Abstract
The functions of the brain—such as sensory perception, memory formation, and behavioral responses—are based on the activity patterns of large numbers of interconnected neurons that form information-processing neuronal circuits. Most brain areas contain diverse types of neurons with specific morphology, gene expression profiles, input/output connectivity, and physiological response profiles. One major goal of neuroscience is to decipher connection patterns among different brain regions and cell types at the scale of the entire brain while keeping synaptic resolution. In this chapter, we first review various circuit tracing methods, and then introduce rabies virus (RV)-mediated transsynaptic tracing methods, which allow one to identify presynaptic neurons of genetically, anatomically, or functionally defined target neurons in a given brain area. This is achieved by genetic control of ‘starter’ cells, from which retrograde transsynaptic spread of RV occurs for only a single synaptic step. We will detail diverse methods that have been developed to restrict starter cells to a unique neuronal type. Following an introduction of RV transsynaptic tracing, the applications of these tools to three diverse biological systems in mice will be discussed: olfaction, neuromodulation, and motor control. From these examples, we will review how RV-mediated transsynaptic tracing has begun to decipher complex circuit architectures throughout the brain and spinal cord, and provides an important link between neuronal connections and circuit function.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
1 Introduction
In the mammalian brain, billions of neurons with highly diversified cell types form neuronal circuits by making trillions of synapses. One major goal of neuroscience is to decipher the basic organization and connection patterns among different brain regions and cell types. Diverse methods have been developed to contribute to this goal. Figure 4.1 shows a schematic comparison of multiple methods in terms of the limit of resolution in the presynaptic side (x-axis) and the postsynaptic side (y-axis).
1.1 Methods for Mapping of Neuronal Circuits at Region-to-Region Resolution
The first category (purple in Fig. 4.1) is characterized by region-to-region resolution in mapping neuronal circuits. For example, axon degeneration [also known as Wallerian degeneration after Waller’s histological finding that sectioning the nerve caused degeneration of axons distal to the injured site (Waller 1850)] has greatly contributed to our understanding of coarse brain organization. It cannot distinguish, however, multiple intermingled cell types on the presynaptic side or pinpoint the postsynaptic partners that the degenerating nerves innervated before induction of the injury.
The second category (cyan) is classical neuronal tracers, which can be injected locally into a brain region, without causing damage, to visualize region-to-region connections in an intact brain (for review, Vercelli et al. 2000; Nassi et al. 2015). Importantly, some of these tracers exhibit direction selectivity: retrograde tracers, such as Fluoro-gold (Schmued and Fallon 1986) and Retrobeads (Waselus et al. 2006), can be preferentially taken up by axons at the injection site and transported back to the cell bodies of these neurons, which may be located in distant brain areas. In contrast, anterograde tracers, represented by biotinylated dextran amine (Veenman et al. 1992) and isotope labeled amino acids (e.g., 3H-leucline; Cowan et al. 1972), can be taken up by cell bodies and dendrites at the injection site and then spread through the neuron to label their axons. These methods do not distinguish between different cell types intermingled at the injection site and therefore are primarily categorized as region-to-region resolution tracers. When combined with other histochemical and electrophysiological methods, however, these tracers can provide additional characterization of labeled neurons. For example, neurons labeled with a retrograde tracer can then be stained with cell type specific markers or have their electrophysiological properties determined, in order to better characterize cell types (for example, see Fig. 4.2a).
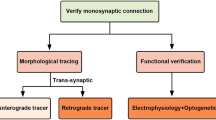
Various examples of mapping methods. a Retrograde labeling of neurons in the basolateral amygdala (BLA) projecting to either nucleus of accumbens (NAc, green) or central amygdala (CeA, red). Two different colors of Retrobeads were used in this study. Researchers analyzed electrophysiological properties of these labeled neurons following fear or reward conditioning. Adapted with permission from Namburi et al. (2015). b Axon projection mapping (Sect. 4.1.2) from serotonin neurons in the dorsal raphe nucleus (DR, B 2 ). In this sample, a Cre-dependent AAV vector (Fig. 4.3c–d) expressing GFP was injected into the DR of Sert-Cre mice in which Cre is expressed in serotonin neurons. Dense axon arborization was visualized throughout the brain, including central amygdala (CeA, B 1 ). Images were taken with permission from Allen Mouse Connectivity Atlas at http://connectivity.brain-map.org/ (sample#114155190). Also see Oh et al. (2014). c An example of rabies virus (RV)-mediated transsynaptic tracing (Sects. 4.1.3 and 4.2) starting from the serotonin neurons in the DR using Sert-Cre mice (C 2 ). Presynaptic neurons were labeled throughout the brain, including Tac2 positive neurons in the CeA (C 1 ). Adapted with permission from Weissbourd et al. (2014). d An example of paired recordings (Sect. 4.1.4) from a synaptically connected mitral cell (MC) and parvalbumin neuron (PVN) in an acute slice of the olfactory bulb. Left an action potential in the MC evokes excitatory postsynaptic current (EPSC) in the PVN. Right in the same pair of cells, an action potential in the PVN evokes an inhibitory postsynaptic current in the MC. This data unambiguously determines that these cells are monosynaptically connected. Adapted with permission from Kato et al. (2013). See Sect. 4.3.1. for details. e An example of ChR2-assisted circuit mapping (CRACM, Sect. 4.1.4) that shows direct monosynaptic excitatory input from the anterior cortical (AC) areas to serotonin and GABAergic neurons in the DR. EPSCs, generated by photostimulation, (1, blue bar) are abolished by application of DNQX (2), an AMPA receptor agonist. Top traces are the average of six trials from the same serotonin neurons. Bottom graph shows the change in EPSC amplitude over time. As exemplified in this case, CRACM determines monosynaptically connected neurons over a distance, with the ability to characterize their synaptic properties. Adapted with permission from Weissbourd et al. (2014). A anterior; P posterior; D dorsal; V ventral; L lateral; M medial. Scale bar in panel C 1 corresponds to 100 μm
1.2 Viral and Genetic Approaches for Axon Mapping
The third category in Fig. 4.1 (yellow) represents viral and genetic methods for mapping neuronal circuits that provide genetic control for cell types from which the tracing is initiated. One method in this category is axon projection mapping, which allows the labeling of entire axonal projections from genetically defined neurons located in a specific brain area. An example shown in Fig. 4.2b represents the axon projection mapping from serotonin neurons located in the dorsal raphe nucleus. GFP-labeled axons from these neurons are visualized throughout the brain, but are enriched in selected nuclei such as the central amygdala (CeA) (for details of serotonin circuit organization, see Sect. 4.4.1).
Utilizing differences in gene expression between cells is a powerful way to define cells types in the brain. To gain access to specific types of neurons expressing a gene X, researchers have generated a large number of transgenic and knock-in mouse lines where expression of a fluorescent protein gene is under the control of the promoter region of gene X (Fig. 4.3a). For example, to visualize inhibitory GABAergic neurons in the brain, GAD2-GFP mice were generated (Tamamaki et al. 2003), where GAD2 stands for glutamate decarboxylase 2 gene, which encodes the specific enzyme that synthesizes GABA. Although powerful for in vivo identification of GABAergic neurons, this mouse alone cannot be used to precisely map axons of individual GABAergic neurons, as it labels millions of neurons throughout the brain. To provide greater spatial resolution, a better approach utilizes local infection by viral vectors (Fig. 4.3b). Tables 4.1 and 4.2 summarize the viral vectors that are often used in circuit mapping. Among them, adeno-associated virus (AAV) is particularly useful, as it can be easily constructed, is safe to handle, and stably drives transgene expression in cells, without apparent cytotoxicity, for months to years. The major drawback of the AAV vector is its limited capacity; an AAV can only accommodate up to 4.7 kb of transgene, including the promoter and transcriptional stop cassette, into its genome. Therefore, in many cases, the limited AAV capacity does not allow inclusion of the full promoter region that is necessary for restricting transgene expression in the defined cell type. How can this problem be solved?
Schematic representation of viral and genetic technology and Cre or tTA dependent AAV vectors. a–c In this simplified neuronal circuit, three brain regions (columns) each containing three neurons are connected by axons (arrows) as indicated. Genetically targeted cells (for example, labeled by a fluorescent protein) are represented in green. Genetic methods can label a specific neuronal type in each brain area, but labeling is often widely distributed in the brain (a). Virus injection can provide better regional resolution, but it is difficult to regulate which cell types in the injection site express the transgene (b). In viral and genetic technology, Cre-dependent adeno-associated virus (AAV) is injected into the target brain region of a transgenic mouse line expressing Cre (shown as blue PacMan symbols). Local injection of AAV provides regional regulation and Cre expression provides the control of cell type-specific labeling. d Cre-dependent AAV using the Flip-Flop Excision (FLEx) switch. Gray and white triangles represent loxP and its mutant lox2272. Cre-mediated recombination between two loxP sites and between two lox2272 sites guarantees unidirectional inversion of a transgene. e Tetracycline transactivator (tTA)-dependent AAV driven by tetracycline responsive element (TRE) promoter. A transgene under the control of TRE promoter is activated by tTA, whose activity is blocked in the presence of doxycycline (Dox)
Researchers found an innovative solution by combining mouse genetics and viral vectors, to create an approach now recognized as viral and genetic technology (Fig. 4.3c). In 2003, the first Flip-Flop Excision (FLEx) switch (Fig. 4.3d) was invented (Schnutgen et al. 2003), where a transgene is initially placed in the opposite direction relative to the promoter activity, rendering it transcriptionally inactive. Only after Cre recombinase irreversibly inverts the FLEx switch can the transgene be expressed. This compact and efficient Cre-dependent switch has been widely used with great success in many applications of AAV vectors.
Typically, for virus-mediated axon projection mapping (Figs. 4.2b, and 4.3c, d), an AAV vector expressing a fluorescent marker protein (e.g., GFP) in a Cre-dependent manner is injected into a target brain area of a transgenic mouse, where Cre is expressed under a specific genetic promoter. Thus, the fluorescent marker will only be expressed in the specific cell types that also contain Cre. In this way, axons of the defined cell type in the defined brain region can be precisely traced throughout the brain. Many brain areas have been mapped in this way using a large number of different Cre lines (for example, see Allen Mouse Brain Connectivity Atlas at http://connectivity.brain-map.org/). Although this method provides specificity for the cell types that are labeled on the presynaptic side of a circuit, it cannot pinpoint the postsynaptic cell types to which the labeled axons connect. How might one further improve this spatial resolution?
1.3 Transsynaptic Tracing with Genetic Control
Transsynaptic tracing offers genetic control of the cells from which the tracing is initiated (called starter cells), and it can label synaptically connected cells throughout the nervous system. Labeled neurons can then be characterized by location, morphology, gene expression patterns, electrophysiological properties, and receptive fields. Figure 4.2c shows an example of rabies virus (RV)-mediated transsynaptic tracing, which will be discussed in greater detail in Sect. 4.2.
Transsynaptic tracing can be classified into two types: protein-based and virus-based methods. Protein-based tracers, represented by Wheat Germ Agglutinin (WGA) (Schwab et al. 1978), can be targeted into genetically defined cells, from which it is transported both to upstream (retrograde) and downstream (anterograde) neurons, presumably via synaptic connections (Yoshihara et al. 1999). As WGA is not toxic to cells, this method allows one to map neuronal circuits in intact animals in vivo with the potential to perform histochemical analyses on traced cell types. The major drawback of this strategy, however, is the fact that the tracer protein is easily diffused below a detection threshold because it is only generated in the starter cells. Also, this method cannot control the direction of labeling (upstream versus downstream). Finally, it is not fully established if the transneuronal transfer of WGA is indeed restricted to the synaptic connections.
The second category of transsynaptic tracing includes viral tracers. Table 4.2 summarizes the most commonly used vectors. Compared with protein-based tracers, viral tracers can be replicated in each step of cell-to-cell spread and therefore do not suffer from decreased labeling intensity due to diffusion. There are five basic properties to be considered when comparing viral tracers: (a) direction preference, (b) synapse specificity, (c) tropism, (d) cytotoxicity, and (e) availability of replication-conditional virus. Similar to classical chemical tracers, some viral tracers exhibit direction preference, that is, they dominantly move in either the anterograde or retrograde direction. For example, RV is a typical retrograde tracer (Ugolini 1995), whose direction preference is determined by the glycoprotein on the surface of the viral particles (Beier et al. 2011). A specific strain of pseudorabies virus (PRV) called Bartha strain also exhibits retrograde direction preference (Card et al. 1992). Although herpes simplex virus type 1 (HSV1) spreads both in retro- and anterograde directions, a specific strain of HSV1, named HSV-H129, is known to travel primarily in the anterograde direction (Zemanick et al. 1991; Sun et al. 1996). When viruses have clear direction specificity, researchers can easily map the labeled cells relative to the starter cells to create precise connection diagrams.
Although viral tracers travel from neuron to neuron (referred to as ‘transneuronal’), this alone does not guarantee synapse specificity of the viral spread, meaning that neurons can be labeled that are not directly connected to starter cells. Several viral vectors have been characterized that vary in terms of their synapse specificities for transneuronal spread. Thus far, synapse specificity of RV spread has been the most intensively characterized in various contexts, including in vivo circuit mapping (see Sects. 4.2.1 and 4.5.1) and ex vivo slice cultures (see Sect. 4.2.2).
When using viral tracers, it is also important to keep in mind that not all neurons in the brain are equally infected by a given viral tracer. The preference of viruses for certain cell types is called viral tropism. Currently the full tropism of transsynaptic viral tracers is not established and therefore caution is required when comparing labeling of different cell types in the brain. Furthermore, unlike AAV, most transsynaptic viral tracers that have been characterized so far are toxic to the infected cells. The cytotoxicity of viral tracers is an important issue if one wants to perform further analyses on infected neurons, such as electrophysiological recording or gene expression analysis, or use infected animals for behavioral experiments. Different viral tracers show a range of cytotoxicity (see Table 4.2). For example, cytotoxicity of RV is low to modest and therefore researchers can conduct Ca2+ imaging of RV-positive neurons several weeks after the infection (Osakada et al. 2011; Reardon et al. 2016). In contrast, HSV or PRV is usually more toxic, with signs of toxicity observed in the infected cells within a few days (Ugolini et al. 1987; Ugolini 2011).
Lastly, replication-conditional viral tracers are very useful for controlling viral spread. Wild-type viral tracers have full capacity to replicate (referred to as replication-competent). For some viral tracers, a single gene has been identified that is necessary for viral replication. This has enabled researchers to delete or transcriptionally inactivate that gene to generate a crippled virus that cannot replicate or propagate. However, replication of the virus can still occur if the missing gene is supplied in trans, or transcriptional inactivation of the gene is removed. This modified virus is called a replication-conditional virus. For example, replication-conditional HSV-H129 (Lo and Anderson 2011) and Bartha strain of PRV (DeFalco et al. 2001) were generated by transcriptional silencing of an essential thymidine kinase (tk) gene. Once the transcription of tk gene from the virus genome is restored by Cre-mediated recombination, the functional virus particles are generated in the Cre+ starter cells and spread in the anterograde (HSV-H129) or retrograde (PRV-Bartha) direction. As we will discuss in Sect. 4.2, replication-conditional RV is generated by deleting a single essential glycoprotein gene from the RV genome. By supplying rabies glycoprotein in trans only in the starter cells, viral replication is restricted to the starter cells, and therefore allows monosynaptic spread of the virus from these cells. Replication-conditional viral tracers offer higher resolution and precision in mapping neuronal connections.
1.4 Electrophysiological Methods for Mapping Neuronal Circuits
In Fig. 4.1, the fourth category (green) represents electrophysiological methods to map circuits. For local circuits, applying patch-clamp methods in brain slice preparations, combined with genetic labeling of neurons, can identify synaptically connected pairs of cells and also provides information regarding their cellular identity. Figure 4.2d shows a paired recording from a parvalbumin positive interneuron that is reciprocally connected to a mitral cell in the mouse olfactory bulb (OB) (Kato et al. 2013). This method allows highly precise mapping of connected neurons, although it can only be applied to a local circuit. In principle, bulk stimulation of axon bundles while recording from the postsynaptic neuron can map long-range connections with region-to-cell resolution. However, this approach cannot distinguish between intermingled axons from different cell types in the presynaptic structure that may be innervating the postsynaptic cell. The development of optogenetics (Deisseroth 2015) has greatly improved the resolution on the presynaptic side. Channelrhodopsin-2 (ChR2), a light activated cation channel isolated from a green alga, can be targeted into genetically defined neurons, for example by injecting Cre-dependent AAVs expressing ChR2 into a transgenic animal where Cre is expressed in specific classes of cells (Fig. 4.3c, d). In this way, the cell bodies and axons of a defined cell type, located in a defined brain region, can be activated by light. Simultaneously, one can monitor electrical responses in the presumed postsynaptic neurons with patch electrodes. If there is a monosynaptic connection between the ChR2-expressing neurons and the neuron being recorded from, an excitatory or inhibitory postsynaptic current should be observed immediately (within several milliseconds) after the onset of the light stimulation. By observing how long it takes the postsynaptic cell to respond after ChR2 activation, researchers can distinguish between monosynaptic and polysynaptic connections. This method, referred to as ChR2-Assisted Circuit Mapping (CRACM) (Petreanu et al. 2007), can identify pre- and postsynaptic partners, and provide additional information regarding their synaptic properties and cell types (defined by gene expression patterns). As an example, Fig. 4.2e shows a direct excitatory monosynaptic connection from frontal cortex pyramidal cells to serotonin neurons in dorsal raphe nucleus (Weissbourd et al. 2014). In this case, the distance between the connected cells is >7 mm. CRACM can also characterize which subcellular structures on the postsynaptic side are targeted by the ChR2-expressing axons, if light activation is restricted to a very small brain region. Given these advantages, CRACM is a standard way to map synaptic connections in many brain regions. One major drawback of CRACM is its low-throughput nature: in a single recording experiment, only one type of neuron can be labeled on the presynaptic side, while electrophysiological methods limit the number of neurons sampled on the postsynaptic side.
1.5 Ultrastructural Method to Map Synaptic Connections
The last categories of Fig. 4.1 (blue) are ultrastructural methods, represented by electron microscopy (EM). EM is a highly reliable method to determine if two cells form synaptic connections. In principle, complete reconstructions of serial EM images can reveal neuronal circuits with synaptic resolution throughout the brain, a method that has been established in nematode Caenorhabditis elegans (White et al. 1986). To generate a 3D reconstruction connection map, thin sections (usually less than 50 nm) are collected and imaged. Then, neuronal structures are extracted from individual images, consecutive images are aligned, and individual neuronal segments across many sections are reconstructed into a 3D volume. Finally, each synaptic contact is assigned to a defined pair of neurons. For large nervous systems such as entire mouse and human brain, these procedures are not only labor intensive, but also require very high precision in each step, as small errors in stacking many thousands of images could accumulate and lead to incorrect connection diagrams. Although this approach is currently only applied to a small piece of brain tissue (up to a few hundred microns, for examples, Helmstaedter et al. 2013; Kasthuri et al. 2015), given rapid advances in computer science, most of the above procedures and quality controls could be fully automated in the future.
2 Rabies Virus-Mediated Transsynaptic Tracing with Genetic Control
In this section, we will focus on transsynaptic tracing methods introduced in Sect. 4.1.3, starting with a brief history. Then we will introduce the principle of monosynaptic restriction of RV tracing, followed by various ways to apply this method to in vivo tracing experiments.
2.1 Development of Replication-Conditional Rabies Tracer
Early in the twentieth century, a limited number of studies suggested that certain types of neurotrophic viruses can travel across neuronal pathways, e.g., Goodpasture and Teague (1923). In the 1980s, researchers started to exploit replication-competent viral tracers to map neuronal pathways from peripheral body tissues to the central brain (for review Vercelli et al. 2000; Nassi et al. 2015; Ugolini 2011). In these studies, HSV1 and PRV (Table 4.2) were widely used in rodents and non-human primates. However, major pitfalls of these viral tracers are that they induce neuronal degeneration and possibly spread via nonspecific spillover of viral particles from infected neurons to nonconnected nearby cells (Ugolini et al. 1987). An important advancement was made in 1995 when a detailed timecourse for RV spread was characterized (Ugolini 1995). In this study, retrograde tracing by RV (challenge virus standard, or CVS strain) was initiated from hypoglossal motor neurons (MNs) that control tongue movement in rat, and viral spread was followed to the brainstem second-order neurons and higher order motor control regions. The results established two important properties of RV: low toxicity and no ‘suspicious’ labeling that would indicate nonsynaptic infection. Initially infected MNs were totally intact in their size and morphology at later stages when third-order neurons were infected in the brain. Also, RV-infected hypoglossal MNs did not spread virus to the inferior olive, which the RV-infected MNs passed through without forming synaptic connections. By contrast, an equivalent experiment using HSV1 did intensively label the inferior olive, presumably through nonspecific spillover of the virus. Together, this pioneering study highlighted that RV is a specific retrograde tracer with a long asymptomatic period.
The next landmark event for transsynaptic tracing techniques was the development of replication-conditional RV, which later allowed genetic control of starter cells and monosynaptic tracing (Wickersham et al. 2007a; Sect. 4.2.2). To understand how this was achieved, it is useful to cover some basic facts of RV virology (Fig. 4.4a). RV is a single-strand negative RNA virus (meaning the genome of RV is complementary to its mRNA) with a small genome (about 12 kb) encoding only five proteins: a nucleoprotein (N), a matrix protein (M), an RNA-dependent RNA polymerase (L), a polymerase cofactor phosphorylated protein (P), and a rabies glycoprotein (RG). The core of the virion consists of helically arranged genomic RNA associated with N, P, and L. These proteins support initial replication of the RV genome in the host cell before RV’s own gene transcription. The core is surrounded by M and lipid bilayers originated from the host cell, on which RG is anchored. Thanks to this simple organization, the functional viral particles can be de novo generated in cultured cells by introducing plasmids containing the RV genome and some of the RV genes listed above (Schnell et al. 1994; Fig. 4.4b, c). This made it possible to genetically manipulate the viral genome: for example, inserting a marker transgene (Mebatsion et al. 1996a) or deleting a rabies gene (Mebatsion et al. 1996b). Using this technique, a complete loss-of-function mutant was generated from an attenuated RV strain called Street Alabama Dufferin (SAD) B19 by deleting the RG gene. Because RG is necessary for efficient budding of RV (Mebatsion et al. 1996b), simple deletion of RG from the RV genome made it difficult to recover viral particles (with naked envelope) from the cultured cells. Researchers, therefore, added back the missing RG in trans via a plasmid transfected into the cultured cells, which allowed the mutant RV genome to be packed into an intact viral envelope (Fig. 4.4d). Hereafter, we will refer to this virus as RVdRG+RG, where the italic dRG represents the genome of the RV with deletion of RG, and +RG indicates the envelope protein supplied in trans. When RVdRG+RG was injected into rat and mouse brains, initial infection occurred normally, since the virus was coated with normal RG, allowing the virus to interact with rabies receptors expressed in the mammalian nervous system (Lafon 2005). However, transsynaptic spread was completely abolished and infected animals were still healthy 11 days after infection, while control animals that had received the same amount of wild-type RV were killed by rabies (Etessami et al. 2000). This experiment clearly demonstrated that RG is necessary for transsynaptic spread of RV and that RVdRG+RG is a valuable nonpathogenic, replication-conditional tracer for in vivo applications.
Generation of replication-conditional RV and the monosynaptic tracing technique. a–f Schematic flow showing how to generate RV mutant virus that is used for monosynaptic tracing. For details, see text. A wild type RV particle is shown in the top left. a Cloning the RV genome into a plasmid. b Genetic modification of the RV genome to delete (del) RG and insert GFP. c Transfection of the mutant RV genome plasmid, as well as plasmids for N, P, L, and RG into a cultured host cell. d Budding of the mutant particle RVdRG-GFP+RG. e Infection of RVdRG-GFP+RG into a BHK-EnvA cell that stably expresses EnvA. f Budding of the pseudotyped RVdRG-GFP+EnvA. This particle is used for monosynaptic tracing. g An example of RV-mediated monosynaptic tracing using ex vivo slice cultures. In this sample, TVA, RG and DsRed2-expressing red neurons in hippocampal slice culture are selectively infected by RVdRG-GFP+EnvA (shown in yellow) to become a starter cell. Many putative presynaptic neurons of starter cells were labeled with GFP. Right schematic of a starter cell. Images are taken with permission from Wickersham et al. (2007a)
2.2 Development of Monosynaptic Tracing Technique
The key idea behind introducing RV as a monosynaptic tracing technique is to provide in vivo trans-complementation of replication-conditional RVdRG, by supplying the missing RG as a transgene. If RG is expressed in neurons of interest, and these neurons are then infected with RVdRG, then they can produce RVdRG+RG particles by trans-complementation (as in cultured cells, see Fig. 4.4d). According to the natural tropism of RG, these particles can spread retrogradely and transsynaptically to the presynaptic neurons, where proliferation of RVdRG occurs. However, if these second order presynaptic neurons do not express RG, RVdRG cannot spread to third-order neurons, because RG is essential for viral spread from neuron to neuron. In this way, this approach can identify monosynaptic connections that are presynaptic to the initially infected neurons of interest.
To make this technique widely applicable, two problems must be solved: unequivocal visualization of RVdRG-infected neurons, and specific introduction of RVdRG into RG-expressing neurons of interest. The first problem was solved by introducing a fluorescent protein gene (e.g., GFP) into the RVdRG genome plasmid, resulting in RVdRG-GFP (Wickersham et al. 2007b; Fig. 4.4b). When coated with RG, these viral particles can efficiently label neurons, including their fine subcellular structures, such as dendritic spines. This allows unambiguous detection of RV-infected neurons in fixed sections, in time-lapse imaging of cultured neurons, or even in vivo via two-photon microscopy. The second problem was also elegantly solved by using a virology technique called pseudotyping, that is, altering the envelope proteins of a virus to change its tropism. The envelope protein of avian sarcoma and leucosis virus, called EnvA, restricts viral infection to cells expressing the corresponding receptor, TVA, a protein which is found in birds but not in mammals (Bates et al. 1993; Young et al. 1993). When a cell line constitutively expressing EnvA is infected with RVdRG-GFP, mutant RV particles coated with EnvA (referred to as RVdRG-GFP+EnvA) are released into the culture medium (Fig. 4.4f). Alone, this virus cannot infect mammalian cells, since they do not express TVA. However, it can infect genetically modified target neurons where expression of a TVA transgene has been transduced.
As a proof-of-principle, transduction of TVA and RG was introduced into a very small number of neurons in hippocampus slice culture, along with a red fluorescent marker, DsRed2. When RVdRG-GFP+EnvA was applied to the culture medium, starter cells that expressed both DsRed2 (a marker for co-expression of RG and TVA) and GFP (from RV) were generated. These neurons, labeled yellow in the culture, specifically promoted de novo production of RVdRG-GFP+RG. Dozens of neurons labeled with GFP alone surrounded the starter cells, suggesting that they were transsynaptically labeled from at least one of the starter cells (Fig. 4.4g; Wickersham et al. 2007a). Along with negative control experiments, these experiments in slice culture neurons established that RVdRG-GFP+EnvA infection is specific to TVA expressing neurons, and that RG is necessary and sufficient for transsynaptic spread of RV. Importantly, monosynaptic restriction of RV spread could be validated electrophysiologically. Using a slice with very sparse starter cells (yellow) and putative presynaptic partners (green), paired recording between these labeled neurons validated that 9 out of the 11 recorded pairs were indeed direct synaptic partners.
Before discussing how this monosynaptic RV tracing technique has been applied to in vivo circuit tracing, let us consider the advantages of monosynaptic tracing over classical multistep tracing. First, monosynaptic tracing allows genetic control of starter cells by selectively expressing TVA receptor and RG in a defined cell type (see Sect. 4.2.3). This specificity is impossible to achieve when tracing with wild type RV, as RV infects multiple types of neurons indiscriminately. Second, monosynaptic tracing technique allows unequivocal identification of direct synaptic partners, whereas classical multistep tracing can only infer the order of connectivity by the timing of the viral spread (Ugolini 1995). Although informative, variability in viral replication in different host cells, the speed and distance of retrograde transport, and the type of synapses that the virus crosses may create timing differences in transsynaptic labeling, compromising the accuracy in which the synaptic order of connected neurons can be identified. Third, RVdRG is less toxic to infected animals compared to classical RV tracing, and therefore allows one to visualize neuronal circuits in relatively healthy animals. This also allows monosynaptic tracing to be combined with other physiological and behavioral experiments that require live animals (for example, see Sect. 4.2.5).
2.3 Cre-dependent Transsynaptic Tracing
A significant advance of monosynaptic RV tracing lies in the ability to genetically control the generation of starter cells. This can be done by selective targeting of RG and TVA to defined types of neurons. Generally, cell types can be characterized by a combination of the following criteria: stereotyped locations, unique gene expression, unique morphology, axonal projection patterns, developmental history, electrophysiological properties, and unique receptive fields. Currently, not all of these criteria can be readily utilized to generate starter cells, but some are available. In this section, we will discuss the methodology behind Cre-dependent tracing methods. Other methods will be discussed in the following sections.
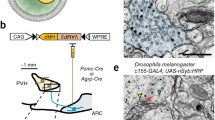
Numerous mouse lines are available that express Cre recombinase in a selective cell type. The viral and genetic technology (see Sect. 4.1.3 and Fig. 4.3c) is a powerful way to generate starter cells of a defined cell type in a defined brain region. Let us use a specific example, such as parvalbumin-positive interneurons (PVNs) in the left primary motor cortex in mice (Wall et al. 2010; Miyamichi et al. 2013). To transsynaptically trace from these cells, we first need a transgenic mouse line where Cre is specifically expressed in the PVNs. Next, we stereotactically inject a small volume (usually 50–300 nl) of a mixture of two Cre-dependent AAVs, expressing RG and TVA fused with a red fluorescent marker mCherry (TVA-mCherry), into the left motor cortex of a PV-Cre mouse (Fig. 4.5a). In 2 weeks, hundreds of PVNs in the left MC will express RG and TVA-mCherry. There are millions of PVNs throughout the mouse brain, but focal injection of the AAVs restricts the location of the starter cells. In the motor cortex, >95% neurons are non-PVNs, but they do not express RG or TVA-mCherry because they do not contain Cre. We then stereotactically deliver a small amount of RVdRG-GFP+EnvA into the same left motor cortex (Fig. 4.5b) to initiate transsynaptic tracing. Finally, 4–7 days after RV infection, we harvest the brain and detect neurons labeled with both GFP and mCherry (starter cells, labeled yellow, Fig. 4.5c) and those labeled with GFP alone (presynaptic partners of the starter cells). In one example, 1012 starter cells were generated close to the injection site, and 8656 GFP+ cells were detected far away from the starter cells in the contralateral motor cortex, ipsilateral somatosensory cortex, and motor-related thalamus. In addition, many thousands of locally infected neurons labeled with GFP were detected near the starter cells (Miyamichi et al. 2013). This tracing strategy has been used with great success in a wide range of brain regions including the olfactory system (Miyamichi et al. 2011, 2013), neuromodulatory systems (Weissbourd et al. 2014; Lerner et al. 2015; Schwarz et al. 2015; Ogawa et al. 2014; Watabe-Uchida et al. 2012; Menegas et al. 2015; Pollak Dorocic et al. 2014; Beier et al. 2015), amygdala (Haubensak et al. 2010), hypothalamus (Krashes et al. 2014), hippocampus (Sun et al. 2014; Kohara et al. 2014), neocortex (Fu et al. 2014; Zhang et al. 2014; Adelson et al. 2014; DeNardo et al. 2015; Kim et al. 2015), striatum (Reardon et al. 2016; Wall et al. 2013), cerebellum (Wall et al. 2010), and spinal cord (Reardon et al. 2016; Ni et al. 2014).
Cre-dependent RV transsynaptic tracing. a Left constructs of two AAV vectors, one expressing TVA-mCherry and one expressing RG in a Cre-dependent manner using FLEx switch (see Fig. 4.3d). WPRE, woodchuck hepatitis virus posttranscriptional regulatory element, which can enhance transgene expression from an AAV vector. The oval in the middle represents a cell expressing Cre (blue PacMan) in which TVA-mCherry and RG are also expressed. Circles on the right represent neurons, one of which corresponds to a Cre-positive cell (with blue PacMan). b RVdRG-GFP+EnvA infects a Cre-positive cell that is expressing TVA and RG, resulting in a yellow starter cell (due to TVA-mCherry from the AAV and GFP from the RVdRG). Note that by mixing two separate AAVs before injection, >80% neurons at the injection site co-express both transgenes (Miyamichi et al. 2013). c If RG is present at the plasma membrane of starter cells (yellow), RVdRG-GFP+RG can transsynaptically spread to presynaptic partners and label them with GFP
At a glance, this method seems complex. Instead of serially injecting AAVs and RV, one could drive expression of RG and TVA via Cre-dependent transgenes introduced into the genome of the animal. In this scenario, all Cre-expressing neurons throughout the brain (for instance all PVNs in a PV-Cre animal) will express RG and TVA. Stereotactic injection of RVdRG-GFP+EnvA into the left motor cortex of these animals will convert PVNs at the injection site into starter cells, and transsynaptic tracing will occur. However, if there are any PVNs located presynaptically to the starter PVNs, they can become ‘secondary’ starter cells because they will also express RG, and their presynaptic partners will also be labeled. Thus, this process can significantly compromise the accuracy of restricted, monosynaptic RV tracing. It is therefore advantageous to spatially restrict the expression of RG using an AAV.
For successful Cre-dependent transsynaptic tracing, two important points should be considered. First, starter cells should be unequivocally labeled to validate their numbers and cell types. Second, Cre-independent, nonspecific labeling of RV should be monitored and controlled. Ideally, Cre-dependent tracing should only originate from Cre-positive starter cells. However, in many cases, it is possible that a small amount of ‘leaky’ expression of TVA and RG can occur. In the example mentioned above (PV-Cre tracing in the motor cortex), negative control experiments omitting Cre still labeled ~70 GFP+ neurons close to the injection site. This Cre-independent GFP expression is likely due to the extremely efficient interaction between TVA receptor and RVdRG-GFP+EnvA. Because these nonspecific GFP+ neurons are indistinguishable from real presynaptic neurons of the starter cells, they compromise the accuracy of tracing within local circuits near the injection site, although usually less than 5% of local GFP+ neurons arise from Cre-independent tracing (Weissbourd et al. 2014; Miyamichi et al. 2013; Schwarz et al. 2015). Note that long-range tracing outside of the injection site is less affected by nonspecific labeling, since leaky expression of RG is not strong enough to support trans-complementation of RVdRG.
To eliminate Cre-independent local labeling, TVA-EnvA interactions can be reduced without affecting TVA-mCherry expression levels. To achieve this, a TVA mutant with 10% affinity to the EnvA-pseudotyped virus (Rong et al. 1998), TVA66T, was used instead of wild-type TVA. The resulting tracing with TVA66T showed no Cre-independent GFP labeling within the brain (Weissbourd et al. 2014; Miyamichi et al. 2013). Although the tracing efficiency of long-range inputs were also reduced (due to a decrease in the number of RVdRG-GFP+EnvA viral particles that initially infect the starter cells), this technique is suitable for analyzing local neuronal circuits. We will see applications of this technique in the OB in Sect. 4.3.
Approaching the end of this section, let us consider a way to control the number of starter cells, in addition to specifying cell types as discussed above. Generally, reducing the titer of AAVs would decrease the number of starter cells. To more empirically control the number of starter cells, a tamoxifen-inducible Cre (CreER) transgenic mouse was crossed with a mouse line conditionally expressing tetracycline transactivator (tTA2) in a Cre-dependent manner, to obtain double transgenic mice. Small amounts of tamoxifen were injected into these mice, resulting in a sparse induction of neurons expressing tTA2 throughout the brain. Instead of using Cre-dependent AAVs, an AAV vector expressing RG and TVA (with a fluorescent marker), driven by tTA2, was injected into a defined brain area (Fig. 4.3e). In the olfactory cortex (Miyamichi et al. 2011), this strategy generated sparse starter cells (samples varied from 4 to 105 starter cells). A ‘lucky’ example in the somatosensory cortex contained a single starter cell, which is useful for analyzing how individual neurons integrate information from different brain regions. Generation of sparse starter cells can also be achieved by single-cell electroporation method (see Sect. 4.2.5).
2.4 Tracing the Relationship Between Input and Output (TRIO)
Axonal projection patterns can be used to define different classes of neurons in the brain. For example, pyramidal neurons of motor cortex located in layer 5 (one of the major output layers of the cortex) can be classified into two major types based on their projection patterns: callosal projection neurons (CPNs) and subcerebral projection neurons (SCPNs) (Greig et al. 2013). CPNs provide callosal projections to the contralateral motor cortex, in addition to bilateral projections to somatosensory cortex and striatum, while SCPNs provide long descending axons to the ipsilateral striatum, medulla and pons, on their way to the spinal cord. Importantly, these two types of neurons are highly intermingled in the cortex, and therefore can potentially be contacted by the same presynaptic neurons that send axons into the motor cortex. Given that their target areas are mostly nonoverlapping, a question arises: do CPNs and SCPNs receive the same input? And if not, what is unique about their presynaptic inputs, and can differences in their input patterns be quantitatively compared? This question can be generalized to many different brain regions, and thus can provide valuable insight into different circuit logics that underlie specific information flow.
Axonal projection patterns have been successfully used to define starter cells in specific cases, such as MNs in the spinal cord and retinal ganglion cells (RGCs) in the eye. Let us analyze these cases, and discuss how to generalize the concept of input–output connectivity for applications in the central brain. In the spinal cord, a group of MNs (a motor pool) innervating the same muscle can be selectively labeled by RV injected into the target muscle. Injecting RVdRG-GFP+RG, along with AAV expressing RG, into the target muscle can convert the MNs of a defined motor pool to starter cells. The RV can then transsynaptically spread to premotor neurons that are presynaptic to that motor pool (for details, see Sect. 4.5.1; Stepien et al. 2010; Tripodi et al. 2011). Note that in this particular case, AAV serotype 6 can efficiently infect MNs retrogradely from their axons at the target muscle only during the neonatal stage. Retrograde infection of AAV in the central brain at adult stages is usually inefficient, though efficiency varies considerably by AAV serotype, titer, lot, and the target brain regions.
In the retina, a specific type of RGCs that convey direction-selective motion were converted to starter cells (Yonehara et al. 2011; Yonehara et al. 2013). This was possible because these RGCs selectively project to the medial terminal nucleus (MTN) in the midbrain. By injecting RVdRG-GFP+RG together with a helper virus (HSV1 or AAV) expressing RG into the MTN of neonatal mice, researchers successfully detected amacrine cells and bipolar cells connecting to the starter RGCs in the retina. These methods that deliver RVdRG-GFP+RG by using the outputs of a neuronal population, however, cannot easily select specific starter cells in the central brain, because many different brain regions and cell types often send convergent projections to the same target. Going back to the motor cortex example, layer 5 SCPNs can be targeted retrogradely from the medulla, but there are millions of neurons throughout the brain that also project to the medulla, including cells in other parts of the cortex, hypothalamus, cerebellum, and spinal cord. To make spatially restricted starter cells, one needs a method to generate starter cells based both on brain region and projection type.
Given that Cre-dependent RV tracing with AAV helper viruses can restrict the location (by AAV injection site) and cell type (by Cre expression) of starter cells (Sect. 4.2.3, Figs. 4.5 and 4.6a), a natural extension of this technique is to supply Cre via the axons of starter cells (Fig. 4.6b). A canine adenovirus 2 (CAV2) driving Cre is well suited for this purpose, as it can efficiently and retrogradely introduce Cre into many types of neurons in the mouse brain (Table 4.1; Soudais et al. 2001; Hnasko et al. 2006). By pairing Cre-dependent RV tracing with CAV2-Cre, (a method called tracing the relationship between input and output, or TRIO), one can determine and compare synaptic inputs to starter cells that have been defined by their location in the brain and output projection patterns (Schwarz et al. 2015). This system was applied to quantitatively analyze presynaptic partners of CPNs and SCPNs in motor cortex layer 5, by injecting CAV2-Cre into contralateral motor cortex versus ipsilateral medulla, respectively. However, a problem with this method is that CPNs are distributed broadly from layer 2/3 to layer 6; the projection pattern alone is not specific enough to restrict starter cells to CPNs only in the layer 5. Note that this is a general problem for many brain regions and cell types. For example, targeting dopamine neurons in the ventral tegmental area (VTA) that project to the striatum would also label GABAergic neurons in the VTA that also project to the same striatum region, ‘contaminating’ the specificity of the starter cell type (see Sect. 4.4.3). What we need here is an additional restriction of starter cells based on the cell type (defined by a genetic marker) in the TRIO method.
Schematic representations of trans-synaptic tracing, TRIO, cTRIO and pseudo-TRIO. In this simplified neuronal circuit, three brain regions (columns of circles), each containing three neurons, are connected by axons (arrows) as indicated. a In Cre-dependent RV trans-synaptic tracing (Miyamichi et al. 2013; Watabe-Uchida et al. 2012; Sect. 4.2.3), starter cells (yellow) are generated in the neurons expressing Cre (represented by blue PacMan symbols) in the target brain region (middle column) regardless of their projection patterns. b In TRIO (Schwarz et al. 2015), CAV2-Cre is injected (blue needle) into one of the output regions to retrogradely supply Cre to the target region. Then, Cre-dependent RV tracing is conducted as in (a). In this case, different cell types within the target regions are not distinguished. c In cTRIO (Schwarz et al. 2015), a TRIO experiment is conducted in transgenic mice in which Cre recombinase is expressed in a specific cell type. The purple PacMan represents Flp recombinase. Note that the cell indicated by the asterisk cannot become a starter cell because it does not express Cre, even though it projects to the same output region. d Schematic representation of pseudo-TRIO. In this experiment using a Cre transgenic mouse, RVdRG-GFP+EnvA is directly injected into one of the output regions, instead of Cre-dependent CAV2-Flp in (c). Cre-dependent AAVs expressing RG and TVA are injected into the target region as in (a). A starter cell, defined by Cre expression and its projection, is generated in the middle of the target region. However, there is another Cre-expressing cell, indicated by a dagger symbol (†) in the target region that also receives the AAV expressing RG. This cell does not project to the RV injection site, but does connect to the starter cell. This cell can become a secondary starter cell, and its presynaptic partners will also be labeled by RV particles (in green). This causes pseudo-positive labeling of input cells (compare input patterns in panel c and d)
This led the development of cell-type-specific TRIO, or cTRIO (Fig. 4.6c), which allows input mapping to genetically defined neuronal populations based on their output patterns (Schwarz et al. 2015). In cTRIO, a transgenic mouse line is used in which Cre is expressed in a specific cell type. Cre-dependent CAV2 driving Flp recombinase is injected into an output region to which the Cre-expressing neurons send their axons. This results in expression of FLP only in the neurons of a defined cell type (by Cre) with a defined projection pattern (by CAV2), which will become the starter cells. Next, Flp-dependent AAVs driving RG and TVA-mCherry are injected directly into the brain region where these neurons are located, followed by RVdRG-GFP+EnvA to initiate transsynaptic tracing. As a proof-of-principle, retinol binding protein 4 (Rbp4)-Cre mice were used to restrict starter cells in layer 5 of cortex (Gerfen et al. 2013), with CAV2-FLEx-Flp injected into contralateral motor cortex or ipsilateral medulla. This time, CPNs and SCPNs in the motor cortex layer 5 were selectively converted to starter cells, and GFP+ input neurons were quantified in the cortex and thalamus. Interestingly, CPNs received proportionally more input from the cortex, while SCPNs obtained more input from the thalamus (Fig. 4.7a). Thus, CPNs and SCPNs in the motor cortex layer 5, albeit with highly intermingled dendritic trees, receive differential cortical versus thalamic input. The functional significance of this organization, as well as the local circuit connectivity of CPNs and SCPNs (Kiritani et al. 2012) remains to be explored.
Example of cTRIO and single-cell-initiated monosynaptic tracing. a Schematic of cTRIO in mouse motor cortex. Cre-dependent CAV2-Flp was injected into contralateral motor cortex (cMC, for CPNs) or medulla (for SCPNs) along with AAVs expressing Flp-dependent TVA-mCherry and RG into the motor cortex, followed by RVdRG-GFP+EnvA. Example coronal sections of motor cortex CPNs starter cells in cTRIO are shown in the middle. Labeled cells include starter cells (yellow, a subset indicated by arrowheads) and local input cells (green), as well as input neurons from the somatosensory cortex (SC) and ventral anterior thalamus (VA). Average fraction of total input neurons in cTRIO is shown in the right graph. Values represent the average fraction of input in each category. Error bars s.e.m. ***, p < 0.001. Scale, 250 µm (middle row), 100 µm (bottom row). Adapted with permission from Schwarz et al. (2015). b Single cell-initiated RV tracing expressing GCaMP6s in the primary visual cortex (V1). Left image represents a 300-μm thick slice containing an electroporated layer 2/3 pyramidal starter cell (yellow) and its local presynaptic neurons (green). Right image shows an example of 3D-reconstruction of the location of a starter cell and its presynaptic neurons. Each filled circle represents a neuron and is colored according to the preferred motion direction (color code is shown at bottom right). Cells that did not respond to the visual stimuli are represented by small black circles. Cells that responded to motion equally in all directions are represented by small gray circles. Images are taken with permission from Wertz et al. (2015)
At first glance, TRIO/cTRIO looks complicated. Indeed, it requires precise targeting of three different viruses into the correct stereotactic coordinates. One may want to eliminate the CAV2 injection and instead generate starter cells in a pathway-selective manner using the fact that RVdRG-GFP can retrogradely transduce starter cells from axons (Fig. 4.6d). This strategy, although used in several influential studies, has a major pitfall regarding input–output specificity. Let us use a specific example from the motor cortex again. In this hypothetical experiment, AAVs conditionally expressing RG and TVA-mCherry are injected into the motor cortex of Rbp4-Cre mice, such that layer 5 CPNs and SCPNs express TVA in their cell bodies and axons. Thanks to the sensitivity of TVA-EnvA interactions, RVdRG-GFP+EnvA injected into the medulla could convert SCPNs in motor cortex layer 5 into starter cells (although in reality this long-range retrograde transduction of EnvA-pseudotyped RV is not very efficient). Presynaptic partners of SCPNs are then labeled by transsynaptic RV spread, resembling cTRIO. However, if any CPNs in motor cortex layer 5 are presynaptic partners to any of the starter SCPNs, they can also become starter cells, because they contain Cre, and therefore can express RG, if infected by AAV (see Fig. 4.6d, ‘secondary’ starter cell indicated by a dagger symbol). This would result in transsynaptic labeling of SCPN and CPN inputs. Therefore, this strategy does not guarantee that starter cells are specific for a defined projection target. Introducing RG based on the projection pattern is crucial for the specificity achieved by TRIO/cTRIO, accomplished through coordinated injections of CAV2 and AAVs.
Finally, a few basic properties of CAV2 that are useful to know for interpreting tracing results must be discussed. The degree of local CAV2 spread at the injection site is important for precisely targeting neurons based on their projections. The olfactory system was used to assess this (Schwarz et al. 2015), thanks to the well-understood organization of mitral cell axonal projections. Mitral cells (MCs) are the projection neurons of the OB, and their axons form a bundle from which thin collaterals innervate cortical pyramidal cells in layer 1a of the piriform cortex. CAV2-Cre was injected into Cre-reporter mice at varied distances from layer 1a in the piriform cortex and labeled MCs in the OB were quantified. This analysis revealed that CAV2 local spread is mostly limited to within 200 μm from the injection needle. However, it was also observed that CAV2 has potential to infect axons in passage, as MCs in the accessory OB, which send myelinated axon bundles through the piriform cortex without making synapses (Shepherd 2004), were also labeled. Careful design of the CAV2-injection site is needed to avoid unwanted labeling from passing axons. Although CAV2 can transduce diverse cell types throughout the brain, the labeling efficiency may vary depending on the brain regions, type of neurons, and distance of retrograde transport. Keeping these cautions in mind, CAV2-based tracing tools are broadly applicable to many brain regions. As no genetically engineered animal is required for TRIO, it can be used in wild type mammals beyond mice. As a proof-of-principle demonstration, presynaptic partners of motor cortex neurons that project to striatum or contralateral motor cortex were visualized in rat (Schwarz et al. 2015). We will also discuss applications of TRIO/cTRIO in neuromodulatory systems (Lerner et al. 2015; Schwarz et al. 2015; Beier et al. 2015) in Sect. 4.4.
2.5 Tracing from Defined Neurons by Developmental History
Utilizing developmental history is a powerful way to define cell types. A salient example is cell birth date, which can be used to target developmentally defined types of neurons associated with a specific brain region, layer, projection pattern, and function. Generation of starter cells based on the birth date of neurons has been used with great success in the research field of ‘adult-born’ neurons. Although most neurons in the brain are generated in a short time window during embryonic development, neurogenesis continues throughout life at two specific locations: the subventricular zone (SVZ) of the lateral ventricles and the subgranular zone (SGZ) of the dentate gyrus (DG) in the hippocampus (Zhao et al. 2008). The SVZ generates OB interneurons that migrate a great distance through the rostral migratory stream, while the SGZ generates local granule cells (GCs) of the DG. To analyze how these neurons are integrated into existing circuits, researchers selectively converted adult-born neurons to starter cells. This was achieved by using a gammaretrovirus that selectively infects dividing cells (Table 4.1), which made it possible to infect neuronal progenitors, but not postmitotic neurons. Gammaretrovirus-expressing TVA and RG constitutively (Deshpande et al. 2013; Vivar et al. 2012) or Cre-dependently (Nakashiba et al. 2012) was injected into the DG of postnatal mice to generate starter cells, and RV tracing labeled their presynaptic neurons. The Cre-dependent gammaretrovirus can be used to further refine the starter cells based on expression of a specific gene. Another strategy is to utilize transgenic mouse lines expressing TVA and RG in a Cre-dependent manner, in conjunction with gammaretrovirus-expressing Cre (Li et al. 2013). Collectively, these studies revealed the timecourse of progressive integration: adult-born neurons receive local connections from multiple types of interneurons before long-range projections are established.
In the case of the OB interneurons, transient induction of a transgene into the SVG can target neurons of a defined birthdate. Several methods have been successfully used to provide TVA and RG to the SVG including electroporation of a plasmid (Arenkiel et al. 2011), infection of gammaretrovirus (Deshpande et al. 2013), or infection of lentivirus-expressing Cre into a transgenic mouse that conditionally expresses TVA and RG (Garcia et al. 2014). These studies revealed previously uncharacterized connectivity in the local OB circuits (see Sect. 4.3), as well as a temporal sequence for the integration of input to the adult-born GCs in the OB. Some basic properties of RV spread were also reported, which have relevancy beyond OB circuits. For example, researchers found that enriched olfactory experience induced a 3-fold increase in the number of RV-labeled local presynaptic partners of newborn GCs (Arenkiel et al. 2011), a phenomenon that was also observed for newborn GCs in the DG after exercise (Deshpande et al. 2013). This can be explained by two scenarios: (1) neuronal activity increased the number of synaptic connections from local neurons to the starter GCs, or (2) RV particles more efficiently spread across active synapses. To distinguish between these two possibilities, neuronal activity was manipulated in cultured OB explants during RV tracing. It was found that blocking SNARE-dependent neurotransmitter release, action potentials, or fast glutamatergic neurotransmission had no significant effect on the number of transsynaptically labeled cells (Arenkiel et al. 2011). Thus, transsynaptic spread of RV is insensitive to changes in neuronal activity, a phenomenon also suggested from in vivo tracing experiments of NMDA receptor knockout starter cells in the neocortex (DeNardo et al. 2015).
Neuronal birth date is also useful to define starter cells beyond just adult-born neurons. In utero electroporation of a Cre-expressing plasmid during specific developmental time windows successfully targeted neurons of defined layers in the neocortex (DeNardo et al. 2015). This method, combined with Cre-dependent RV tracing (Sect. 4.2.3), allowed researchers to map local and long-distance input to the layer 2/3 neurons in the somatosensory cortex, which were compared to inputs of layer 5 starter cells (generated using an Rbp4-Cre mouse). In principle, in utero electroporation-based methods can be used in many animals beyond mice to define cell types or layers of starter cells.
2.6 Tracing from Defined Neurons by Electrophysiological Properties
Other useful characteristics for defining starter cells include their electrophysiological properties and their receptive fields. To achieve this, electroporation of plasmids into a single cell in vivo was performed after whole-cell patch-clamp recording (Rancz et al. 2011), which promoted transgene expression in the recorded cells after patching. In this way, TVA and RG were introduced into layer 5 pyramidal cells with defined orientation selectivity in mouse primary visual cortex. RV transsynaptic tracing resulted in the generation of a single starter cell and labeled on average 346 presynaptic neurons. A follow-up study (Velez-Fort et al. 2014) demonstrated that corticocortical projection neurons in the visual cortex layer 6 are broadly tuned in their orientation selectivity and predominantly receive input from deep layers of local visual cortex, whereas cortical-thalamic projection neurons in the same area are sharply tuned to orientation and direction information, and receive more long-range input from higher cortical areas. Thus, similar to CPNs and SCPNs in the motor cortex layer 5 analyzed by cTRIO (Sect. 4.2.4), visual cortex layer 6 projection neurons with distinct output specificities integrate different contextual and stimulus-related information within and outside of the cortical network. Thanks to advancements in engineering-modified RV variants (Osakada et al. 2011; Reardon et al. 2016), RV-expressing Ca2+-indicator GCaMP6 was used in single cell-initiated monosynaptic tracing in visual cortex (Wertz et al. 2015). Two-photon Ca2+ imaging revealed motion direction preferences of individual presynaptic neurons labeled from a single visual cortex layer 2/3 neuron of a defined direction preference (Fig. 4.7b). This allowed a systematic comparison of the direction preference of each presynaptic neuron with its postsynaptic partner across all layers of cortex. The result revealed rather unexpected properties: (1) Presynaptically labeled neurons within each layer exhibited similar direction preference; and (2) only one-third of visual cortex layer 2/3 neurons received presynaptic input with similar preferred direction selectivity. In other words, two-thirds of visual cortex layer 2/3 neurons receive ‘untuned’ presynaptic input that cannot be distinguished from a pool of neurons with random direction selectivity. How the postsynaptic receptive field is correctly tuned, as well as the significance of such an organization, remains to be explored. Collectively, these pioneering studies uncovered a link between the neuronal connectivity, electrophysiological properties, and receptive fields of each neuron at single-cell resolution in vivo.
In addition to technical advances and biological findings, these studies also provide a useful insight with regards to the number of presynaptic neurons that are labeled by RV from a single starter cell. An important limitation of RV tracing is that it only labels a subset of inputs, presumably due to the limited number of RV particles that each starter cell can generate. The labeling efficiency of RV can be enhanced by increasing the amount of RG expressed in the starter cells for trans-complementation and increasing the number of RVdRG particles entering the starter cells (Miyamichi et al. 2013). The strain of RV also has an influence on efficiency (CVS strain is more efficient than SAD) (Reardon et al. 2016). As the actual number of inputs per cell in the brain is unknown, it is impossible to determine the exact efficiency of RV tracing. However, based on a few simple assumptions (Miyamichi et al. 2011), cortical pyramidal cells in mice are estimated to collect input from 400–1600 presynaptic neurons. The number of RV-labeled cells per a single starter cell observed in the single-cell tracing sample was about 250 [somatosensory cortex layer 5 neuron (Miyamichi et al. 2011)], about 350 [V1 layer 5 neuron (Rancz et al. 2011)], about 380 [cortical-striatal projection neurons in V1 layer 6 (Velez-Fort et al. 2014)] and about 420 [V1 layer 2/3 neuron (Wertz et al. 2015)]. Thus, the RV tracing is estimated to visualize 20–100% of putative presynaptic partners in these cases. With improvements to the RV genome, strains of RV, and methods to drive RG, the efficiency of RV tracing will be further increased in the future.
3 Transsynaptic Tracing in the Olfactory System
In this section, we will discuss how RV tracing has been used to identify new components of a neuronal circuit, and to analyze the topography of the neuronal map. The mouse olfactory system provides examples for these applications of RV tracing. First, let us summarize the basic knowledge of the organization of the mouse olfactory system (Mori and Sakano 2011). The sense of smell is the primary means for most animals to find food, mates, and predators from a distance. Airborne odorants in the environment are initially detected by odorant receptors (ORs), a diverse family of G-protein-coupled receptors expressed in the olfactory epithelium. The olfactory coding in the epithelium is combinatorial: each OR is activated by multiple odorants, and each odorant is recognized by a unique combination of ORs. This enables the olfactory system to distinguish many more odorants than the number of OR genes. Two important rules underlie the transformation of this combinatorial OR code in the nose into neuronal activity patterns by which the brain can interpret the identity, concentration, and in some cases, the meanings of odorants. (1) One neuron–one receptor rule, that is, individual olfactory receptor neurons (ORNs), the sensory neurons in the olfactory epithelium, express a single type of OR. By this rule, the combinatorial code of OR activation is converted into population neuronal activities of ORNs on a one-to-one basis. (2) One OR–one projection site rule, that is, thousands of ORNs expressing the same OR-type project their axons to a common projection site, called a glomerulus, in the OB (Fig. 4.8a). In mice, there are ~1000 intact OR genes, and therefore ~1000 ORN types. Their projections form ~2000 glomeruli, with each ORN type corresponding to approximately two glomeruli on the surface of the OB. This organization allows the transformation of ORNs’ population activities in the epithelium into discrete patterns of glomerular activities (an odorant map) in the OB. How does the odorant map inform higher olfactory centers? RV transsynaptic tracing has been used in several efforts to answer this question.
Basic organization of mouse olfactory system and RV trans-synaptic tracing in the OB. a Side view of the mouse brain highlighting the organization of the olfactory system. Two types of ORNs (out of ~1000 types) are shown in two colors. ORNs of the same type send convergent axonal projections to the target glomerulus. In the OB, each mitral (blue, red) and tufted (green) cell sends an apical dendrite to a single glomerulus, where it receives input from a single type of ORN. MCs project long-distance axons to many olfactory cortical areas, whereas tufted cells innervate only anterior parts of the olfactory cortices (Igarashi et al. 2012). © 2015 from Principles of Neurobiology by Luo. Reproduced by permission of Garland Science/Taylor & Francis Group LLC. b Left a coronal section of the OB showing one of starter MCs and presynaptically labeled cells with GFP in the OB. PVNs are visualized by anti-PV antibodies in magenta. Note that most GFP+ cells in the external plexiform layer (EPL) are PV+ (indicated by yellow arrowheads). Right a 3D-reconstructed OB sample (lateral view) showing the spatial distribution of starter MTCs (yellow) and their presynaptic PVNs located in the EPL (magenta). Images are obtained with permission from Miyamichi et al. (2013). c Top, schematic drawing of OB local circuits. Three MCs are shown in three colors. Each PVN in the EPL (represented by a magenta cell) receives input from, and sends output to, widely distributed MTCs (for example, green and cyan MCs) for broad lateral inhibition of MTC output. In contrast, the feedback loop mediated by GCs (represented by a red cell) connects nearby MCs (blue and cyan MCs) for local lateral inhibition. GL glomerular layer; MCL mitral cell layer; IPL internal plexiform layer; GCL granule cell layer. d Schematic summary of pharmacogenetic inactivation of PVNs to analyze odorant response profiles of MCs. The graph represents tuning curves of MCs to seven different odorants before (blue) and after (red) inactivation of nearby PVNs. Adapted with permission from Kato et al. (2013). A anterior; P posterior; D dorsal; V ventral; L lateral; M medial
3.1 Transsynaptic Tracing in the Olfactory Bulb
Within each glomerulus in the OB, ORN axons form glutamatergic synapses with the primary dendrites of MCs and tufted cells (TCs), the two types of projection neurons of the OB (Fig. 4.8a; Shepherd 2004). Hereafter, these two types of projection neurons are collectively referred to as MTCs. Individual MTCs send a single primary dendrite to a single glomerulus. Importantly, MTCs are not just relaying olfactory information from the epithelium to the cortex, as there are diverse local interneurons in the OB. The precise function of these local networks remains elusive because of the heterogeneity of these interneurons, their diverse physiological properties and their complex synaptic connectivity. By using Cre-dependent RV transsynaptic tracing with reduced nonspecific local background (Sect. 4.2.3) and a transgenic mouse line driving Cre exclusively in the MTCs (Pcdh21-Cre), the presynaptic neurons to MTCs within the local OB circuits were determined (Miyamichi et al. 2013). In addition to GCs, which are known to provide feedback inhibition to MCs (Shepherd 2004), an unexpected presynaptic partner of MTCs were labeled via these tracing studies: cells within the external plexiform layer. About 40% of GFP+ neurons presynaptic to the starter MTCs were located in this layer, ~90% of which were parvalbumin positive interneurons (PVNs, Fig. 4.8b). 3D-reconstruction of labeled GFP+ cells showed that MTCs receive input from widely distributed PVNs (~300 μm in distance), which was in sharp contrast to the narrowly organized input from GCs (<100 μm in distance). To characterize the electrophysiological properties and odorant response profiles of these PVNs in the OB, individual PVNs in the dorsal surface of the OB were recorded by in vivo two-photon targeted patch-clamp method, while the animals received odorant stimulations. This revealed that PVNs were activated less-selectively by many odorants. Because PVNs do not directly receive input from the ORNs, the source of this broad odorant receptive range was unclear. RV transsynaptic tracing was therefore applied to PVNs (using PV-Cre mice), which revealed that PVNs received broad MTC input. Together, these data uncovered a previously unknown feedback loop in the OB, with MTCs → PVNs → MTCs, by which a single MTC can inhibit hundreds of MTCs at a distance (Fig. 4.8c).
Before dissecting the function of this circuit, let us confirm that RV tracing reports precise synaptic connectivity between neurons. Although synapse specificity of RV spread has been supported in various contexts (Ugolini 1995; Reardon et al. 2016; Miyamichi et al. 2011; Wickersham et al. 2007a), OB interneurons are unique in that they form dendrodendritic synapses: PVNs and GCs send dendrites to the EPL where they receive excitatory input from the secondary dendrites of MTCs, and send inhibitory output to the dendrites of the same or other MTCs. It was not clear if RV transsynaptic tracing works in such connections. RV tracing data in the OB was therefore compared with the connection diagram obtained from an independent mapping method, paired recording in the OB slice (Kato et al. 2013). Paired recording revealed that MTC → PVN connections are about tenfold more frequent than MTC → CG connections. This ratio is very similar to what was observed in the RV tracing, supporting the precision of these methods. Thus, RV tracing can map synaptic partners connected by dendrodendritic synapses.
What would be the function of PVNs in processing olfactory information in the OB? To answer this question, researchers combined in vivo Ca2+ imaging of MCs (Kato et al. 2012) with pharmacogenetic silencing (Magnus et al. 2011) of PVNs. Upon inactivation of local PVNs, the odorant responses of MTCs were elevated almost linearly without changing their tuning curve (Fig. 4.8d; Kato et al. 2013). This linear transformation of odorant tuning is consistent with the model that PVNs provide output gain control to MTCs, that is, scaling odorant response intensities of individual MTCs to allow intensity-invariant information processing.
Another example of using RV tracing to find an uncharacterized circuit component is the identification of local corticotropin releasing hormone (CRH)-expressing inhibitory interneurons in the EPL of the OB that connect to adult born GCs (Garcia et al. 2014; also see Sect. 4.2.5). Although CRH-expressing neurons in the paraventricular nucleus of the hypothalamus (PVN) are well known for mediating systemic stress response, the function of CRH in the OB was completely unknown. Therefore researchers analyzed gene knockout mice that lack CRH, and found that these animals had a reduced number of surviving adult born GCs. In addition, conditional loss of the CRH receptor in the adult-born GCs decreased their survival. Overexpression of a constitutively active form of the CRH receptor in the adult born GCs induced an increase in dendritic arborization and expression of synaptic proteins. Together, these data suggest that EPL interneurons provide CRH to newborn GCs and facilitate synaptogenesis in the OB. As CRH+ neurons in the EPL significantly overlap with PVNs, which are tuned to a wild range of odorants, this CRH-mediated assembly of newborn GCs can partially explain why an odorant-enriched environment increases the number of GCs (Arenkiel et al. 2011) in the OB. As highlighted in these studies, RV tracing can identify a previously uncharacterized circuit component and provide anatomical basis for dissecting its function. In the OB local circuits, many types of neurons remain to be functionally analyzed in future investigations.
3.2 Transsynaptic Tracing from the Olfactory Cortex
Olfactory information, after being processed in the OB local circuits, is then transferred to various regions of the olfactory cortex via long axonal projections of MCs and TCs (Fig. 4.8a). How the spatial glomerular map in the OB is anatomically represented in the olfactory cortex is a fundamental question that would help us to ultimately understand the nature of olfactory perception. Therefore, RV tracing was applied to generate a small number of starter cells in the different parts of the olfactory cortex, and the spatial organization of RV-labeled MCs and their corresponding glomeruli were analyzed in a 3D-reconstructed OB model (Miyamichi et al. 2011). This study revealed three principles that define cortical representations of the OB input.
First, individual cortical neurons receive direct input from MCs representing at least four glomeruli. This convergence of multiple MC inputs enables cortical neurons to integrate information from discrete olfactory channels. In addition, layer 1 GABAergic neurons in the piriform cortex receive input from a greater number of glomeruli than layer 2/3 pyramidal cells, consistent with slice physiology data (Poo and Isaacson 2009; Stokes and Isaacson 2010). This organization allows layer 1 GABAergic neurons to integrate broad olfactory information and provide global feedforward inhibition to the cortical pyramidal neurons. Second, neurons restricted to small olfactory cortical regions (as few as 2–4 neurons that are less than 100 μm apart) receive input from glomeruli that are broadly distributed in the OB (Fig. 4.9). This organization suggests that, in contrast to visual and somatosensory systems where cortical neurons maintain a peripheral topographic map, olfactory cortex discards the fine spatial map used in the OB. Third, different cortical areas receive differentially organized OB input. For example, the cortical amygdala (CoA) preferentially receives dorsal OB input, whereas the piriform cortex samples the whole OB without obvious bias. The lack of spatial organization in MC → piriform cortex connections revealed by RV tracing is also supported by fine axon tracing experiments from defined glomeruli (Sosulski et al. 2011) and individual MCs (Igarashi et al. 2012), supporting the reliability of RV tracing in the olfactory cortex.
Organization and function of olfactory cortical areas. Patterns of glomeruli retrogradely labeled by RV tracing from piriform cortex (red anterior; magenta posterior) and cortical amygdala (three samples combined, green). Left schematic of injection sites. Right standard OB glomeruli model and superimposed glomeruli map visualized by RV tracing after reconstruction into 3D space based on serial sections. A anterior; P posterior; D dorsal; V ventral. Adapted from with permission from Miyamichi et al. (2011). © 2015 from Principles of Neurobiology by Luo. Reproduced by permission of Garland Science/Taylor & Francis Group LLC
In vivo Ca2+ imaging experiments demonstrated that individual piriform cortex neurons activated by specific odorants were distributed broadly across the piriform cortex without obvious spatial patterns (Stettler and Axel 2009). Seemingly random input from MCs to the piriform cortex provides an anatomical basis for this physiological data, together with intensive association fibers that connect broadly distributed piriform neurons (Franks et al. 2011). This organization allows piriform cortex neurons to receive a large number of combinations of different olfactory channels, and can serve as a basis on which olfactory experiences plastically adjust the strength of connections to create odorant representations based on an individual’s experience. In contrast, MC axons originating from defined glomeruli exhibited stereotyped broad patches of innervation in the CoA (Sosulski et al. 2011), and many neurons in this structure received biased input that favors glomeruli in the dorsal OB (Miyamichi et al. 2011), which is known to mediate odorants with innately defined meanings (Kobayakawa et al. 2007).
To decipher the principles of information processing in each olfactory cortical area, more integrated connection diagrams are needed among olfactory cortical areas, which receive input from higher brain centers and also send outputs to various brain areas in the hypothalamus, frontal cortex, hippocampus, and ventral striatum. These maps should include detailed information about the types of pre- and postsynaptic neurons that are connected, and their synaptic properties. RV tracing and TRIO will be valuable tools to draw such connection diagrams in the future.
4 Transsynaptic Tracing in Neuromodulatory Systems
So far, the RV tracing studies we have described were performed on neuronal circuits with fairly specific functions in the nervous system. For instance, to relay detection of a specific odorant, MCs in the OB project to brain areas that are highly relevant for olfaction (see Sect. 4.3). But not all neuronal circuits in the brain have such a narrow function with regards to the information they relay, the brain areas they project to, or the behaviors they regulate. Frequently, it is important for the brain to adjust the activity of many neuronal circuits simultaneously, depending on internal and external cues, to modify behaviors accordingly. The process it uses to accomplish this is called neuromodulation (Marder 2012).
The specific cells that mediate this process are called neuromodulatory neurons, and they differ in several important ways from other types of neurons. First of all, whereas most neurons in the brain communicate via the rapid release of neurotransmitters, such as glutamate or GABA, at specific synaptic sites, neuromodulatory neurons can release their neurotransmitters more diffusely into the extracellular space. This bulk release of neurotransmitter allows neuromodulatory neurons to broadly activate both synaptic and extra-synaptic receptors (most commonly G-protein coupled receptors) that are located on target cells. This method of release allows neuromodulatory neurons to signal over longer distances, and slower and longer time courses, than synaptic communication. Another key difference is that classes of neuromodulatory neurons release different types of neurotransmitter, including monoamines [dopamine, serotonin, histamine, and norepinephrine (NE)], acetylcholine, and a variety of neuropeptides. Of note, it was long thought that different classes of neuromodulatory neurons release distinct neurotransmitters, but recent studies indicate that the neurotransmitter profiles of certain neuromodulatory neurons are actually quite diverse, and can contain combinations of fast neurotransmitters and neuromodulators (Stuber et al. 2010; Liu et al. 2013; Hnasko et al. 2010; Saunders et al. 2015). Overall though, the majority of neurons that express and secrete different types of neuromodulators can be found in largely segregated areas of the brain (Fig. 4.10a).
Transsynaptic tracing studies in the serotonin system. a Schematic of cell body locations for the major neuromodulatory systems in the brain. b 3D reconstructions of whole brain monosynaptic inputs to 5-HT neurons in the DR (left) or MR (right). Starter cells generated in the DR or MR of Sert-Cre mice are labeled in red. RVdRG-GFP infected input neurons are labeled in green. Adapted with permission from Pollak Dorocic et al. (2014). c Examples of RV transsynaptic spread from either 5-HT+ or Gad2+ starter cells within the DR. DR-5HT neurons receive much more input from prefrontal cortical areas than DR-GABA neurons. D dorsal; V ventral; L lateral; M medial. Adapted with permission from Weissbourd et al. (2014). d RV tracing (green) was combined with in situ hybridization labeling (red) to identify specific cell types within the PVN that provide input to DR-5HT neurons. Adapted with permission from Weissbourd et al. (2014). Abbreviations: DR dorsal raphe; LC locus coeruleus; LDT laterodorsal tegmental nucleus; MR median raphe; MS medial septal nucleus; NBM nucleus basalis of Meynert; PVN paraventricular nucleus of the hypothalamus; PPT pedunculopontine nucleus; TM tuberomammilary nucleus; SNc substantia nigra compacta; VTA ventral tegmental nucleus. Scale bars: 250 µm (c)
Now that the concept of neuromodulation has been introduced, let us quickly discuss the unique challenges that researchers face when studying the organization and function of these systems. Unlike many of the circuits that we have discussed so far, which have specialized roles, neuromodulatory neurons participate in a huge assortment of complex behaviors related to motivation, mood, attention, movement, sleep, and memory (to name a few!). Thus, these neurons must communicate with many anatomically and functionally distinct areas of the brain and spinal cord, making their anatomical organization exceedingly complex. Additionally, the location of the neuromodulatory neurons themselves presents several challenges. As mentioned above, the majority of neurons for each of the neuromodulatory systems can be found in specific regions of the brain (Fig. 4.10a). However, these regions are located far below the surface of the brain, and they often lack clear anatomical landmarks or boundaries, making them difficult to target with high specificity. In addition, neuromodulatory neurons within these brain areas are often interspersed with other cell types, such as excitatory or inhibitory neurons. This heterogeneity limits the accuracy with which neuromodulatory neurons can be specifically manipulated for experiments, while not affecting neighboring cells.
Thus, genetically regulated RV tracing provides several advantages for studying the anatomical organization of neuromodulatory systems. The robustness of the RV spread allows inputs to be visualized throughout the entire brain, which is ideal for neuromodulatory neurons that likely receive inputs from diverse brain regions. Also, the ability to precisely control which neurons become starter cells is beneficial for neuromodulatory circuit tracing, due to the challenges discussed above. Because of these advantages, RV tracing of neuromodulatory systems has produced many new insights into how the anatomy of these systems contributes to their functional diversity in the brain.
4.1 Transsynaptic Tracing in the Serotonin System
The main source of serotonin (5-HT) in the brain comes from two adjacent areas at the midline of the midbrain and brainstem, called the dorsal and median raphe (DR and MR, Fig. 4.10a). Neurons that express and release 5-HT (~20,000 in the rat brain) are heterogeneously distributed throughout these structures, along with large populations of glutamatergic, GABAergic, and peptidergic neurons (Bang and Commons 2012; Hioki et al. 2010). In fact, it is estimated that only a quarter of the neurons within the DR and MR express 5-HT (Steinbusch et al. 1980)! Release of 5-HT from DR and MR neurons can regulate mood, arousal, food seeking, sexual behavior, pain modulation, and aggression. Dysfunction in 5-HT release is also strongly correlated with several severe neurological disorders, such as depression and anxiety, and drugs that change brain levels of 5-HT are the most commonly prescribed treatment for these diseases (Müller and Jacobs 2010). Therefore, identifying the organizational principles controlling delivery of 5-HT throughout the brain has both physiological and clinical implications.
While inputs to the DR and MR regions of the brain have been classically characterized (Aghajanian and Wang 1977), the variability of cell types within these structures made it impossible to discretely determine which inputs were connected to which cell types. Thus, the ability to restrict the transsynaptic spread of RV from specific starter cells was a huge advancement for determining input connectivity within the 5-HT system. Utilizing the Cre-dependent transsynaptic tracing system (Sect. 4.2.3), several groups recently characterized the inputs received by specific populations of neurons within the DR and MR, shedding light on how these neurons may achieve their functional diversity (Weissbourd et al. 2014; Ogawa et al. 2014; Pollak Dorocic et al. 2014). Overall, the DR received more inputs than the MR, but the densest inputs into both regions came from neurons within the somatomotor cortices, amygdala, lateral hypothalamus, striatum, pallidum, inferior colliculus, and sensory and motor-related areas of the pons and medulla (Fig. 4.10b). MR-5HT neurons received less input from the prefrontal cortex, but had a preference for inputs from the hypothalamus and brainstem, compared to inputs targeting DR-5HT neurons. Meanwhile, DR-5HT neurons received more input from the CeA. Of importance, RV tracing illuminated several inputs onto DR-5HT neurons that had previously been disputed in the field; inputs arising from the prefrontal cortex and lateral habenula (LHb) (Fig. 4.10c). Using a combination of electrophysiology and optogenetics, the authors were able to functionally verify these anatomical projections, supporting the ability of RV reagents to accurately identify specific connections throughout the brain (Weissbourd et al. 2014; Pollak Dorocic et al. 2014; see also Fig. 4.2e).
While these studies comprehensively described the brain-wide inputs received by 5-HT neurons in the DR and MR, it is important to remember that extensive diversity also exists within these structures, which could contribute to differences in connectivity. To address this question, RV tracing was used to characterize the inputs received by two different cell types (5-HT and GABA) within the DR (Weissbourd et al. 2014). Overall, these neuronal subsets had similar input profiles, with the densest number of inputs to both populations contributed by the hypothalamus, amygdala, medulla, and cortex. However, differences were also observed, such as the DR-5HT neurons receive twice as much input from the cortex, while DR-GABA neurons receive greater input from the CeA and BNST. Cortical to DR connections were also functionally verified using CRACM strategies (see Sect. 4.1.4). Another important advance of broad interest from this work was the pairing of RV tracing with in situ labeling methods to validate the cell type identity (glutamatergic, GABAergic, or peptidergic) for many of the inputs received by DR neuron populations (Fig. 4.10d). These experiments highlighted that differences in neuronal circuit anatomy exist on multiple levels: starter cells can receive input from different brain areas, and also receive input from different cell types within the same brain area.
4.2 Transsynaptic Tracing in the Norepinephrine System
Next, we will discuss the organization of another neuromodulatory system with significant functional overlap to the 5-HT system, specifically, the NE system. Structurally, the NE system is very different from many of the other neuromodulatory systems, in that the neurons supplying almost all of the brain’s NE are restricted to a very tiny nucleus in the hindbrain, called the Locus Coeruleus (LC) (Fig. 4.10a). Also unlike other neuromodulatory structures, the LC is thought to be homogeneous in that all cells contained within it express NE (~1500 neurons per LC) (Swanson and Hartman 1975). This small population of neurons sends projections to almost all areas of the brain and spinal cord to release NE. LC-NE neurons are activated by a huge range of diverse behaviors broadly related to arousal, such as wakefulness, attention, memory formation, and stress response (Berridge and Waterhouse 2003). Thus, similar to the 5-HT system, an important question for LC-NE neurons is how they might be organized to mediate these different behavioral responses. One hypothesis is that subsets of LC neurons may be connected with distinct brain areas that participate in different arousal behaviors. Traditionally, this has been especially challenging to address for the NE system, as the small size of the LC makes it difficult to specifically target for anatomical studies. However, RV tracing and TRIO methodologies (see Sect. 4.2.4) have revealed new organizational properties regarding the brain-wide connectivity of NE neurons. RV tracing indicates that LC neurons receive inputs from over one hundred different brain areas (Schwarz et al. 2015), which is a much more diverse input profile than previously reported (Aston-Jones et al. 1986; Cedarbaum and Aghajanian 1978). However, TRIO and cTRIO studies illuminated that the NE system is highly integrative. Specifically, LC neurons projecting to diverse areas of the brain (OB, auditory cortex, hippocampus, cerebellum, or medulla) received similar distributions of inputs from widespread areas of the brain (Fig. 4.11a). Additionally, LC neurons were found to collateralize their axons extensively between many brain regions, further emphasizing a broad anatomical organization (Schwarz et al. 2015; Nagai et al. 1981; Room et al. 1981). This is in contrast to the dopamine system, where several specificities with regards to the input–output connectivity of these neurons have been observed (see Sect. 4.4.3 and Fig. 4.11c, d). The functional significance of this broad organization, and how LC neurons promote distinct arousal behaviors, have yet to be determined, but the pairing of viral-genetic tools with functional manipulations should provide additional insight.
Transsynaptic tracing studies in the Catecholamine systems. a Schematic of TRIO results from subpopulations of LC neurons projecting to different regions of the brain. Overall, it was observed that populations of LC neurons received similar input regardless of the brain regions that they projected to. Adapted with permission from Schwarz et al. (2015). b Unlike the norepinephrine system, differences were observed in the amount of RV-labeled input neurons for different populations of DA neurons. For instance, VTA-DA neurons received more input from the PLH (top), while SNc-DA neurons received more input from the STh (bottom). Scale bar = 1 mm. Adapted with permission from Watabe-Uchida et al. (2012). c TRIO identified a specific population of VTA-DA neurons that receive increased input from the Ant Ctx, and send specific projections to the lateral NAc. To identify a role of this circuit, researchers expressed ChR2 in Ant Ctx neurons while implanting an optical fiber over the VTA. Mice were subjected to an intracranial self-stimulation procedure, where nosepokes into different ports induced different frequencies of photostimulation. Experimental mice developed a strong preference for the 20 Hz stimulation, suggesting that Ant Ctx to VTA projections are reinforcing. Adapted with permission from Beier et al. (2015). d TRIO performed on populations of SNc-DA neurons also uncovered specificities. Anatomically, DMS- and DLS-projecting neurons make strong reciprocal connections (left). These populations of SNc-DA neurons also responded differently to appetitive (sucrose) or aversive (shock) stimuli (right). Adapted with permission from Lerner et al. (2015). Abbreviations: Ant Ctx anterior prefrontal cortex; CeL lateral division of the central amygdala; cp cortical peduncle; DLS dorsolateral striatum; DMS dorsomedial striatum; DR dorsal raphe; DS dorsal striatum; LC locus coeruleus; NAc nucleus accumbens; PLH peduncular part of the lateral hypothalamus; PSTh parasubthalamic nucleus; SNc substantia nigra compacta; STh subthalamic nucleus; VTA ventral tegmental nucleus; ZI zona incerta
4.3 Transsynaptic Tracing and Functional Analysis in the Dopamine System
Unlike the 5-HT and NE systems discussed above, dopaminergic (DA) neurons have traditionally been thought to have a more specific role in the brain: modulation of goal-directed behavior. DA neurons in the VTA and substantia nigra compacta (SNc) are activated by rewards, or cues indicating a reward is coming (Mirenowicz and Schultz 1996). This is supported by classic neuroanatomical data showing that midbrain DA neurons send strong projections to brain areas involved in goal-directed behavior, such as the striatum, nucleus accumbens (NAc), and frontal cortex (Beckstead et al. 1979). Studies have also indicated that several drugs of abuse increase DA signaling in these output areas to enhance their addictive properties (Bassareo et al. 1996). However, other studies demonstrate that the function of DA neurons is likely more complex, as they can also encode saliency, valence, aversion, memory formation, and movement coordination (Bromberg-Martin et al. 2010). Further supporting a more diverse role for DA neurons, subsets of DA neurons within the VTA and SNc differ in their electrophysiological properties and their projection targets (Lammel et al. 2011; Lammel et al. 2008; Margolis et al. 2008). Therefore, while it is increasingly clear that DA neurons have many roles in the brain, their organizational properties (e.g., who do they receive input from, and where do they project to?) are less obvious. To comprehensively address this question, several groups have utilized RV strategies.
The first study to do this performed transsynaptic RV tracing from DA neurons in the VTA and SNc to visualize the brain-wide inputs received by these neurons (Watabe-Uchida et al. 2012). Several important advances arose from this work. First of all, the authors introduced Cre-mediated RV transsynaptic tracing (see Sect. 4.2.3). Furthermore, the authors performed a brain-wide analysis of RV-labeled inputs to VTA and SNc-DA neurons to compare the synaptic input profiles received by these two populations. Their comprehensive analysis revealed that DA neurons in both the VTA and SNc receive direct synaptic inputs from many more brain areas than previously thought (such as many areas of the cortex), emphasizing the improved sensitivity of RV tracing for detecting anatomical connectivity of neurons over traditional tracing reagents (discussed in Sect. 4.1). While most of the input received by these two DA populations arose from similar brain areas, specific differences were also observed. For instance, DA neurons in the VTA received more input from the lateral hypothalamus, while SNc-DA neurons received preferential input from the subthalamic nucleus and somatosensory and motor cortices (Fig. 4.11B). The authors hypothesized that these input differences may underlie the preference of different DA neurons to encode saliency (cortical/thalamic inputs) versus value (hypothalamic inputs). RV tracing also resolved an ongoing debate in the field regarding which neurons in the SNc receive input from the striatum, as input tracing from DA neurons (using DAT-Cre) or GABA neurons (using Vgat-Cre) labeled distinct subsets of neurons in the striatum (Chuhma et al. 2011; Xia et al. 2011).
Thus, this study lays an important anatomical groundwork for how differences in the anatomical connectivity of DA circuits may affect their function. However, it focused on identifying inputs received by DA neurons, but it is also important to consider where DA neurons send their projections to. Also, to make the leap between anatomy and function, it is necessary to expand upon the observations made from anatomical studies with other technologies, such as electrophysiology, optogenetics, or pharmacology. As a first step towards this, non-pseudotyped RV labeling was paired with other labeling and electrophysiology methods to make several findings about connectivity differences for subsets of DA neurons. It was found that DA neurons located in the lateral VTA preferentially project to the NAc and receive input from the laterodorsal tegmentum (LDT) (Lammel et al. 2012). Meanwhile, medially located VTA-DA neurons project strongly to the prefrontal cortex (PFC) and receive input from the LHb. To explore this functionally, RV expressing channelrhodopsin-2 (RV-ChR2) was injected into the VTA, to retrogradely infect neurons in the LDT or LHb, and an optical fiber was implanted into the LDT or LHb. Phasic stimulation of LDT → VTA neurons in vivo induced robust conditioned place preference (CPP), meaning that the mice strongly preferred the side of a behavioral arena that was paired with light stimulation. Remarkably, activation of the LHb → VTA neurons had the opposite effect: strong conditioned place aversion (CPA) was induced in these animals. Overall, this study nicely highlights the power of combining RV tracing with other methods to uncover important specificities within seemingly similar neuronal populations (DA neurons of the VTA). However, experiments were focused on a small number of brain areas connected to the VTA. Furthermore, tracing was limited to DA neurons within the VTA. Yet, neurons within the VTA are known to receive input from many brain areas, and are molecularly heterogeneous. Therefore, it is likely that greater diversity exists within the DA system beyond the specific circuits characterized here (Watabe-Uchida et al. 2012; Margolis et al. 2012; Hnasko et al. 2012).
Toward this, two groups recently performed input–output anatomical and connectivity studies on a greater scale (Fig. 4.11c, d). Specifically, one study used RV tracing and TRIO methodologies to compare the inputs received by populations of VTA-DA neurons projecting to the lateral or medial shell of the NAc, the medial prefrontal cortex (mPFC), or the amygdala (Beier et al. 2015). The largest differences in connectivity were observed for the NAc-projecting populations, with VTA-DA neurons projecting to the lateral NAc receiving more input from the anterior cortex, dorsal striatum, and NAc, but less input from the dorsal raphe. Meanwhile, VTA-DA neurons projecting to the medial shell receive more medial shell input (forming a strong reciprocal connection), but much less striatal input. But how would the activation of these specific circuits affect the behavior of the mouse? Focusing on the anterior cortex → VTA → lateral NAc, the authors introduced ChR2 into the anterior cortex while placing an optical fiber over the VTA (Fig. 4.11c). The animals were then trained on intracranial self-stimulation protocol, where nose pokes induced stimulation of different frequencies (1–20 Hz). Overall, mice had a strong preference for the 20 Hz stimulation, indicating that anterior cortex → VTA connections could be strongly rewarding.
Finally, could there also be differences in the connectivity of SNc-DA neurons that further explain the diverse roles of DA release in the brain? Such a hypothesis seems plausible since it was observed that DA subtypes had differences in their anatomical connectivity (Watabe-Uchida et al. 2012). In addition, subsets of SNc-DA neurons vary in their firing rates (Schiemann et al. 2012). However, others have argued that SNc neurons may be functionally similar, as manipulating SNc-DA neurons projecting to either the dorsal medial striatum (DMS) or dorsal lateral striatum (DLS) similarly affects cognitive processes (Darvas and Palmiter 2010; Darvas and Palmiter 2009). To explore these potentially disparate results, TRIO was applied to trace the brain-wide inputs of either DMS or DLS-projecting SNc-DA neurons (Lerner et al. 2015). Whereas DMS-projecting SNc-DA neurons received strong input from NAc and DMS, DLS-projecting SNc-DA neurons received strong input from DLS, indicating a robust reciprocal connectivity for these SNc sub-circuits (Fig. 4.11d). But how might these anatomical differences relate to the animal’s behavior? To address this, GCaMP6 was expressed in either DMS or DLS-projecting SNc neurons, and an optical fiber was implanted into SNc. Using fiber photometry, the activity of these neuronal subsets was monitored while the mice received either appetitive or aversive stimuli. DMS and DLS-projecting SNc-DA neurons showed similar activation patterns when the outcome was rewarding (sucrose delivery), but did not respond if sucrose was withheld. However, these neuron populations had opposing responses to aversive stimuli (foot shock), with DMS-projecting neurons showing a dip in their activity during the shock, while DLS-projecting neurons increased their activity (Fig. 4.11d). Together, these experiments emphasize that discrete circuits exist through the SNc to relay a variety of information related not only to reward, but also for aversive cues.
An independent study of the input–output organization of DA circuits additionally observed that a subset of VTA-DA neurons, projecting to the very posterior end of the striatum, had the greatest differences from other VTA-DA neurons with regards to the inputs they received (Menegas et al. 2015). Of note, this study failed to observe some of the differences in input–output connectivity reported in previous papers, though this could be explained by differences in technical aspects of the tracing methodologies. Still, this collection of papers nicely emphasize that RV-based anatomical studies provide important insight to guide more functional studies of relevant brain circuits.
4.4 Transsynaptic Tracing in Peptidergic Circuits
Beyond the main neuromodulatory systems discussed above, RV tracing has also been useful for determining the connectivity of complex peptidergic circuits in the brain. Many of these circuits are contained within the hypothalamus, which is an incredibly diverse brain region, both in terms of the many different cell types that are present there, and the many brain regions that it connects to. Thus, RV tracing has allowed researchers to target several intermingled peptidergic circuits within the hypothalamus with an accuracy and resolution that has not been possible with classic anatomical tools. For instance, neurons within the arcuate nucleus (ARC) of the hypothalamus express the neuropeptide called agouti-related peptide (AgRP) and regulate feeding behaviors (Fig. 4.12a). These neurons become activated by caloric deficiency, and their stimulation induces hunger and an increase in food intake (Atasoy et al. 2012; Krashes et al. 2011; Aponte et al. 2011). However, knowing which neurons upstream to AgRP cells are driving their activation is important for understanding how hunger and appetite are regulated in the brain. RV tracing from the ARC in AgRP-Cre animals showed that these neurons receive most of their input from neurons locally in the ARC, as well as from the PVN and dorsal medial hypothalamus (DMH) (Fig. 4.12b; Krashes et al. 2014). These anatomical connections were verified using CRACM methods. Furthermore, researchers were able to functionally link specific subsets of AgRP inputs to feeding behavior using Designer Receptor Exclusively Activated by Designer Drugs (DREADD) methodologies (Roth 2016). DREADD activation of PVN subpopulations (Pacap- or TRH-expressing neurons) induced robust feeding in animals, an effect that could be blocked if AgRP neurons in the ARC were simultaneously silenced (Fig. 4.12c; Krashes et al. 2014).
Anatomical and functional characterization of peptidergic feeding circuits in the hypothalamus. a The ARC contains a diverse population of cell types, including POMC+ and AgRP+ neurons, whose activity have opposing effects on feeding behaviors (Atasoy et al. 2012; Krashes et al. 2011; Aponte et al. 2011; Zhan et al. 2013). b Monosynaptic spread of RVdRG-GFP from AgRP+ starter cells in the ARC identified inputs locally within the ARC, as well as inputs in distinct hypothalamic nuclei, such as the DMH and PVH. Adapted with permission from Krashes et al. (2014). c Using DREADD technology, it was found that specific cell types within the PVH send projections to AgRP+ neurons in ARC to mediate feeding. hM3Dq-mediated activation of PVH-TRH or PVH-PACAP neurons increased feeding behaviors in mice, an effect that could be blocked by simultaneous inhibition of ARC-AgRP neurons. Inhibition of PVH-TRH neurons also reduced normal food intake. Adapted with permission from Krashes et al. (2014). Abbreviations: AgRP agouti-related peptide; ARC arcuate nucleus; DMH dorsal medial hypothalamus; PACAP pituitary adenylate cyclase-activating polypeptide; POMC pro-opiomelanocortin; PVH paraventricular nucleus of the hypothalamus; TRH thyrotropin-releasing hormone
Additional studies have further characterized the projections of the ARC-AgRP neurons (Betley et al. 2013). Optogenetic stimulation of AgRP+ terminals (expressing ChR2) in different brain areas led to the interesting observation that activation of only some of the AgRP projections caused increased feeding. Anatomical experiments using RV to infect ARC-AgRP+ neurons from their axonal terminals suggested that the projections to different brain areas were discrete, with minimal collateralization between regions, implying that subsets of ARC-AgRP neurons have different roles in the brain.
More recently, RV has also been used to explore the connectivity of another cell type important for feeding behavior, the pro-opiomelanocortin (POMC) neurons. These cells are located both in the ARC (along with the AgRP neurons) (Fig. 4.12a), as well as in the nucleus tractus solitarius (NTS) (Cowley et al. 2001). Unlike AgRP neurons, stimulation of POMC neurons in the ARC or NTS suppresses feeding (Aponte et al. 2011; Zhan et al. 2013). Thus, comparing the circuit organization of these two cell types, especially within the ARC where their cell bodies are intermingled, greatly enhances our understanding of how these systems are coordinated to regulate feeding behaviors. Overall, RV tracing revealed that input patterns for POMC and AgRP neurons in the ARC were largely similar, which is perhaps surprising since these neurons have opposing functions (Wang et al. 2015). Such a finding might suggest that the anatomical organization of these circuits plays a minor role in mediating their activity. Instead, their function may be more directly controlled by other means, such as neuromodulation or hormonal signaling. Alternatively, while the distribution of GFP+ neurons received by ARC-POMC and ARC-AgRP neurons was similar, it could be that differences exist in the cell type identity of these inputs. In contrast, the inputs received by POMC neurons in the NTS were very different, originating from several nuclei in the pons and brainstem. This result indicates that distinct circuits exist in the brain (ARC-POMC, NTS-POMC) to suppress feeding behaviors.
Together, the studies described in this section highlight the huge advances that RV reagents have provided for uncovering the anatomical connectivity of complex circuits in the brain. In addition, we hope that we have emphasized how viral tracing can be used in parallel with other methods to discern the function of specific neuronal populations to regulate diverse behaviors.
5 Transsynaptic Tracing in the Spinal Cord
In the motor system, each skeletal muscle is controlled by a pool of MNs, the cell bodies of which are clustered in stereotypic positions in the ventral horn of the spinal cord (Fig. 4.13a). MNs integrate information from multiple sources, including input from diverse spinal premotor neurons, descending commands from the brainstem motor-related areas and motor cortex, and sensory feedback from proprioceptive neurons. Coherent movements are achieved by the spatiotemporally appropriate recruitment of MNs through the activity of these diverse presynaptic neurons (Arber 2012; Goulding 2009). Understanding their connectivity rules will provide mechanistic insights into the coordinated activation of muscles. We discuss in this section how RV transsynaptic tracing has begun to reveal complex yet specific connection patterns in the spinal cord and brainstem underlying motor controls.
Transsynaptic tracing studies in the spinal cord. a Schematic representation of the muscle coinjection method. Coinjection of RVdRG-GFP+RG and a helper AAV expressing RG into the target muscle can retrogradely label MNs and convert them to starter cells (one of which is shown in yellow). Their presynaptic partners are labeled in green. Adapted with permission from Stepien et al. (2010). b Schematic demonstration of synapse specificity of RV transsynaptic spread using sensory-motor circuits. Starter cells that are genetically restricted to the ankle muscle had transsynaptically labeled proprioceptive neurons in the dorsal root ganglion (DRG) connecting to the same GS (but not antagonistic TA) muscles. Note that in this experiment dsRed expressing RV was used. Adapted with permission from Reardon et al. (2016). c Left premotor neurons labeled by two distinct MN pools that regulate antagonistic muscles in the ankle, TA and GS. Right transverse projections of digital reconstructions showing distribution of GS and TA premotor neurons in the spinal cord. Dots indicate labeled neurons. Scale bar, 70 μm. Images are taken with permission from Tripodi et al. (2011). d Premotor neurons labeled from the contralateral MN pools innervating TA muscles in wild type (left) and in Lbx1-lineage specific EphA4 conditional knockout mouse (right). Images are taken with permission from Satoh et al. (2016)
5.1 Methods to Generate Starter Cells in Motor Neurons
As introduced in Sect. 4.2.4, starter cells can be generated in the MNs belonging to a specific motor pool by coinjecting RVdRG+RG and a helper AAV expressing RG into a target skeletal muscle during the neonatal period in mice (Stepien et al. 2010) (hereafter we will call this scheme ‘muscle coinjection method’ for simplicity, Fig. 4.13a). There are three major advantages in the muscle coinjection method. First, just by changing a target muscle, researchers can easily generate starter cells in distinct MN pools. This enabled systematic investigation of the structure of premotor landscapes that regulate antagonistic movements of a joint (Tripodi et al. 2011), and coordinated movements of different limbs (Esposito et al. 2014; Goetz et al. 2015), as we will discuss in Sect. 4.5.2. Second, because the muscle coinjection method does not utilize mouse genetics for generating starter cells, it can be combined with various Cre-mediated viral and genetic tools, such as cell-type specific labeling, lineage tracing, axon projection mapping, and gene conditional knockout. Third, in principle, it can be applied to many mammalian species beyond mice in which Cre transgenic animals are currently unavailable. However, there is also a caveat in this method, with regards to the specificity of RV retrograde tracing. In the central nervous system, RV exhibits remarkable retrograde direction specificity (Ugolini 2011; Miyamichi et al. 2011; Zampieri et al. 2014). An important exception to this general rule was found in the peripheral sensory neurons. RV particles can directly infect proprioceptive sensory neurons in the dorsal root ganglion (DRG) (Tsiang et al. 1989) and ORNs in the olfactory epithelium (Astic et al. 1993), and can then anterogradely and transsynaptically spread to the postsynaptic neurons in the spinal cord and in the OB (Zampieri et al. 2014). Although the cellular mechanisms underlying this sensory neuron-specific anterograde RV spread remain elusive, this property of RV should be carefully considered when using the muscle coinjection method, as it can also generate starter cells in the proprioceptive sensory neurons in the DRG that connect to a target muscle.
At the neuromuscular junction, MNs release acetylcholine (ACh) as a neurotransmitter. To genetically restrict the starter cells to MNs, researchers utilized a transgenic mouse line expressing Cre under the promoter of choline acetyltransferase (ChAT) gene, which encodes the specific enzyme that synthesizes ACh. To avoid potential ‘contamination’ of ChAT positive non-MN local neurons in the spinal cord (Stepien et al. 2010), researchers injected Cre-dependent AAVs expressing RG and TVA into a target muscle, and RVdRG+EnvA into the spinal cord of ChAT-Cre mice (Fig. 4.13b; Reardon et al. 2016). In this configuration, proprioceptive sensory neurons in the DRG are genetically excluded from the starter cells because they do not express ChAT. With this ‘clean’ condition, synapse specificity of RV retrograde spread was elegantly demonstrated in known sensory-motor circuits controlling the ankle joint. Two distinct muscles in the ankle, the gastrocnemius (GS) and tibialis anterior (TA) muscles, work antagonistically: GS muscles act as extensors (whose contraction increases the angle of the ankle joint) and TA muscles act as flexors (whose contraction decreases the angle) (Fig. 4.13b). Proprioceptive sensory neurons monitoring GS muscles connect to the MNs that innervate the same GS muscles (forming a monosynaptic loop), but not to the MNs inverting antagonistic TA muscles, despite the fact that dendrites of these two types of MNs are intermingled in the spinal cord (Frank and Westerfield 1983). If RV can nonsynaptically spread to nearby neurons, retrograde tracing from the MNs innervating GS muscles will mislabel proprioceptive sensory neurons connecting to TA muscles. It turned out that RV-positive sensory endings in the GS muscles were clearly observed, but not in the TA muscles (Reardon et al. 2016), demonstrating a tightly controlled synaptic spread of RV particles in vivo (Fig. 4.13b).
Although utilizing ChAT-Cre mice and Cre-dependent AAVs injected into a muscle provides better restriction of starter cells to a defined MN pool, this method compromises the utility of Cre-mediated tools for other labeling and genetic manipulations beyond generating starter cells. The muscle coinjection method may be beneficial if RV anterograde tracing from the proprioceptive starter cells are limited in the experimental condition. Researchers, therefore, compared patterns of the spinal premotor neurons labeled from three defined MN pools by two methods: the muscle coinjection method and ChAT-Cre-mediated genetic control of starter MNs (Goetz et al. 2015). Both methods showed very similar spatial distributions of labeled premotor neurons in the spinal cord that were highly characteristic to the starter MN pools. Thus, muscle coinjection method can be reliably used for the MNs tested in this study, and presumably for other MNs. However, careful control experiments should be conducted when the experimental conditions are changed. In the following sections, we will discuss biological findings obtained mainly by using the muscle coinjection method.
5.2 Organization of Presynaptic Neurons of a Defined Motor Neuron Pool
The muscle coinjection method allows researchers to assess global 3D distributions of presynaptic neurons that connect to a defined MN pool. Labeled premotor neurons are bilaterally distributed (Fig. 4.13a) across many spinal cord segments, and contain known premotor populations (Stepien et al. 2010; Levine et al. 2014). These premotor neurons consist of multiple subtypes that use different neurotransmitters including glutamate, GABA, glycine, and ACh. Overall distributions of spinal premotor neuron connectivity to an individual MN pool exhibit a high degree of reproducibility across animals. In contrast, analysis of premotor neurons connecting to MN pools with distinct function in motor behavior reveals striking differences in distribution (Tripodi et al. 2011; Goetz et al. 2015). As a remarkable example, Fig. 4.13c shows the spatially segregated distributions of premotor neurons that connect to MNs innervating the antagonistic ankle muscles, extensor GS and flexor TA, along the medial-lateral axis of the spinal cord. Analysis of developmental history revealed that these premotor interneuron populations were derived from common progenitor domains, but segregated by birthdate of neurons. Further, proprioceptive sensory feedback via the DRG was preferentially targeted to medial premotor populations connecting to the extensor MN pools. Together, RV tracing reveals the structural basis for controlling functionally distinct muscles at the premotor circuit level.
The complexity of spinal premotor neurons, including their diverse developmental origins, neurotransmitter types, and connection specificity, poses a major challenge for researchers to functionally dissect them in specific regulations of motor coordination. Here we focus on one example of motor control, the sequential stepping of left and right limbs during walking. This relatively simple locomotor behavior is generated by the rhythmic activity of MNs under the control of spinal neuronal networks known as central pattern generators (CPGs) that comprise of multiple interneuron cell types (Goulding 2009). A gene knockout study demonstrated that EphA4, a tyrosine kinase axon guidance receptor expressed by many excitatory spinal neurons, played an important role in normal walking in mice. Wild-type mice alternate their left and right limbs during walking, while the mutant mice synchronize their limbs (rabbit-like hopping rather than walking). What would be the underlying circuit alterations responsible for walking abnormality in the mutant mice? Researchers have begun to shed light on this issue by using the muscle coinjection method. Labeling of TA flexor premotor neurons in EphA4 mutant mice led to the identification of abnormal synaptic input to the MNs from contralateral dorsal interneurons, a population defined by a transient expression of a transcription factor Lbx1 during development (Satoh et al. 2016). In an EphA4 conditional knockout (cKO) mouse where EphA4 was removed specifically from Lbx1+ neurons during development, approximately seven-times more TA premotor interneurons were labeled in the dorsal contralateral quadrant compared with WT control (Fig. 4.13d). As these Lbx1-lineage spinal interneurons directly receive proprioceptive sensory information, abnormal connectivity in EphA4 mutant mice can transmit the sensory feedback signals to both sides of the spinal cord, while in wild type mice they are dominantly transmitted to the ipsilateral side. As a result, the cKO mice displayed different locomotor patterns depending on the strength of sensory feedback. While they showed normal alternating gait during walking (with strong sensory feedback), the same mice exhibited synchronous gait strokes during swimming (without strong sensory feedback), akin to a butterfly-like swimming style, in sharp contrast to a crawling-like swimming style with limb alternation observed in wild-type mice. Thus, Lbx1-lineage specific cKO of EphA4 induces specific connectivity changes in sensory-relay interneurons that influence the robustness of gait choice during locomotion. This work also demonstrates the power of combining the muscle coinjection method with mouse genetics for the analysis of neuronal circuits in specific gene loss-of-function models.
The Lbx1-lineage spinal interneurons are only a part of the CPG network for walking. A homeobox transcription factor Dbx1, which defines the fate of spinal interneurons called V0 interneurons during development, also impacts the left–right alternations during walking (Lanuza et al. 2004). Genetic and cell-type specific ablation of V0 interneurons caused a similar abnormal hopping phenotype at all locomotion speeds, while more selective ablation of inhibitory or excitatory V0 interneurons showed abnormal hopping at a specific range of speed (Talpalar et al. 2013). More comprehensive connection studies in the future should reveal a full picture of spinal pattern generators and their modulators that underlie locomotion in mice.
5.3 The Brainstem Nuclei for Motor Controls
In addition to the local spinal premotor interneurons, RV tracing also labels long-distance brainstem nuclei that are directly presynaptic to a defined MN pool in the spinal cord. These nuclei may bridge between the higher brain areas that command motor controls and spinal networks for motor output. A salient example is the identification of premotor neurons in a brainstem nucleus called medullary reticular formation ventral part (MdV), which preferentially innervate MNs controlling forelimb over those regulating hindlimb (Fig. 4.14a; Esposito et al. 2014). MdV premotor neurons are glutamatergic (vGluT2+) and innervate a specific subset of forelimb MNs as well as spinal interneurons. To decipher their function in motor control, two lines of experiments were performed. First, Cre-dependent RV transsynaptic tracing using vGluT2-Cre mice demonstrated that glutamatergic neurons in the MdV integrated various inputs from upper motor centers including motor cortex, deep cerebellar nuclei (which convey the output signals of the cerebellum), superior colliculus, and other brainstem nuclei. Second, cell-type specific ablation or pharmacogenetic silencing of the MdV glutamatergic neurons markedly impaired skilled motor behaviors, such as accelerating rotarod task, without affecting a simple motor behavior such as running around the home cage. Thus, while simple locomotion seems to be regulated in the spinal pattern generators, skilled behavior requires higher brain centers.
Neuronal circuits in the brainstem motor control areas by RV transsynaptic tracing. a Left images distribution of brainstem premotor neurons that innervate the forelimb (left) or hindlimb (right) muscles, revealed by the muscle coinjection method. Dots represent premotor neurons, which are differentially colored according to their locations in specific brainstem nuclei. Yellow area vestibular nuclei; green area inferior olive. Distributions of forelimb and hindlimb premotor neurons are comparable in most brainstem nuclei, except in MdV, which contains almost exclusively forelimb premotor neurons. Right schematic summarizes the connection diagram of MdV, which collects broad motor-related inputs from higher brain centers and innervates forelimb MNs (yellow circle), as well as excitatory (green circle) and inhibitory (red circle) spinal premotor neurons. Adapted with permission from Esposito et al. (2014). b Schematic summary of vestibular feedback signals to MNs via lateral vestibular nucleus (LVe) in the brainstem. LVe neurons preferentially innervate extensor over flexor MN pools, and within the extensor MN pools, slow over fast-fatigable MNs. Adapted with permission from Basaldella et al. (2015). c Schematic summary of spinal premotor projection neurons that send copy signals to the lateral reticular nucleus (LRN) in the brainstem, which relays the copy signals to the cerebellum via mossy fibers. Spinal premotor projection neurons, colored according to different progenitor origins, send projections to distinct spatial domains in the LRN. Adapted with permission from Pivetta et al. (2014)
Another important brainstem area that is presynaptic to MNs is the lateral vestibular nucleus (LVe). Neurons in this region convey information from the peripheral vestibular system in the inner ear about movement and orientation of the head. The vestibular feedback signals can be used for continuous postural adjustments during movement, which is necessary for reliable motor behaviors. Researchers applied the muscle coinjection method to limb muscles and found direct premotor neurons in the LVe (Fig. 4.14b; Basaldella et al. 2015). To analyze the type of MNs that LVe neurons provide vestibular feedback signals to, axon projection mapping was used in the LVe, with putative postsynaptic MNs labeled retrogradely from the target muscles. Two layers of specificity for LVe connectivity to MNs were highlighted. LVe axons preferentially contacted extensor (GS) over flexor (TA) motor pools. In addition, LVe axons targeted slow MN subtypes over fast-fatigable subtypes within the extensor pools. What is the importance of this target specificity? Individual MNs vary in size and small MNs innervate relatively few muscle fibers to form motor units that generate small forces (called slow motor units). This is especially important for sustained muscle contraction, such as the posture maintenance. In contrast, the larger MNs innervate larger muscle fibers that generate more forces, but are easily fatigued (called fast-fatigable motor units), which play important roles in brief exertions of large forces such as running or jumping (Kanning et al. 2010). Thus, LVe neurons are specifically programmed such that the vestibular feedback signals are preferentially conveyed to the subtype of MNs that regulate muscle movements for posture maintenance.
So far we have discussed brainstem to spinal cord connections, but it is known that the opposite flow, from spinal cord to brainstem, also exists. For example, the lateral reticular nucleus (LRN) in the brainstem receives axonal projections from spinal premotor neurons and relays their activities to the cerebellum via mossy fibers. This signal strongly correlates with ongoing spinal intrinsic information (‘copy signals’) and therefore enables the cerebellum to compare planned actions and ongoing performance of motor control in the spinal cord. To analyze this circuit in detail, researchers applied the muscle coinjection method and detected axonal projections of spinal premotor neurons in specific areas of the LRN (Pivetta et al. 2014). To directly link the spinal input to the cerebellum, TRIO strategy (Sect. 4.2.4) was used and revealed that LRN monosynaptically relayed a diverse set of premotor input associated with functionally distinct MN pools to the cerebellum (Fig. 4.14c). Then, axon projection mapping was used to analyze the topography, the neurotransmitter types, and developmental origin of spinal premotor neurons that project to the LRN. This analysis revealed that both excitatory and inhibitory spinal inputs were highly co-localized within the LRN, but neurons in the different spinal cord domains (cervical or lumbar) innervated distinct LRN domains, showing topographic organization. Also, spinal neurons of different progenitor origins (marked by a transient expression of a specific transcription factor during development) established distinct axonal terminations in the LRN. Collectively, these data provide the anatomical basis upon which functional dissection studies of individual copy signals (Azim et al. 2014; Fink et al. 2014) can be integrated to decipher the principles of accurate motor task execution.
The brainstem also contains MNs regulating cranial muscles. For example, whisking (bilaterally coordinated rhythmic sweeping movements of whiskers to detect the texture, shape, and location of objects) is mediated by a specific type of MNs called vibrissal facial motor neurons (vFMNs) in the lateral facial nucleus in the brainstem. Studies of vFMN premotor pools provide an interesting example of utilizing RV tracing to analyze the developmental timecourse of circuit assembly. Researchers injected RVdRG-GFP+RG into the whisker-controlling muscles of a transgenic mouse where all MNs were expressing RG (Takatoh et al. 2013). This strategy permits a side-by-side comparison of labeled premotor neurons before (postnatal day 8) and after (postnatal day 15) the developmental onset of coordinated whisking behaviors. It turned out that the emergence of whisking was associated with the addition of new sets of bilateral excitatory inputs to vFMNs from neurons located in a brainstem nucleus called LPGi (lateral paragigantocellularis). Axon projection mapping indicated that neurons in the LPGi relayed motor commands from the motor cortex to the vFMNs. Therefore, this study shed light on how neuronal circuit assembly during development facilitates the bilateral coordination and cortical control of whisking.
Together, these studies of premotor neurons highlight the rapid advances that RV tracing have provided for uncovering complex yet elegant connection patterns in the spinal cord and brainstem underlying motor controls. These studies also nicely combined RV tracing with other viral and genetic tools, such as lineage tracing, cell-type specific axon projection mapping, gene conditional knockout, and manipulation of neuronal activities. Many techniques and ideas developed in the motor control system should facilitate anatomical, developmental, and functional studies of other neuronal circuits based on RV tracing.
6 Future Perspectives
As we have discussed in the above sections, RV-mediated transsynaptic tracing has vastly expanded the scope of neuroanatomy by providing precise information of presynaptic partners to defined neuron populations throughout the brain and spinal cord. We will now end by briefly considering several future directions of transsynaptic tracing techniques.
First, the range of species and developmental stages to which RV tracing can be applied should be extended. Currently, RV tracing has been most successfully used in adult mice. Just like comparative genomics has greatly expanded our understanding of the evolution of gene families, comparative connectomics are essential to illuminate the evolution of neuronal circuits. For example, neuronal circuits regulating social behaviors may drastically change as animals develop in more sophisticated communities. Likewise, circuits regulating feeding and energy expenditure might be altered when animals evolve a specific feeding habit. To achieve these studies, it is necessary to establish RV tracing methods in various mammalian species. To further expand tracing studies beyond mammalian species, new viral tools would be required. For example, a recent study showed that VSV (see Table 4.2) can be a better tracer than RV in some bird and fish species (Mundell et al. 2015). RV tracing also has a potential to reveal developmental processes (Takatoh et al. 2013), but current AAV-based methods require a ~2 week incubation period before RV tracing is initiated. Future technical advances are needed to fully utilize RV tracing in embryonic and early postnatal animals, to decipher the developmental assembly of neuronal circuits in the central brain. In addition to extending retrograde tracing techniques, developing a more efficient and less toxic anterograde transsynaptic tracing system will further facilitate the tracing studies, as currently available anterograde viral vectors are either limited to specific cell types [e.g., sensory neurons (Zampieri et al. 2014; Astic et al. 1993)] or highly toxic to the infected neurons (see Table 4.2).
Second, a standard data acquisition and analysis platform is necessary to compare tracing data from various brain regions and cell types obtained from multiple laboratories. Typically, the acquisition of tracing data has been achieved by serially sectioning the brain, imaging individual sections, manually defining regional boundaries in each section, and counting the number of cells in each region (Weissbourd et al. 2014; Schwarz et al. 2015; Watabe-Uchida et al. 2012; Beier et al. 2015). These processes are not only labor-intensive and require significant expertise of brain anatomy, but also are prone to human-based errors and biases. Ideally, tracing data should be acquired with fully automatic brain imaging methods, such as serial sectioning 2-photon microscopy (Velez-Fort et al. 2014) or optical clearing methods followed by light-sheet microscopy (Lerner et al. 2015; Menegas et al. 2015; Niedworok et al. 2012). An automated data analysis platform for the classification of labeled cells into distinct brain regions should also be established. Often, fine regional boundaries between brain sub-nuclei are not apparent based on the gross morphology of the brain, and remain a subject of dispute among anatomy researchers. Rapid advances in the field of machine learning, as well as improvements in automatic brain imaging methods, may provide powerful solutions to this issue. Once precise methods to image brains, to extract information of labeled cells, and to register them into a common reference brain are established, it will become possible to quantitatively compare input neurons to various starter cell types throughout the brain. A powerful algorithm to compare the ‘similarity’ of labeled patterns among samples, similar to BLAST in the field of genomics, will be highly useful. This will permit systematic comparisons of connection diagrams in a variety of contexts; for example, male versus female, young versus aged, healthy versus diseased, and before and after animals acquire new tasks. The impacts of genetic modifications (such as conditional gene knockout as pioneered by DeNardo et al. 2015, Satoh et al. 2016) on neuronal circuit assembly and integrity can also be systematically investigated.
Lastly, RV tracing should directly link connectivity to circuit function. One important advantage of RV over other neurotrophic viruses is that neurons survive much longer following viral infection. This enables functional imaging and perturbation of neurons that have been identified based on their connectivity. Indeed, researchers have developed various RV vectors for expressing molecules that manipulate and measure neuronal activity, or express a recombinase (Cre or Flp) to induce expression of a second transgene (Osakada et al. 2011; Reardon et al. 2016). For example, RV-mediated ChR2-expressing cortical neurons elicited light-induced action potentials a month after infection, and GCaMP6f-expressing hippocampal projection neurons showed neuronal burst activities in behaving mice at least two weeks after infection (Reardon et al. 2016). Many of these, or analogous constructs, have begun to dissect the complex neuronal circuits in the brain (see Sect. 4.2.6; Wertz et al. 2015, Yamawaki and Shepherd 2015) and in retina (Yonehara et al. 2013). Future studies will greatly expand this direction to dissect how circuit function is orchestrated by various input neurons in the brain and spinal cord.
References
Adelson JD, Sapp RW, Brott BK, Lee H, Miyamichi K, Luo L, Cheng S, Djurisic M, Shatz CJ (2016) Developmental sculpting of intracortical circuits by MHC Class I H2-Db and H2-Kb. Cereb Cortex 26(4):1453–1463. doi:10.1093/cercor/bhu243
Aghajanian GK, Wang RY (1977) Habenular and other midbrain raphe afferents demonstrated by a modified retrograde tracing technique. Brain Res 122(2):229–242
Aponte Y, Atasoy D, Sternson SM (2011) AGRP neurons are sufficient to orchestrate feeding behavior rapidly and without training. Nat Neurosci 14(3):351–355. doi:10.1038/nn.2739
Arber S (2012) Motor circuits in action: specification, connectivity, and function. Neuron 74(6):975–989. doi:10.1016/j.neuron.2012.05.011
Arenkiel BR, Hasegawa H, Yi JJ, Larsen RS, Wallace ML, Philpot BD, Wang F, Ehlers MD (2011) Activity-induced remodeling of olfactory bulb microcircuits revealed by monosynaptic tracing. PLoS ONE 6(12):e29423. doi:10.1371/journal.pone.0029423
Astic L, Saucier D, Coulon P, Lafay F, Flamand A (1993) The CVS strain of rabies virus as transneuronal tracer in the olfactory system of mice. Brain Res 619(1–2):146–156
Aston-Jones G, Ennis M, Pieribone VA, Nickell WT, Shipley MT (1986) The brain nucleus locus coeruleus: restricted afferent control of a broad efferent network. Science 234(4777):734–737
Atasoy D, Betley JN, Su HH, Sternson SM (2012) Deconstruction of a neural circuit for hunger. Nature 488(7410):172–177. doi:10.1038/nature11270
Azim E, Jiang J, Alstermark B, Jessell TM (2014) Skilled reaching relies on a V2a propriospinal internal copy circuit. Nature 508(7496):357–363. doi:10.1038/nature13021
Bang SJ, Commons KG (2012) Forebrain GABAergic projections from the dorsal raphe nucleus identified by using GAD67-GFP knock-in mice. J Comp Neurol 520(18):4157–4167. doi:10.1002/cne.23146
Basaldella E, Takeoka A, Sigrist M, Arber S (2015) Multisensory signaling shapes vestibulo-motor circuit specificity. Cell 163(2):301–312. doi:10.1016/j.cell.2015.09.023
Bassareo V, Tanda G, Petromilli P, Giua C, Di Chiara G (1996) Non-psychostimulant drugs of abuse and anxiogenic drugs activate with differential selectivity dopamine transmission in the nucleus accumbens and in the medial prefrontal cortex of the rat. Psychopharmacology 124(4):293–299
Bates P, Young JAT, Varmus HE (1993) A receptor for subgroup-a rous-sarcoma virus is related to the low-density-lipoprotein receptor. Cell 74(6):1043–1051. doi:10.1016/0092-8674(93)90726-7
Beckstead RM, Domesick VB, Nauta WJ (1979) Efferent connections of the substantia nigra and ventral tegmental area in the rat. Brain Res 175(2):191–217
Beier KT, Saunders A, Oldenburg IA, Miyamichi K, Akhtar N, Luo L, Whelan SP, Sabatini B, Cepko CL (2011) Anterograde or retrograde transsynaptic labeling of CNS neurons with vesicular stomatitis virus vectors. Proc Natl Acad Sci USA 108(37):15414–15419. doi:10.1073/pnas.1110854108
Beier KT, Steinberg EE, DeLoach KE, Xie S, Miyamichi K, Schwarz L, Gao XJ, Kremer EJ, Malenka RC, Luo L (2015) Circuit architecture of VTA dopamine Neurons revealed by systematic input-output mapping. Cell 162(3):622–634. doi:10.1016/j.cell.2015.07.015
Berridge CW, Waterhouse BD (2003) The locus coeruleus-noradrenergic system: modulation of behavioral state and state-dependent cognitive processes. Brain Res Brain Res Rev 42(1):33–84
Betley JN, Cao ZF, Ritola KD, Sternson SM (2013) Parallel, redundant circuit organization for homeostatic control of feeding behavior. Cell 155(6):1337–1350. doi:10.1016/j.cell.2013.11.002
Bromberg-Martin ES, Matsumoto M, Hikosaka O (2010) Dopamine in motivational control: rewarding, aversive, and alerting. Neuron 68(5):815–834. doi:10.1016/j.neuron.2010.11.022
Callaway EM, Luo L (2015) Monosynaptic circuit tracing with glycoprotein-deleted rabies viruses. J Neurosci Off J Soc Neurosci 35(24):8979–8985. doi:10.1523/JNEUROSCI.0409-15.2015
Card JP, Whealy ME, Robbins AK, Enquist LW (1992) Pseudorabies virus envelope glycoprotein gI influences both neurotropism and virulence during infection of the rat visual system. J Virol 66(5):3032–3041
Cedarbaum JM, Aghajanian GK (1978) Afferent projections to the rat locus coeruleus as determined by a retrograde tracing technique. J Comp Neurol 178(1):1–16. doi:10.1002/cne.901780102
Chuhma N, Tanaka KF, Hen R, Rayport S (2011) Functional connectome of the striatal medium spiny neuron. J Neurosci Off J Soc Neurosci 31(4):1183–1192. doi:10.1523/JNEUROSCI.3833-10.2011
Cowan WM, Gottlieb DI, Hendrickson AE, Price JL, Woolsey TA (1972) The autoradiographic demonstration of axonal connections in the central nervous system. Brain Res 37(1):21–51
Cowley MA, Smart JL, Rubinstein M, Cerdan MG, Diano S, Horvath TL, Cone RD, Low MJ (2001) Leptin activates anorexigenic POMC neurons through a neural network in the arcuate nucleus. Nature 411(6836):480–484. doi:10.1038/35078085
Darvas M, Palmiter RD (2009) Restriction of dopamine signaling to the dorsolateral striatum is sufficient for many cognitive behaviors. Proc Natl Acad Sci USA 106(34):14664–14669. doi:10.1073/pnas.0907299106
Darvas M, Palmiter RD (2010) Restricting dopaminergic signaling to either dorsolateral or medial striatum facilitates cognition. J Neurosci Off J Soc Neurosci 30(3):1158–1165. doi:10.1523/JNEUROSCI.4576-09.2010
DeFalco J, Tomishima M, Liu H, Zhao C, Cai X, Marth JD, Enquist L, Friedman JM (2001) Virus-assisted mapping of neural inputs to a feeding center in the hypothalamus. Science 291(5513):2608–2613. doi:10.1126/science.1056602
Deisseroth K (2015) Optogenetics: 10 years of microbial opsins in neuroscience. Nat Neurosci 18(9):1213–1225. doi:10.1038/nn.4091
DeNardo LA, Berns DS, DeLoach K, Luo L (2015) Connectivity of mouse somatosensory and prefrontal cortex examined with trans-synaptic tracing. Nat Neurosci 18(11):1687–1697. doi:10.1038/nn.4131
Deshpande A, Bergami M, Ghanem A, Conzelmann KK, Lepier A, Gotz M, Berninger B (2013) Retrograde monosynaptic tracing reveals the temporal evolution of inputs onto new neurons in the adult dentate gyrus and olfactory bulb. Proc Natl Acad Sci USA 110(12):E1152–E1161. doi:10.1073/pnas.1218991110
Esposito MS, Capelli P, Arber S (2014) Brainstem nucleus MdV mediates skilled forelimb motor tasks. Nature 508(7496):351–356. doi:10.1038/nature13023
Etessami R, Conzelmann KK, Fadai-Ghotbi B, Natelson B, Tsiang H, Ceccaldi PE (2000) Spread and pathogenic characteristics of a G-deficient rabies virus recombinant: an in vitro and in vivo study. J Gen Virol 81:2147–2153
Fink AJ, Croce KR, Huang ZJ, Abbott LF, Jessell TM, Azim E (2014) Presynaptic inhibition of spinal sensory feedback ensures smooth movement. Nature 509(7498):43–48. doi:10.1038/nature13276
Frank E, Westerfield M (1983) Development of sensory-motor synapses in the spinal cord of the frog. J Physiol 343:593–610
Franks KM, Russo MJ, Sosulski DL, Mulligan AA, Siegelbaum SA, Axel R (2011) Recurrent circuitry dynamically shapes the activation of piriform cortex. Neuron 72(1):49–56. doi:10.1016/j.neuron.2011.08.020
Fu Y, Tucciarone JM, Espinosa JS, Sheng N, Darcy DP, Nicoll RA, Huang ZJ, Stryker MP (2014) A cortical circuit for gain control by behavioral state. Cell 156(6):1139–1152. doi:10.1016/j.cell.2014.01.050
Garcia I, Quast KB, Huang L, Herman AM, Selever J, Deussing JM, Justice NJ, Arenkiel BR (2014) Local CRH signaling promotes synaptogenesis and circuit integration of adult-born neurons. Dev Cell 30(6):645–659. doi:10.1016/j.devcel.2014.07.001
Gerfen CR, Paletzki R, Heintz N (2013) GENSAT BAC cre-recombinase driver lines to study the functional organization of cerebral cortical and basal ganglia circuits. Neuron 80(6):1368–1383. doi:10.1016/j.neuron.2013.10.016
Goetz C, Pivetta C, Arber S (2015) Distinct limb and trunk premotor circuits establish laterality in the spinal cord. Neuron 85(1):131–144. doi:10.1016/j.neuron.2014.11.024
Goodpasture EW, Teague O (1923) Transmission of the virus of herpes febrilis along nerves in experimentally infected rabbits. J Med Res 44(2):139–184 (137)
Goulding M (2009) Circuits controlling vertebrate locomotion: moving in a new direction. Nat Rev Neurosci 10(7):507–518. doi:10.1038/nrn2608
Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, Macklis JD (2013) Molecular logic of neocortical projection neuron specification, development and diversity. Nat Rev Neurosci 14(11):755–769. doi:10.1038/nrn3586
Haubensak W, Kunwar PS, Cai H, Ciocchi S, Wall NR, Ponnusamy R, Biag J, Dong HW, Deisseroth K, Callaway EM, Fanselow MS, Luthi A, Anderson DJ (2010) Genetic dissection of an amygdala microcircuit that gates conditioned fear. Nature 468(7321):270–276. doi:10.1038/nature09553
Helmstaedter M, Briggman KL, Turaga SC, Jain V, Seung HS, Denk W (2013) Connectomic reconstruction of the inner plexiform layer in the mouse retina. Nature 500(7461):168–174. doi:10.1038/nature12346
Hioki H, Nakamura H, Ma YF, Konno M, Hayakawa T, Nakamura KC, Fujiyama F, Kaneko T (2010) Vesicular glutamate transporter 3-expressing nonserotonergic projection neurons constitute a subregion in the rat midbrain raphe nuclei. J Comp Neurol 518(5):668–686. doi:10.1002/cne.22237
Hnasko TS, Perez FA, Scouras AD, Stoll EA, Gale SD, Luquet S, Phillips PE, Kremer EJ, Palmiter RD (2006) Cre recombinase-mediated restoration of nigrostriatal dopamine in dopamine-deficient mice reverses hypophagia and bradykinesia. Proc Natl Acad Sci USA 103(23):8858–8863. doi:10.1073/pnas.0603081103
Hnasko TS, Chuhma N, Zhang H, Goh GY, Sulzer D, Palmiter RD, Rayport S, Edwards RH (2010) Vesicular glutamate transport promotes dopamine storage and glutamate corelease in vivo. Neuron 65(5):643–656. doi:10.1016/j.neuron.2010.02.012
Hnasko TS, Hjelmstad GO, Fields HL, Edwards RH (2012) Ventral tegmental area glutamate neurons: electrophysiological properties and projections. J Neurosci Off J Soc Neurosci 32(43):15076–15085. doi:10.1523/JNEUROSCI.3128-12.2012
Igarashi KM, Ieki N, An M, Yamaguchi Y, Nagayama S, Kobayakawa K, Kobayakawa R, Tanifuji M, Sakano H, Chen WR, Mori K (2012) Parallel mitral and tufted cell pathways route distinct odor information to different targets in the olfactory cortex. J Neurosci Off J Soc Neurosci 32(23):7970–7985. doi:10.1523/JNEUROSCI.0154-12.2012
Kanning KC, Kaplan A, Henderson CE (2010) Motor neuron diversity in development and disease. Annu Rev Neurosci 33:409–440. doi:10.1146/annurev.neuro.051508.135722
Kasthuri N, Hayworth KJ, Berger DR, Schalek RL, Conchello JA, Knowles-Barley S, Lee D, Vazquez-Reina A, Kaynig V, Jones TR, Roberts M, Morgan JL, Tapia JC, Seung HS, Roncal WG, Vogelstein JT, Burns R, Sussman DL, Priebe CE, Pfister H, Lichtman JW (2015) Saturated reconstruction of a volume of neocortex. Cell 162(3):648–661. doi:10.1016/j.cell.2015.06.054
Kato HK, Chu MW, Isaacson JS, Komiyama T (2012) Dynamic sensory representations in the olfactory bulb: modulation by wakefulness and experience. Neuron 76(5):962–975. doi:10.1016/j.neuron.2012.09.037
Kato HK, Gillet SN, Peters AJ, Isaacson JS, Komiyama T (2013) Parvalbumin-expressing interneurons linearly control olfactory bulb output. Neuron 80(5):1218–1231. doi:10.1016/j.neuron.2013.08.036
Kim EJ, Juavinett AL, Kyubwa EM, Jacobs MW, Callaway EM (2015) Three types of cortical layer 5 neurons that differ in brain-wide connectivity and function. Neuron 88(6):1253–1267. doi:10.1016/j.neuron.2015.11.002
Kiritani T, Wickersham IR, Seung HS, Shepherd GM (2012) Hierarchical connectivity and connection-specific dynamics in the corticospinal-corticostriatal microcircuit in mouse motor cortex. J Neurosci Off J Soc Neurosci 32(14):4992–5001. doi:10.1523/JNEUROSCI.4759-11.2012
Kobayakawa K, Kobayakawa R, Matsumoto H, Oka Y, Imai T, Ikawa M, Okabe M, Ikeda T, Itohara S, Kikusui T, Mori K, Sakano H (2007) Innate versus learned odour processing in the mouse olfactory bulb. Nature 450(7169):503–508. doi:10.1038/nature06281
Kohara K, Pignatelli M, Rivest AJ, Jung HY, Kitamura T, Suh J, Frank D, Kajikawa K, Mise N, Obata Y, Wickersham IR, Tonegawa S (2014) Cell type-specific genetic and optogenetic tools reveal hippocampal CA2 circuits. Nat Neurosci 17(2):269–279. doi:10.1038/nn.3614
Krashes MJ, Koda S, Ye C, Rogan SC, Adams AC, Cusher DS, Maratos-Flier E, Roth BL, Lowell BB (2011) Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. J Clin Investig 121(4):1424–1428. doi:10.1172/JCI46229
Krashes MJ, Shah BP, Madara JC, Olson DP, Strochlic DE, Garfield AS, Vong L, Pei H, Watabe-Uchida M, Uchida N, Liberles SD, Lowell BB (2014) An excitatory paraventricular nucleus to AgRP neuron circuit that drives hunger. Nature 507(7491):238–242. doi:10.1038/nature12956
Lafon M (2005) Rabies virus receptors. J Neurovirol 11(1):82–87. doi:10.1080/13550280590900427
Lammel S, Hetzel A, Hackel O, Jones I, Liss B, Roeper J (2008) Unique properties of mesoprefrontal neurons within a dual mesocorticolimbic dopamine system. Neuron 57(5):760–773. doi:10.1016/j.neuron.2008.01.022
Lammel S, Ion DI, Roeper J, Malenka RC (2011) Projection-specific modulation of dopamine neuron synapses by aversive and rewarding stimuli. Neuron 70(5):855–862. doi:10.1016/j.neuron.2011.03.025
Lammel S, Lim BK, Ran C, Huang KW, Betley MJ, Tye KM, Deisseroth K, Malenka RC (2012) Input-specific control of reward and aversion in the ventral tegmental area. Nature 491(7423):212–217. doi:10.1038/nature11527
Lanuza GM, Gosgnach S, Pierani A, Jessell TM, Goulding M (2004) Genetic identification of spinal interneurons that coordinate left-right locomotor activity necessary for walking movements. Neuron 42(3):375–386
Lerner TN, Shilyansky C, Davidson TJ, Evans KE, Beier KT, Zalocusky KA, Crow AK, Malenka RC, Luo L, Tomer R, Deisseroth K (2015) Intact-brain analyses reveal distinct information carried by SNc dopamine subcircuits. Cell 162(3):635–647. doi:10.1016/j.cell.2015.07.014
Levine AJ, Hinckley CA, Hilde KL, Driscoll SP, Poon TH, Montgomery JM, Pfaff SL (2014) Identification of a cellular node for motor control pathways. Nat Neurosci 17(4):586–593. doi:10.1038/nn.3675
Li Y, Stam FJ, Aimone JB, Goulding M, Callaway EM, Gage FH (2013) Molecular layer perforant path-associated cells contribute to feed-forward inhibition in the adult dentate gyrus. Proc Natl Acad Sci USA 110(22):9106–9111. doi:10.1073/pnas.1306912110
Liu S, Plachez C, Shao Z, Puche A, Shipley MT (2013) Olfactory bulb short axon cell release of GABA and dopamine produces a temporally biphasic inhibition-excitation response in external tufted cells. J Neurosci Off J Soc Neurosci 33(7):2916–2926. doi:10.1523/JNEUROSCI.3607-12.2013
Lo L, Anderson DJ (2011) A cre-dependent, anterograde transsynaptic viral tracer for mapping output pathways of genetically marked neurons. Neuron 72(6):938–950. doi:10.1016/j.neuron.2011.12.002
Magnus CJ, Lee PH, Atasoy D, Su HH, Looger LL, Sternson SM (2011) Chemical and genetic engineering of selective ion channel-ligand interactions. Science 333(6047):1292–1296. doi:10.1126/science.1206606
Marder E (2012) Neuromodulation of neuronal circuits: back to the future. Neuron 76(1):1–11. doi:10.1016/j.neuron.2012.09.010
Margolis EB, Mitchell JM, Ishikawa J, Hjelmstad GO, Fields HL (2008) Midbrain dopamine neurons: projection target determines action potential duration and dopamine D(2) receptor inhibition. J Neurosci Off J Soc Neurosci 28(36):8908–8913. doi:10.1523/JNEUROSCI.1526-08.2008
Margolis EB, Toy B, Himmels P, Morales M, Fields HL (2012) Identification of rat ventral tegmental area GABAergic neurons. PLoS ONE 7(7):e42365. doi:10.1371/journal.pone.0042365
Mebatsion T, Schnell MJ, Cox JH, Finke S, Conzelmann KK (1996a) Highly stable expression of a foreign gene from rabies virus vectors. Proc Natl Acad Sci USA 93(14):7310–7314. doi:10.1073/pnas.93.14.7310
Mebatsion T, Konig M, Conzelmann KK (1996b) Budding of rabies virus particles in the absence of the spike glycoprotein. Cell 84(6):941–951. doi:10.1016/S0092-8674(00)81072-7
Menegas W, Bergan JF, Ogawa SK, Isogai Y, Umadevi Venkataraju K, Osten P, Uchida N, Watabe-Uchida M (2015) Dopamine neurons projecting to the posterior striatum form an anatomically distinct subclass. eLife 4:e10032. doi:10.7554/eLife.10032
Mirenowicz J, Schultz W (1996) Preferential activation of midbrain dopamine neurons by appetitive rather than aversive stimuli. Nature 379(6564):449–451. doi:10.1038/379449a0
Miyamichi K, Amat F, Moussavi F, Wang C, Wickersham I, Wall NR, Taniguchi H, Tasic B, Huang ZJ, He Z, Callaway EM, Horowitz MA, Luo L (2011) Cortical representations of olfactory input by trans-synaptic tracing. Nature 472(7342):191–196. doi:10.1038/nature09714
Miyamichi K, Shlomai-Fuchs Y, Shu M, Weissbourd BC, Luo L, Mizrahi A (2013) Dissecting local circuits: parvalbumin interneurons underlie broad feedback control of olfactory bulb output. Neuron 80(5):1232–1245. doi:10.1016/j.neuron.2013.08.027
Mori K, Sakano H (2011) How is the olfactory map formed and interpreted in the mammalian brain? Annu Rev Neurosci 34:467–499. doi:10.1146/annurev-neuro-112210-112917
Müller CP, Jacobs BL (2010) Handbook of the behavioral neurobiology of serotonin. Elsevier, Amsterdam
Mundell NA, Beier KT, Pan YA, Lapan SW, Goz Ayturk D, Berezovskii VK, Wark AR, Drokhlyansky E, Bielecki J, Born RT, Schier AF, Cepko CL (2015) Vesicular stomatitis virus enables gene transfer and transsynaptic tracing in a wide range of organisms. J Comp Neurol 523(11):1639–1663. doi:10.1002/cne.23761
Nagai T, Satoh K, Imamoto K, Maeda T (1981) Divergent projections of catecholamine neurons of the locus coeruleus as revealed by fluorescent retrograde double labeling technique. Neurosci Lett 23(2):117–123
Nakashiba T, Cushman JD, Pelkey KA, Renaudineau S, Buhl DL, McHugh TJ, Barrera VR, Chittajallu R, Iwamoto KS, McBain CJ, Fanselow MS, Tonegawa S (2012) Young dentate granule cells mediate pattern separation, whereas old granule cells facilitate pattern completion. Cell 149(1):188–201. doi:10.1016/j.cell.2012.01.046
Namburi P, Beyeler A, Yorozu S, Calhoon GG, Halbert SA, Wichmann R, Holden SS, Mertens KL, Anahtar M, Felix-Ortiz AC, Wickersham IR, Gray JM, Tye KM (2015) A circuit mechanism for differentiating positive and negative associations. Nature 520(7549):675–678. doi:10.1038/nature14366
Nassi JJ, Cepko CL, Born RT, Beier KT (2015) Neuroanatomy goes viral! Front Neuroanat 9:80. doi:10.3389/fnana.2015.00080
Ni Y, Nawabi H, Liu X, Yang L, Miyamichi K, Tedeschi A, Xu B, Wall NR, Callaway EM, He Z (2014) Characterization of long descending premotor propriospinal neurons in the spinal cord. J Neurosci Off J Soc Neurosci 34(28):9404–9417. doi:10.1523/JNEUROSCI.1771-14.2014
Niedworok CJ, Schwarz I, Ledderose J, Giese G, Conzelmann KK, Schwarz MK (2012) Charting monosynaptic connectivity maps by two-color light-sheet fluorescence microscopy. Cell reports 2(5):1375–1386. doi:10.1016/j.celrep.2012.10.008
Ogawa SK, Cohen JY, Hwang D, Uchida N, Watabe-Uchida M (2014) Organization of monosynaptic inputs to the serotonin and dopamine neuromodulatory systems. Cell Rep 8(4):1105–1118. doi:10.1016/j.celrep.2014.06.042
Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, Wang Q, Lau C, Kuan L, Henry AM, Mortrud MT, Ouellette B, Nguyen TN, Sorensen SA, Slaughterbeck CR, Wakeman W, Li Y, Feng D, Ho A, Nicholas E, Hirokawa KE, Bohn P, Joines KM, Peng H, Hawrylycz MJ, Phillips JW, Hohmann JG, Wohnoutka P, Gerfen CR, Koch C, Bernard A, Dang C, Jones AR, Zeng H (2014) A mesoscale connectome of the mouse brain. Nature 508(7495):207–214. doi:10.1038/nature13186
Osakada F, Mori T, Cetin AH, Marshel JH, Virgen B, Callaway EM (2011) New rabies virus variants for monitoring and manipulating activity and gene expression in defined neural circuits. Neuron 71(4):617–631. doi:10.1016/j.neuron.2011.07.005
Oyibo HK, Znamenskiy P, Oviedo HV, Enquist LW, Zador AM (2014) Long-term Cre-mediated retrograde tagging of neurons using a novel recombinant pseudorabies virus. Front Neuroanat 8:86. doi:10.3389/fnana.2014.00086
Petreanu L, Huber D, Sobczyk A, Svoboda K (2007) Channelrhodopsin-2-assisted circuit mapping of long-range callosal projections. Nat Neurosci 10(5):663–668. doi:10.1038/nn1891
Pivetta C, Esposito MS, Sigrist M, Arber S (2014) Motor-circuit communication matrix from spinal cord to brainstem neurons revealed by developmental origin. Cell 156(3):537–548. doi:10.1016/j.cell.2013.12.014
Pollak Dorocic I, Furth D, Xuan Y, Johansson Y, Pozzi L, Silberberg G, Carlen M, Meletis K (2014) A whole-brain atlas of inputs to serotonergic neurons of the dorsal and median raphe nuclei. Neuron 83(3):663–678. doi:10.1016/j.neuron.2014.07.002
Poo C, Isaacson JS (2009) Odor representations in olfactory cortex: “sparse” coding, global inhibition, and oscillations. Neuron 62(6):850–861. doi:10.1016/j.neuron.2009.05.022
Rancz EA, Franks KM, Schwarz MK, Pichler B, Schaefer AT, Margrie TW (2011) Transfection via whole-cell recording in vivo: bridging single-cell physiology, genetics and connectomics. Nat Neurosci 14(4):527–532. doi:10.1038/nn.2765
Reardon TR, Murray AJ, Turi GF, Wirblich C, Croce KR, Schnell MJ, Jessell TM, Losonczy A (2016) Rabies virus CVS-N2c strain enhances retrograde synaptic transfer and neuronal viability. Neuron 89:1–14. doi:10.1016/j.neuron.2016.01.004
Rong L, Gendron K, Strohl B, Shenoy R, Wool-Lewis RJ, Bates P (1998) Characterization of determinants for envelope binding and infection in Tva, the subgroup A avian sarcoma and leukosis virus receptor. J Virol 72(6):4552–4559
Room P, Postema F, Korf J (1981) Divergent axon collaterals of rat locus coeruleus neurons: demonstration by a fluorescent double labeling technique. Brain Res 221(2):219–230
Roth BL (2016) DREADDs for neuroscientists. Neuron 89(4):683–694. doi:10.1016/j.neuron.2016.01.040
Satoh D, Pudenz C, Arber S (2016) Context-dependent gait choice elicited by EphA4 Mutation in Lbx1 spinal interneurons. Neuron 89:1–13
Saunders A, Granger AJ, Sabatini BL (2015) Corelease of acetylcholine and GABA from cholinergic forebrain neurons. eLife 4. doi:10.7554/eLife.06412
Schiemann J, Schlaudraff F, Klose V, Bingmer M, Seino S, Magill PJ, Zaghloul KA, Schneider G, Liss B, Roeper J (2012) K-ATP channels in dopamine substantia nigra neurons control bursting and novelty-induced exploration. Nat Neurosci 15(9):1272–1280. doi:10.1038/nn.3185
Schmued LC, Fallon JH (1986) Fluoro-Gold: a new fluorescent retrograde axonal tracer with numerous unique properties. Brain Res 377(1):147–154
Schnell MJ, Mebatsion T, Conzelmann KK (1994) Infectious rabies viruses from cloned cDNA. EMBO J 13(18):4195–4203
Schnutgen F, Doerflinger N, Calleja C, Wendling O, Chambon P, Ghyselinck NB (2003) A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. Nat Biotechnol 21(5):562–565. doi:10.1038/nbt811
Schwab ME, Javoy-Agid F, Agid Y (1978) Labeled wheat germ agglutinin (WGA) as a new, highly sensitive retrograde tracer in the rat brain hippocampal system. Brain Res 152(1):145–150
Schwarz LA, Miyamichi K, Gao XJ, Beier KT, Weissbourd B, DeLoach KE, Ren J, Ibanes S, Malenka RC, Kremer EJ, Luo L (2015) Viral-genetic tracing of the input-output organization of a central noradrenaline circuit. Nature 524(7563):88–92. doi:10.1038/nature14600
Shepherd GM (2004) The synaptic organization of the brain, 5th edition, 5th edn. Oxford University Press, Oxford
Sosulski DL, Bloom ML, Cutforth T, Axel R, Datta SR (2011) Distinct representations of olfactory information in different cortical centres. Nature 472(7342):213–216. doi:10.1038/nature09868
Soudais C, Laplace-Builhe C, Kissa K, Kremer EJ (2001) Preferential transduction of neurons by canine adenovirus vectors and their efficient retrograde transport in vivo. FASEB J Off Publ Federation Am Soc Exp Biol 15(12):2283–2285. doi:10.1096/fj.01-0321fje
Spaete RR, Frenkel N (1982) The herpes simplex virus amplicon: a new eucaryotic defective-virus cloning-amplifying vector. Cell 30(1):295–304
Steinbusch HW, van der Kooy D, Verhofstad AA, Pellegrino A (1980) Serotonergic and non-serotonergic projections from the nucleus raphe dorsalis to the caudate-putamen complex in the rat, studied by a combined immunofluorescence and fluorescent retrograde axonal labeling technique. Neurosci Lett 19(2):137–142
Stepien AE, Tripodi M, Arber S (2010) Monosynaptic rabies virus reveals premotor network organization and synaptic specificity of cholinergic partition cells. Neuron 68(3):456–472. doi:10.1016/j.neuron.2010.10.019
Stettler DD, Axel R (2009) Representations of odor in the piriform cortex. Neuron 63(6):854–864. doi:10.1016/j.neuron.2009.09.005
Stokes CC, Isaacson JS (2010) From dendrite to soma: dynamic routing of inhibition by complementary interneuron microcircuits in olfactory cortex. Neuron 67(3):452–465. doi:10.1016/j.neuron.2010.06.029
Stuber GD, Hnasko TS, Britt JP, Edwards RH, Bonci A (2010) Dopaminergic terminals in the nucleus accumbens but not the dorsal striatum corelease glutamate. J Neurosci Off J Soc Neurosci 30(24):8229–8233. doi:10.1523/JNEUROSCI.1754-10.2010
Sun N, Cassell MD, Perlman S (1996) Anterograde, transneuronal transport of herpes simplex virus type 1 strain H129 in the murine visual system. J Virol 70(8):5405–5413
Sun Y, Nguyen AQ, Nguyen JP, Le L, Saur D, Choi J, Callaway EM, Xu X (2014) Cell-type-specific circuit connectivity of hippocampal CA1 revealed through Cre-dependent rabies tracing. Cell reports 7(1):269–280. doi:10.1016/j.celrep.2014.02.030
Swanson LW, Hartman BK (1975) The central adrenergic system. An immunofluorescence study of the location of cell bodies and their efferent connections in the rat utilizing dopamine-beta-hydroxylase as a marker. J Comp Neurol 163(4):467–505. doi:10.1002/cne.901630406
Takatoh J, Nelson A, Zhou X, Bolton MM, Ehlers MD, Arenkiel BR, Mooney R, Wang F (2013) New modules are added to vibrissal premotor circuitry with the emergence of exploratory whisking. Neuron 77(2):346–360. doi:10.1016/j.neuron.2012.11.010
Talpalar AE, Bouvier J, Borgius L, Fortin G, Pierani A, Kiehn O (2013) Dual-mode operation of neuronal networks involved in left-right alternation. Nature 500(7460):85–88. doi:10.1038/nature12286
Tamamaki N, Yanagawa Y, Tomioka R, Miyazaki J, Obata K, Kaneko T (2003) Green fluorescent protein expression and colocalization with calretinin, parvalbumin, and somatostatin in the GAD67-GFP knock-in mouse. J Comp Neurol 467(1):60–79. doi:10.1002/cne.10905
Tripodi M, Stepien AE, Arber S (2011) Motor antagonism exposed by spatial segregation and timing of neurogenesis. Nature 479(7371):61–66. doi:10.1038/nature10538
Tsiang H, Lycke E, Ceccaldi PE, Ermine A, Hirardot X (1989) The anterograde transport of rabies virus in rat sensory dorsal root ganglia neurons. J Gen Virol 70(Pt 8):2075–2085. doi:10.1099/0022-1317-70-8-2075
Ugolini G (1995) Specificity of rabies virus as a transneuronal tracer of motor networks: transfer from hypoglossal motoneurons to connected second-order and higher order central nervous system cell groups. J Comp Neurol 356(3):457–480. doi:10.1002/cne.903560312
Ugolini G (2011) Rabies virus as a transneuronal tracer of neuronal connections. Adv Virus Res 79:165–202. doi:10.1016/B978-0-12-387040-7.00010-X
Ugolini G, Kuypers HG, Simmons A (1987) Retrograde transneuronal transfer of herpes simplex virus type 1 (HSV 1) from motoneurones. Brain Res 422(2):242–256
Veenman CL, Reiner A, Honig MG (1992) Biotinylated dextran amine as an anterograde tracer for single- and double-labeling studies. J Neurosci Methods 41(3):239–254
Velez-Fort M, Rousseau CV, Niedworok CJ, Wickersham IR, Rancz EA, Brown AP, Strom M, Margrie TW (2014) The stimulus selectivity and connectivity of layer six principal cells reveals cortical microcircuits underlying visual processing. Neuron 83(6):1431–1443. doi:10.1016/j.neuron.2014.08.001
Vercelli A, Repici M, Garbossa D, Grimaldi A (2000) Recent techniques for tracing pathways in the central nervous system of developing and adult mammals. Brain Res Bull 51(1):11–28
Vivar C, Potter MC, Choi J, Lee JY, Stringer TP, Callaway EM, Gage FH, Suh H, van Praag H (2012) Monosynaptic inputs to new neurons in the dentate gyrus. Nat Commun 3:1107. doi:10.1038/ncomms2101
Wall NR, Wickersham IR, Cetin A, De La Parra M, Callaway EM (2010) Monosynaptic circuit tracing in vivo through Cre-dependent targeting and complementation of modified rabies virus. Proc Natl Acad Sci USA 107(50):21848–21853. doi:10.1073/pnas.1011756107
Wall NR, De La Parra M, Callaway EM, Kreitzer AC (2013) Differential innervation of direct- and indirect-pathway striatal projection neurons. Neuron 79(2):347–360. doi:10.1016/j.neuron.2013.05.014
Waller A (1850) Experiments on the section of the glossopharyngeal and hypoglossal nerves of the frog, and observations of the alterations produced thereby in the structure of their primitive fibres. Phil Trans R Soc Lond 140:423–429
Wang D, He X, Zhao Z, Feng Q, Lin R, Sun Y, Ding T, Xu F, Luo M, Zhan C (2015) Whole-brain mapping of the direct inputs and axonal projections of POMC and AgRP neurons. Front Neuroanat 9:40. doi:10.3389/fnana.2015.00040
Waselus M, Galvez JP, Valentino RJ, Van Bockstaele EJ (2006) Differential projections of dorsal raphe nucleus neurons to the lateral septum and striatum. J Chem Neuroanat 31(4):233–242. doi:10.1016/j.jchemneu.2006.01.007
Watabe-Uchida M, Zhu L, Ogawa SK, Vamanrao A, Uchida N (2012) Whole-brain mapping of direct inputs to midbrain dopamine neurons. Neuron 74(5):858–873. doi:10.1016/j.neuron.2012.03.017
Weissbourd B, Ren J, DeLoach KE, Guenthner CJ, Miyamichi K, Luo L (2014) Presynaptic partners of dorsal raphe serotonergic and GABAergic neurons. Neuron 83(3):645–662. doi:10.1016/j.neuron.2014.06.024
Wertz A, Trenholm S, Yonehara K, Hillier D, Raics Z, Leinweber M, Szalay G, Ghanem A, Keller G, Rozsa B, Conzelmann KK, Roska B (2015) PRESYNAPTIC NETWORKS. Single-cell-initiated monosynaptic tracing reveals layer-specific cortical network modules. Science 349(6243):70–74. doi:10.1126/science.aab1687
White JG, Southgate E, Thomson JN, Brenner S (1986) The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci 314(1165):1–340
Wickersham IR, Lyon DC, Barnard RJ, Mori T, Finke S, Conzelmann KK, Young JA, Callaway EM (2007a) Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron 53(5):639–647. doi:10.1016/j.neuron.2007.01.033
Wickersham IR, Finke S, Conzelmann KK, Callaway EM (2007b) Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat Methods 4(1):47–49. doi:10.1038/nmeth999
Xia Y, Driscoll JR, Wilbrecht L, Margolis EB, Fields HL, Hjelmstad GO (2011) Nucleus accumbens medium spiny neurons target non-dopaminergic neurons in the ventral tegmental area. J Neurosci Off J Soc Neurosci 31(21):7811–7816. doi:10.1523/JNEUROSCI.1504-11.2011
Yamawaki N, Shepherd GM (2015) Synaptic circuit organization of motor corticothalamic neurons. J Neurosci Off J Soc Neurosci 35(5):2293–2307. doi:10.1523/JNEUROSCI.4023-14.2015
Yonehara K, Balint K, Noda M, Nagel G, Bamberg E, Roska B (2011) Spatially asymmetric reorganization of inhibition establishes a motion-sensitive circuit. Nature 469(7330):407–410. doi:10.1038/nature09711
Yonehara K, Farrow K, Ghanem A, Hillier D, Balint K, Teixeira M, Juttner J, Noda M, Neve RL, Conzelmann KK, Roska B (2013) The first stage of cardinal direction selectivity is localized to the dendrites of retinal ganglion cells. Neuron 79(6):1078–1085. doi:10.1016/j.neuron.2013.08.005
Yoshihara Y, Mizuno T, Nakahira M, Kawasaki M, Watanabe Y, Kagamiyama H, Jishage K, Ueda O, Suzuki H, Tabuchi K, Sawamoto K, Okano H, Noda T, Mori K (1999) A genetic approach to visualization of multisynaptic neural pathways using plant lectin transgene. Neuron 22(1):33–41
Young JA, Bates P, Varmus HE (1993) Isolation of a chicken gene that confers susceptibility to infection by subgroup A avian leukosis and sarcoma viruses. J Virol 67(4):1811–1816
Zampieri N, Jessell TM, Murray AJ (2014) Mapping sensory circuits by anterograde transsynaptic transfer of recombinant rabies virus. Neuron 81(4):766–778. doi:10.1016/j.neuron.2013.12.033
Zemanick MC, Strick PL, Dix RD (1991) Direction of transneuronal transport of herpes simplex virus 1 in the primate motor system is strain-dependent. Proc Natl Acad Sci USA 88(18):8048–8051
Zhan C, Zhou J, Feng Q, Zhang JE, Lin S, Bao J, Wu P, Luo M (2013) Acute and long-term suppression of feeding behavior by POMC neurons in the brainstem and hypothalamus, respectively. J Neurosci Off J Soc Neurosci 33(8):3624–3632. doi:10.1523/JNEUROSCI.2742-12.2013
Zhang S, Xu M, Kamigaki T, Hoang Do JP, Chang WC, Jenvay S, Miyamichi K, Luo L, Dan Y (2014) Selective attention. Long-range and local circuits for top-down modulation of visual cortex processing. Science 345(6197):660–665. doi:10.1126/science.1254126
Zhao C, Deng W, Gage FH (2008) Mechanisms and functional implications of adult neurogenesis. Cell 132(4):645–660. doi:10.1016/j.cell.2008.01.033
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this chapter
Cite this chapter
Miyamichi, K., Schwarz, L.A. (2017). Connectivity and Circuit Architecture Using Transsynaptic Tracing in Vertebrates. In: Çelik, A., Wernet, M. (eds) Decoding Neural Circuit Structure and Function. Springer, Cham. https://doi.org/10.1007/978-3-319-57363-2_4
Download citation
DOI: https://doi.org/10.1007/978-3-319-57363-2_4
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-57362-5
Online ISBN: 978-3-319-57363-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)