoma. J Cancer 7(15):2280–2289
Pulling LC, Divine KK, Klinge DM, Gilliland FD, Kang T, Schwartz AG et al (2003) Promoter hypermethylation of the O6- methylguanine-DNA methyltransferase gene: more common in lung adenocarcinomas from never-smokers than smokers and associated with tumor progression. Cancer Res 63(16):4842–4848
Qin H, Zhu J, Zeng Y et al (2017) Aberrant promoter methylation of hOGG1 may be associated with increased risk of non-small cell lung cancer. Oncotarget 8(5):8330–8341
Radhakrishnan VM, Jensen TJ, Cui H, Futscher BW, Martinez JD (2011) Hypomethylation of the 14-3-3sigma promoter leads to increased expression in non-small cell lung cancer. Genes Chromosom Cancer 50:830–836
Raji OY, Duffy SW, Agbaje OF et al (2012) Predictive accuracy of the Liverpool lung project risk model for stratifying patients for computed tomography screening for lung cancer: a case–control and cohort validation study. Ann Intern Med 157(4):242–250
Ramakrishnan S, Pokhrel S, Palani S, Pflueger C, Parnell TJ, Cairns BR et al (2016) Counteracting H3K4 methylation modulators Set1 and Jhd2 co-regulate chromatin dynamics and gene transcription. Nat Commun 7:11949
Ramirez JL, Rosell R, Taron M et al (2005) 14-3-3sigma methylation in pretreatment serum circulating DNA of cisplatin-plus-gemcitabine-treated advanced non-small-cell lung cancer patients predicts survival: the Spanish Lung Cancer Group. J Clin Oncol 23:9105–9112
Rattray NJ, Hamrang Z, Trivedi DK, Goodacre R, Fowler SJ (2014) Taking your breath away: metabolomics breathes life in to personalized medicine. Trends Biotechnol 32(10):538–548
Rauch T, Wang Z, Zhang X et al (2007) Homeobox gene methylation in lung cancer studied by genome-wide analysis with a microarray-based methylated CpG island recovery assay. Proc Natl Acad Sci U S A 104:5527–5532
Rauch TA, Zhong X, Wu X et al (2008) High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc Natl Acad Sci U S A 105:252–257
Renaud S, Pugacheva EM, Delgado MD et al (2007) Expression of the CTCF-paralogous cancer-testis gene, brother of the regulator of imprinted sites (BORIS), is regulated by three alternative promoters modulated by CpG methylation and by CTCF and p53 transcription factors. Nucl Acids Res 35:7372–7388
Richardson F, Young GD, Sennello R, Wolf J, Argast GM, Mercado P et al (2012) The evaluation of E-Cadherin and vimentin as biomarkers of clinical outcomes among patients with non-small cell lung cancer treated with erlotinib as second- or third-line therapy. Anticancer Res 32(2):537–552
Russo AL, Thiagalingam A, Pan H, Califano J, Cheng KH, Ponte JF et al (2005) Differential DNA hypermethylation of critical genes mediates the stage-specific tobacco smoke-induced neoplastic progression of lung cancer. Clin Cancer Res 11(7):2466–2470
Salazar F, Molina MA, Sanchez-Ronco M et al (2011) First-line therapy and methylation status of CHFR in serum influence outcome to chemotherapy versus EGFR tyrosine kinase inhibitors as second-line therapy in stage IV non-small-cell lung cancer patients. Lung Cancer 72:84–91
Sasaki H, Moriyama S, Nakashima Y et al (2004) Histone deacetylase 1 mRNA expression in lung cancer. Lung Cancer 46:171–178
Schiffmann I, Greve G, Jung M, Lübbert M (2016) Epigenetic therapy approaches in non-small cell lung cancer: update and perspectives. Epigenetics 11(12):858–870
Schlensog M, Magnus L, Heide T, Eschenbruch J, Steib F, Tator M et al (2016) Epigenetic loss of putative tumor suppressor SFRP3 correlates with poor prognosis of lung adenocarcinoma patients. Epigenetics
Schmidt B, Beyer J, Dietrich D et al (2015) Quantification of cell-free mSHOX2 plasma DNA for therapy monitoring in advanced stage non-small cell (NSCLC) and small-cell lung cancer (SCLC) patients. PLoS One 10:e0118195
Schmiemann V, Bocking A, Kazimirek M et al (2005) Methylation assay for the diagnosis of lung cancer on bronchial aspirates: a cohort study. Clin Cancer Res 11:7728–7734
Schuebel KE, Chen W, Cope L et al (2007) Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet 3:1709–1723
Selamat SA, Galler JS, Joshi AD et al (2011) DNA methylation changes in atypical adenomatous hyperplasia, adenocarcinoma in situ, and lung adenocarcinoma. PLoS One 6:e21443
Seligson DB, Horvath S, McBrian MA et al (2009) Global levels of histone modifications predict prognosis in different cancers. Am J Pathol 174:1619–1628
Seng TJ, Currey N, Cooper WA et al (2008) DLEC1 and MLH1 promoter methylation are associated with poor prognosis in non-small cell lung carcinoma. Br J Cancer 99:375–382
Seo SK, Jin HO, Woo SH et al (2011) Histone deacetylase inhibitors sensitize human non-small cell lung cancer cells to ionizing radiation through acetyl p53-mediated c-myc down-regulation. J Thorac Oncol 6:1313–1319
Shackelford RE, Kaufmann WK, Paules RS (1999) Cell cycle control, checkpoint mechanisms, and genotoxic stress. Environ Health Perspect 107(Suppl 1):5–24
Shames DS, Girard L, Gao B et al (2006) A genome-wide screen for promoter methylation in lung cancer identifies novel methylation markers for multiple malignancies. PLoS Med 3:e486
Shen X, Zhuang Z, Zhang Y, Chen Z, Shen L, Pu W et al (2015) JARID1B modulates lung cancer cell proliferation and invasion by regulating p53 expression. Tumour Biol 36:7133–7142
Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA et al (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119:941–953
Shiba-Ishii A, Noguchi M (2012) Aberrant stratifin overexpression is regulated by tumor-associated CpG demethylation in lung adenocarcinoma. Am J Pathol 180:1653–1662
Shinjo K, Okamoto Y, An B et al (2012) Integrated analysis of genetic and epigenetic alterations reveals CpG island methylator phenotype associated with distinct clinical characters of lung adenocarcinoma. Carcinogenesis 33:1277–1285
Shoji F, Haro A, Yoshida T et al (2010) Prognostic significance of intratumoral blood vessel invasion in pathologic stage IA non-small cell lung cancer. Ann Thorac Surg 89:864–869
Siegel R, Naishadham D, Jemal A (2013) Cancer statistics, 2013. CA Cancer J Clin 63:11–30
Simkin M, Abdalla M, El-Mogy M, Haj-Ahmad Y (2012) Differences in the quantity of DNA found in the urine and saliva of smokers versus nonsmokers: implications for the timing of epigenetic events. Epigenomics 4:343–352
Singh A, Settleman J (2010) EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene 29(34):4741–4751
Smith LT, Lin M, Brena RM et al (2006) Epigenetic regulation of the tumor suppressor gene TCF21 on 6q23-q24 in lung and head and neck cancer. Proc Natl Acad Sci U S A 103:982–987
Soini Y, Kosma VM, Pirinen R (2015) KDM4A, KDM4B and KDM4C in non-small cell lung cancer. Int J Clin Exp Pathol 8(10):12922–12928
Strauss GM, Herndon JE 2nd, Maddaus MA et al (2008) Adjuvant paclitaxel plus carboplatin compared with observation in stage IB non-small-cell lung cancer: CALGB 9633 with the Cancer and Leukemia Group B, Radiation Therapy Oncology Group, and North Central Cancer Treatment Group Study Groups. J Clin Oncol 26:5043–5051
Su Y, Fang HB, Jiang F (2016) Integrating DNA methylation and microRNA biomarkers in sputum for lung cancer detection. Clin Epigenetics 8:109
Suga Y, Miyajima K, Oikawa T, Maeda J, Usuda J, Kajiwara N et al (2008) Quantitative p16 and ESR1 methylation in the peripheral blood of patients with non-small cell lung cancer. Oncol Rep 20(5):1137–1142
Suzuki M, Hao C, Takahashi T et al (2005) Aberrant methylation of SPARC in human lung cancers. Br J Cancer 92:942–948
Suzuki M, Mohamed S, Nakajima T et al (2008) Aberrant methylation of CXCL12 in non-small cell lung cancer is associated with an unfavorable prognosis. Int J Oncol 33:113–119
Tang X, Khuri FR, Lee JJ et al (2000) Hypermethylation of the deathassociated protein (DAP) kinase promoter and aggressiveness in stage I non-small-cell lung cancer. J Natl Cancer Inst 92:1511–1516
Tekpli X, Skaug V, Bæra R, Phillips DH, Haugen A, Mollerup S (2016) Estrogen receptor expression and gene promoter methylation in non-small cell lung cancer – a short report. Cell Oncol (Dordr) 39(6):583–589
Thiery JP, Acloque H, Huang RY et al (2009) Epithelial-mesenchymal transitions in development and disease. Cell 139(5):871–890
Thinnes CC, England KS, Kawamura A, Chowdhury R, Schofield CJ, Hopkinson RJ (2014) Targeting histone lysine demethylases-progress, challenges, and the future. Biochim Biophys Acta 1839:1416–1432
Toyooka S, Maruyama R, Toyooka KO et al (2003) Smoke exposure, histologic type and geography-related differences in the methylation profiles of non-small cell lung cancer. Int J Cancer 103:153–160
Toyooka S, Suzuki M, Tsuda T et al (2004) Dose effect of smoking on aberrant methylation in non-small cell lung cancers. Int J Cancer 110:462–464
Toyooka S, Mitsudomi T, Soh J et al (2011) Molecular oncology of lung cancer. Gen Thorac Cardiovasc Surg 59:527–537
Toyota M, Ahuja N, Ohe-Toyota M et al (1999) CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A 96:8681–8686
Tsou JA, Galler JS, Siegmund KD, Laird PW, Turla S, Cozen W et al (2007) Identification of a panel of sensitive and specific DNA methylation markers for lung adenocarcinoma. Mol Cancer 6:70
Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P et al (2006) Histone demethylation by a family of JmjC domain-containing proteins. Nature 439:811–816
Usadel H, Brabender J, Danenberg KD et al (2002) Quantitative adenomatous polyposis coli promoter methylation analysis in tumor tissue, serum, and plasma DNA of patients with lung cancer. Cancer Res 62:371–375
Vaissiere T, Hung RJ, Zaridze D et al (2009) Quantitative analysis of DNA methylation profiles in lung cancer identifies aberrant DNA methylation of specific genes and its association with gender and cancer risk factors. Cancer Res 69:243–252
Van Den Broeck A, Brambilla E, Moro-Sibilot D et al (2008) Loss of histone H4K20 trimethylation occurs in preneoplasia and influences prognosis of non-small cell lung cancer. Clin Cancer Res 14:7237–7245
Vasilatos SN, Katz T, Oesterreich S, Wan Y, Davidson NE, Huang Y (2013) Crosstalk between lysine-specific demethylase 1 (LSD1) and histone deacetylases mediates antineoplastic efficacy of HDAC inhibitors in human breast cancer cells. Carcinogenesis 34:1196–1207
Villalba M, Diaz-Lagares A, Redrado M, de Aberasturi AL, Segura V, Bodegas ME et al (2016) Epigenetic alterations leading to TMPRSS4 promoter hypomethylation and protein overexpression predict poor prognosis in squamous lung cancer patients. Oncotarget 7(16):22752–22769
Wang YC, Lu YP, Tseng RC, Lin RK, Chang JW, Chen JT et al (2003) Inactivation of hMLH1 and hMSH2 by promoter methylation in primary non-small cell lung tumors and matched sputum samples. J Clin Invest 111(6):887–895
Wang J, Lee JJ, Wang L et al (2004) Value of p16INK4a and RASSF1A promoter hypermethylation in prognosis of patients with resectable non-small cell lung cancer. Clin Cancer Res 10:6119–6125
Wang M, Vikis HG, Wang Y et al (2007) Identification of a novel tumor suppressor gene p34 on human chromosome 6q25.1. Cancer Res 67:93–99
Wang H, Zhang B, Chen D et al (2015) Real-time monitoring efficiency and toxicity of chemotherapy in patients with advanced lung cancer. Clin Epigenetics 7:119
Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL et al (2005) Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet 37(8):853–862
Weinberg RA (1995) The retinoblastoma protein and cell cycle control. Cell 81:323–330
Weiner A, Hsieh TH, Appleboim A, Chen HV, Rahat A, Amit I et al (2015) High-resolution chromatin dynamics during a yeast stress response. Mol Cell 58:371–386
Xiang Y, Zhu Z, Han G, Ye X, Xu B, Peng Z et al (2007) JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. Proc Natl Acad Sci U S A 104:19226–19231
Xiao D, He J (2010) Epithelial mesenchymal transition and lung cancer. J Thorac Dis 2(3):154–159
Xiao P, Chen JR, Zhou F, Lu CX, Yang Q, Tao GH et al (2014) Methylation of P16 in exhaled breath condensate for diagnosis of non-small cell lung cancer. Lung Cancer 83(1):56–60
Xing J, Stewart DJ, Gu J, Lu C, Spitz MR, Wu X (2008) Expression of methylation-related genes is associated with overall survival in patients with non-small cell lung cancer. Br J Cancer 98:1716–1722
Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, Erdjument-Bromage H et al (2007) PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell 25:801–812
Yan W, Xu N, Han X, Zhou XM, He B (2016) The clinicopathological significance of FHIT hypermethylation in non-small cell lung cancer, a meta-analysis and literature review. Sci Rep 6:19303
Yanagawa N, Tamura G, Oizumi H et al (2003) Promoter hypermethylation of tumor suppressor and tumor-related genes in non-small cell lung cancers. Cancer Sci 94(7):589–592
Yanagawa N, Tamura G, Oizumi H et al (2007) Promoter hypermethylation of RASSF1A and RUNX3 genes as an independent prognostic prediction marker in surgically resected non-small cell lung cancers. Lung Cancer 58:131–138
Yanagawa N, Tamura G, Oizumi H, Endoh M, Sadahiro M, Motoyama T (2011) Inverse correlation between EGFR mutation and FHIT, RASSF1A and RUNX3 methylation in lung adenocarcinoma: relation with smoking status. Anticancer Res 31:1211–1214
Yano M, Toyooka S, Tsukuda K et al (2005) Aberrant promoter methylation of human DAB2 interactive protein (hDAB2IP) gene in lung cancers. Int J Cancer 113:59–66
Yoshino M, Suzuki M, Tian L et al (2009) Promoter hypermethylation of the p16 andWif-1 genes as an independent prognostic marker in stage IA non-small cell lung cancers. Int J Oncol 35:1201–1209
Zhang F, Zhang T, Teng ZH et al (2009) Sensitization to gamma-irradiation-induced cell cycle arrest and apoptosis by the histone deacetylase inhibitor trichostatin A in nonsmall cell lung cancer (NSCLC) cells. Cancer Biol Ther 8:823–831
Zhang Y, Wang R, Song H, Huang G, Yi J, Zheng Y et al (2011) Methylation of multiple genes as a candidate biomarker in non-small cell lung cancer. Cancer Lett 303(1):21–28
Zhang X, Yang X, Wang J, Liang T, Gu Y, Yang D (2015) Down-regulation of PAX6 by promoter methylation is associated with poor prognosis in non small cell lung cancer. Int J Clin Exp Pathol 8(9):11452–11457
Zhang J, Fu J, Pan Y, Zhang X, Shen L (2016) Silencing of miR-1247 by DNA methylation promoted non-small-cell lung cancer cell invasion and migration by effects of STMN1. Onco Targets Ther 9:7297–7307
Zhou Q, Agoston AT, Atadja P et al (2008) Inhibition of histone deacetylases promotes ubiquitin-dependent proteasomal degradation of DNA methyltransferase 1 in human breast cancer cells. Mol Cancer Res 6:873–883
Zochbauer-Muller S, Minna JD, Gazdar AF (2002) Aberrant DNA methylation in lung cancer: biological and clinical implications. Oncologist 7:451–457
Acknowledgements
This manuscript was supported by the IMI contract no. 115749 CANCER-ID (E.L, S.M).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this chapter
Cite this chapter
Mastoraki, S., Lianidou, E. (2017). DNA and Histone Methylation in Lung Cancer. In: Kaneda, A., Tsukada, Yi. (eds) DNA and Histone Methylation as Cancer Targets. Cancer Drug Discovery and Development. Humana Press, Cham. https://doi.org/10.1007/978-3-319-59786-7_15
Download citation
DOI: https://doi.org/10.1007/978-3-319-59786-7_15
Published:
Publisher Name: Humana Press, Cham
Print ISBN: 978-3-319-59784-3
Online ISBN: 978-3-319-59786-7
eBook Packages: MedicineMedicine (R0)