Abstract
Augmented Reality (AR) guidance systems are currently being developed to help laparoscopic surgeons locate hidden structures such as tumours and major vessels. This can be achieved by registering pre-operative 3D data such as CT or MRI with the laparoscope’s live video. For soft organs this is very challenging, and quantitative evaluation is both difficult and limited in the literature. It has been done previously by measuring registration accuracy using retrospective (non-live) data. However a performance evaluation of a real-time system in live use has not been presented. The clinical benefit has therefore not been measured. We describe an AR guidance system based on an existing one with several important improvements, that has been evaluated in an ex-vivo pre-clinical study for guiding tumour resections with porcine kidneys. The main improvement is a considerably better way to visually guide the surgeon, by showing them how to access the tumour with an incision tool. We call this Tool Access Visualisation. Performance was measured with the negative margin rate across 59 resected pseudo-tumours. This was 85.2% with AR guidance and 41.9% without, showing a very significant improvement (\(p=0.0010\), two-tailed Fisher’s exact test).
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction and Background
There is much ongoing research to develop AR guidance systems to improve laparosurgery. One important goal is to visualise hidden internal structures such as tumours and major vessels by augmenting optical images from a laparoscope with pre-operative 3D radiological data from MRI or CT. Despite considerable research, robust systems capable of handling soft tissue deformation do not yet exist. To achieve this three main challenges must be overcome. The first is to build a segmented deformable model of the organ and its internal structures from the radiological data. This process is the least time-critical because it can be done before intervention. The second challenge is real-time registration, where the goal is to transform the model to the laparoscope’s coordinate frame using live visual information present in the laparoscope’s image. The third challenge is visualisation, where the goal is to augment the laparoscope’s image with data from the organ model in order to guide the surgeon. This paper focuses on the registration and visualisation problems, for which a number of approaches have been proposed. Registration algorithms have been described and tested with various organs including the liver [6, 7, 11], uterus [3, 4] and kidney [9, 12, 13]. To date most previous approaches have been developed for robotic surgery with stereo laparoscopes. However there are a few that work with monocular laparoscopes [3, 4, 6, 9, 12]. The challenges with monocular laparoscopes are far greater due to the lack of depth information. However they are used in the majority of laparosurgery due to several factors including cost, image resolution and smaller port size. Of these, the systems which are capable of robust registration over long durations are [4, 12]. These work by first performing an initial registration, to align the model to one [12] or several [4] reference laparoscopic images. Then 2D texture features are detected in the reference images with e.g. SURF [2] and mapped onto the model’s surface. Once done the model is automatically registered to new laparoscopic images using feature-based tracking. An important difference between [4, 12] is that in [12] the initial registration was done manually with a rigid model, whereas in [4] it was done semi-automatically with a deformable model. Therefore [4] could handle deformation due to insufflation and other factors, which is required for accurate registration of soft organs.
A main shortcoming of the above approaches is the lack of thorough quantitative evaluation, and the lack of live usage tests. In the above papers all quantitative results were presented using retrospective videos from pre-recorded surgeries. Therefore evaluating the practical benefit of the system for surgical guidance was not done. From a technical standpoint, moving from pre-recorded videos to live surgery is far from trivial. The main issues are time constraints: any time for manual stages becomes significant and parameter tuning is not really possible. Furthermore, when processing live videos the effective frame-rate is limited by the AR system’s speed, which is usually well below the frame-rate of a pre-recorded video (typically 30–50 fps). Therefore inter-frame motion is more significant, and depending on the algorithm can severely affect performance.
We present an AR guidance system with monocular laparoscopes, based on [4], which runs in real-time and which has been tested live with a systematic pre-clinical user study. This evaluation measured the benefit of AR guided tumour resection using ex-vivo porcine kidneys. To achieve this a number of improvements were made to [4]. These were for generalising the approach to general biomechanical organ models, reducing manual processing time and a much better approach to AR visualisation, which we call Tool Access Visualisation.
2 AR Guidance System
We first describe the system’s inputs (Sect. 2.1) and give a global overview of the registration algorithm (Sect. 2.2). We then describe the two main components of the registration algorithm, which are the initial registration and tracking stages (Sects. 2.3 and 2.4 respectively). Lastly we present Tool Access Visualisation (Sect. 2.5).
2.1 System Inputs
The system requires a segmented pre-operative biomechanical 3D model, which has surface meshes for the organ and internal structures that are to be visualised. Here the internal structures are tumours and their safe tissue margin. A safe tissue margin is a border around the tumour of healthy tissue which should also be removed, whose thickness w depends on the risk factor of the particular tumour. We require two functions from the biomechanical model. The first is the transform function \(f(\mathbf {p};\mathbf {x}_t): \varOmega \rightarrow \mathbb {R}^3\), which transforms a 3D point \(\mathbf {p}\) in the model’s 3D domain \(\varOmega \subset \mathbb {R}^3\) to the laparoscope’s coordinates frame, where the vector \(\mathbf {x}_t\) denotes the model’s parameters at time t. The second is an internal energy function \(E_{internal}(\mathbf {x}_{t}):\mathbb {R}^{d}\rightarrow \mathbb {R}^{+}\) which gives the internal energy for transforming the organ with \(\mathbf {x}_t\), where d is the dimensionality of \(\mathbf {x}_{t}\). Both f and \(E_{internal}\) must be continuous and at least first-order differentiable. We also require the laparoscope to be intrinsically calibrated.
We describe here the input models used in the presented experiments to make things concrete for the reader. These came from T2 weighted MRI with segmentation done semi-automatically using MITK [14]. The deformation models were tetrahedral Finite Element Models (FEMs) built with a 3D vertex grid (6 mm spacing) cropped to the organ. Therefore \(\mathbf {x}_t\) held the unknown 3D positions of the FEM’s vertices in laparoscope coordinates. Trilinear interpolation was used to compute \(f(\mathbf {p};\mathbf {x}_t)\). For \(E_{internal}\) the Saint Venant-Kirchoff strain energy was used, with rough generic values for the Young’s modulus E and Poisson’s ratio \(\nu \). These were for healthy kidney tissue \(E=7\,\)kPa, \(\nu =0.43\) [5], healthy uterus tissue \(E=96\,\)kPa, \(\nu =0.45\) [10] and myomas \(E=532\,\)kPa, \(\nu =0.48\) [10]. Note that in the registration problem there is always a balancing weight between the internal energy and energy coming from image cues (which have no real physical meaning). Therefore only the relative values of E are important to us (with respect to the balancing weight), rather than their absolute values.
2.2 Overview of the Registration Algorithm
The registration problem is to determine \(\mathbf {x}_t\) for a given live laparoscopic image streamed at time t. We break this down into two stages. The first is a non-live stage which is called the initial registration stage. The second is a live-stage which is called the tracking stage. The purpose of the initial registration stage is two-fold. Firstly, it is to determine the change of shape of the organ between its pre-operative state and an intra-operative reference state. Secondly, it is used to associate texture with the organ’s surface, which is required in the tracking stage. To achieve high robustness we make a simplifying assumption in the tracking stage, which is that the organ does not deform significantly during this stage. Therefore the tracking stage can be modelled with rigid update transforms, which can be estimated far more quickly and robustly than deformable transforms. In practice this assumption is reasonable by asking the surgeon to not physically deform the organ during the tracking stage (i.e. the period that they wish to use live AR guidance). Formally, the two stages break down \(f(\mathbf {p};\mathbf {x}_t)\) as \(f(\mathbf {p};\mathbf {x}_{t})=M(f(\mathbf {p};\mathbf {x});\mathbf {R}_{t},\mathbf {t}_{t})\). Here \(\mathbf {x}\) denotes the organ’s interventional reference state. The function \(M(\cdot ;\mathbf {R}_{t},\mathbf {t}_{t}):\mathbb {R}^3\rightarrow \mathbb {R}^3\) denotes a rigid update transform at time t, parameterised by a rotation \(\mathbf {R}_{t}\in \mathcal {SO}_{3}\) and translation \(\mathbf {t}_{t}\in \mathbb {R}^3\). Thus the initial registration stage is for estimating \(\mathbf {x}\) and the tracking stage is for estimating \((\mathbf {R}_{t},\mathbf {t}_{t})\). To have live AR, only the tracking stage needs to be real-time. The time to solve the initial registration stage is a delay period before AR can run. With our current implementation this takes approximately three minutes with non-optimised C++/Matlab code on a standard workstation PC (approximately two minutes for manual pre-processing and one minute for optimisation). The tracking stage is an implementation of [3] in optimised C++/CUDA and runs at approximately 16 fps.
2.3 The Initial Registration Stage
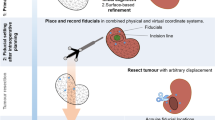
Overview. We illustrate the problem setup in Fig. 1 using a human uterus with a deep myoma. This stage is more challenging to solve than the tracking stage because (i) we have no texture information associated with the organ’s surface (because it comes from a radiological image segmentation), and (ii) it is a deformable registration. We tackle it similarly to [4] using a 3D point cloud reconstruction of the interventional scene by running rigid Structure-from-Motion (SfM), for which mature methods exist. To do this we require some sample images of the organ, known as keyframes, observing it from different viewpoints and distances. In our experiments typically no more than 10 are required, which are gathered as the surgeon performs an initial exploration of the organ (referred to as the exploratory phase). Note that a SfM reconstruction is up to an unknown global scale factor s. We resolve this jointly with registration.
We formulate the problem as a non-linear energy-minimisation problem, with energies coming from prior and data terms. The prior term encodes the model’s internal energy, which is used to regularise the problem. The data terms are illustrate in Fig. 1. The first involves the point cloud reconstruction, and encourages the organ’s surface mesh to fit to the point cloud. Importantly, this is a robust data term, which accounts for the fact that the reconstruction may contain outliers or some residual background and/or occluding structures. The second data term complements the first and uses contour cues. Specifically, it is a silhouette contour term which makes the organ model’s silhouette align to its associated contours in the keyframe images. This is complementary to the point cloud data term for two reasons. Firstly, we never usually reconstruct points reliably at these regions due to occlusions. Secondly, it anchors the organ’s silhouette to the contour fragments, which is a strong constraint. We implement this data term with some manual assistance, similarly to [4]. Specifically, a human operator marks in a keyframe where they can confidently see the organ’s silhouette contours (Fig. 1, first row). We call these contour fragments. The operator does not need to mark contour fragments in all keyframes (it can be done with only one). If more keyframes are marked then we have more constraints, but it takes more time. We use a default of four keyframes. They also do not need to be contiguous. Optionally, an anatomical landmark data term can also be included [4], which is used to align distinct landmarks that can be found on the organ’s surface in the pre-operative and laparoscopic images. In [4] for the uterus, these were the Fallopian tube/uterus junctions, which were manually located. Landmarks help guide registration if the initialisation is poor or there is very strong deformation. In the presented experiments we do not use them in the optimisation process, so are omitted from the energy function.
The main improvements over [4] are summarised as follows:
-
In [4] the deformable model used was a 3D affine model. This was shown to be sufficient for simple deformations of the human uterus but is not sufficient in general. We extend the approach to work with general biomechanical models. This requires changing the core energy function to include the model’s internal energy (without this the problem is highly under-constrained), and to correct the interventional reconstruction’s scale factor s. Note that in [4], s could be absorbed into the affine model’s coefficients. This is generally not possible with biomechanical models because a change of scale affects the internal energy.
-
In [4] the organ’s surface was assumed to have disc topology. We generalise this to arbitrary fixed topologies.
-
We have sped up the process of marking contour fragments considerably with a touch-screen interface, where the operator marks them with rough finger strokes. These strokes then guide an automatic refinement method based on intelligent scissors [8]. This reduces manual effort, typically taking less than 10 s for a single keyframe.
Interventional 3D reconstruction. During the exploratory phase the laparoscopic video is saved to disk, then N keyframes \(\{K_1,\dots K_N\}\) are extracted. We index these with \(i\in [1,N]\). This is done by uniformly sampling the video into N intervals with a default \(N=12\). For each interval i we take the keyframe \(K_i\) to be the one with the lowest optical motion, using the Sum-of-Absolute Difference (SAD) as the metric computed between consecutive frame pairs. This is done to improve the quality of the reconstruction because SfM work best with sharper images. We then run a state-of-the-art dense SfM algorithm (currently Photoscan [1]) to compute a dense point cloud reconstruction \(\mathcal {Q}\overset{\mathrm {def}}{=}\{\mathbf {q}_{1},\dots \mathbf {q}_{M}\}\), \(\mathbf {q}_{j}\in \mathbb {R}^{3}\), and the keyframe camera pose matrices \(\mathbf {M}_i \in \mathcal {SE}_4\). These hold the laparoscope’s rotation matrix \(\mathbf {R}_{i}\in \mathcal {SO}_3\) and translation vector \(\mathbf {t}_{i}\in \mathbb {R}^3\) relative to the point cloud. Recall that \(\mathcal {Q}\) and \(\mathbf {t}_i\) are defined up to the unknown scale factor \(s\in \mathbb {R}^+\). We chose Photoscan because it has been shown to work well on laparoscopic data [3] and can produce far denser reconstructions than purely feature-based methods. There may exist some keyframes whose pose is not computable, due to e.g. insufficient visual overlap. We currently deal with this by simply removing the keyframe. The point cloud \(\mathcal {Q}\) may contain background and/or foreground structures that partially occlude the organ. We currently deal with this by a human operator cropping them using a fast lasso-based user interface [1]. To reduce time we do not require the cropping to be perfect. We allow some non-organ points to remain in \(\mathcal {Q}\) and we deal with them by making the associated data term robust (see below). In some instances SfM may fail, which typically occurs when the keyframe overlap is insufficient. This can usually be resolved by extracting more keyframes by doubling the number of intervals and re-running SfM. In rare events SfM may still fail, due to very weak texture. In these cases we find image enhancement such as Storz’s CLARA can help. Alternatively, SLAM could be tried because unlike SfM it exploits temporal continuity.
Initialisation. We initialise \(\mathbf {x}\) with a rigid transform, denoted by \(\mathbf {M}\in \mathcal {SE}_4\). In some cases \(\mathbf {M}\) can be considered known a priori, for example if the laparoscope is assumed to be in a canonical position. When this cannot be assumed, we compute it with a small amount of manual interaction as follows. A small number of point correspondences (at least four) are selected on the organ’s surface model and one of the keyframe images. Without loss of generality let this be the first keyframe \(K_1\). We then compute a rigid transform \(\mathbf {M}_a\) from model coordinates to laparoscope coordinates, by fitting the correspondences using OpenCV’s PnP method. The point correspondences are computed using an interactive user interface, where the model can be freely rotated to present it from a similar viewpoint as the keyframe’s viewpoint. This significantly eases the operator’s task. We then initialise s as follows. First we transform the model by \(\mathbf {M}_a\) and render it using OpenGL with the same intrinsic parameters as the laparoscope. This generates a depth map d(x, y), and we compute s by comparing depths in d to the depths of \(\mathcal {Q}\). Specifically, let \(\tilde{d}_j\) be the depth of \(\mathbf {q}_j\) to the laparoscope in keyframe 1, and \((x_j,y_j)\) be its 2D position in the keyframe’s image. We can then estimate s by \(s\approx d(x_j,y_j)/\tilde{d}_j\). Note that only points that project within the render’s silhouette can be used. To compute s robustly, we compute it as the median value from all such points. Finally, the transform \(\mathbf {M}\) is given by the composition \(\mathbf {M} = \mathbf {M}_s\,\mathbf {M}^{-1}_i\,\mathbf {M}^{-1}_s\,\mathbf {M}_a\), where \(\mathbf {M}_s\) is an isotropic scaling by s.
The energy function. To improve clarity we define all image points in normalised pixel coordinates, which is possible given the intrinsic calibration. The energy function \(E(\mathbf {x},s)\in \mathbb {R}^+\) consists of the point cloud data term \(E_{point}\), which encourages the organ’s surface to fit to \(\mathcal {Q}\), the contour data tern \(E_{contour}\), which encourages the organ’s silhouette contours to fit to the contour fragments, and the prior term, which is the model’s internal energy \(E_{internal}(\mathbf {x})\):
where \(\lambda _{contour}\) and \(\lambda _{internal}\) are scalar weights specific to the organ category. The set \(\mathcal {C}_i\) denotes all pixels on the contour fragments in keyframe i.
We construct \(E_{point}\) using an Iterative Closest Point (ICP)-based energy. This works using a set of virtual point correspondences \(\mathcal {P}=\{\mathbf {p}_1,\dots \mathbf {p}_M\}\) with \(\mathbf {p}_j\in \partial \varOmega \) denoting the unknown position of point j on the organ’s surface mesh. For a given \((\mathbf {x},s)\) we compute \(E_{point}\) as follows. First we transform the organ’s surface mesh according to \(f(\cdot ;\mathbf {x})\) and rescale the point cloud with \(\hat{\mathbf {q}}_{j}\leftarrow s\,\mathbf {q}_{j}\). We then set \(\mathbf {p}_j\) as the closest point to \(\hat{\mathbf {q}}_{j}\) on the surface’s mesh. We define \(E_{point}\) using a robust point-to-plane distance function, which is inspired by point-to-plane ICP with rigid objects. This allows the model to slide over the point cloud without resistance, and is defined as follows:
The function \(v_{j}(\mathbf {x})\in \mathbb {R}^4\) gives the organ surface’s tangent plane at \(f(\mathbf {p}_{j})\). The function \(d_{plane}(\mathbf {v},\mathbf {q})\) gives the signed distance between a plane \(\mathbf {v}\) and a 3D point \(\mathbf {q}\). The function \(\rho :\mathbb {R}\rightarrow \mathbb {R}^{+}\) is an M-estimator and is crucial to achieve robust registration. Its purpose is to align the reconstructed point \(\hat{\mathbf {q}}_{j}\) with the organ’s surface, but to do so robustly to account for non-organ points being in \(\mathcal {Q}\) or poorly reconstructed points. The model should not align these points, and the M-estimator facilitates this by reducing the influence of their alignment error on E. We have tested various types and good results are obtained with pseudo-L1 \(\rho ({x})\overset{\mathrm {def}}{=}\sqrt{{x}^{2}+\epsilon }\) with \(\epsilon = 10^{-3}\) being a small constant, which is used to make \(\rho \) differentiable everywhere.
We construct \(E_{contour}\) similarly with virtual point correspondences. Specifically, for a given pair \((\mathbf {x},s)\) and a given keyframe i we construct a set of virtual correspondences \(\mathcal {R}_i=\{\mathbf {r}_1,\dots ,\mathbf {r}_{C(i)}\}\) where \(\mathbf {r}_k\in \partial \varOmega \) denotes the unknown position of the \(k^{th}\) contour fragment pixel \(\mathbf {c}_k\) on the model’s surface. The virtual correspondences are points on the organ surface mesh’s occluding contours, and are computed as follows. First we transform the organ’s surface mesh according to \(f(\cdot ;\mathbf {x})\), then transform it to laparoscope coordinates using \((\mathbf {R}_{i}, s\,\mathbf {t}_{i})\). We then render the surface mesh using OpenGL, using the same intrinsic parameters as the laparoscope. We then take the render’s silhouette and extract all the pixels on the render’s boundary, which is put into a set \(\mathcal {B}\). For each contour fragment pixel \(\mathbf {c}_k\in \mathcal {C}_j\) we compute its closest point \(\mathbf {b}_k\in \mathcal {B}\) and form a correspondence with it. We then compute the 3D position of \(\mathbf {b}_k\) in model coordinates, which is easy to do with an OpenGL shader, and assign it to \(\mathbf {r}_k\). We then evaluate \(E_{contour}\) as the alignment error from all correspondences:
where \(\pi ([x,y,z]^{\top })\overset{\mathrm {def}}{=}1/z[x,y]^{\top }\) is the perspective projection function and C is the total number of contour fragment pixels. Here the M-estimator \(\rho \) is also used to handle the fact that some of the contour fragment pixels may be erroneous, which can sometimes occur if the intelligent scissoring fails to snap correctly at low-contrast edges.
Optimisation. We optimise E by iterative non-linear optimization using a stiff-to-flexible strategy. This is used to improve convergence by starting with a stiff model, optimising E, then reducing the stiffness to account for more and more deformation. We use a default of \(l=6\) stiffness levels with \(\lambda _{internal}(l)=2\,\lambda _{internal}(l-1)\). For each level we alternate between computing the virtual correspondence sets (\(\mathcal {R}_i\) and \(\mathcal {P}\)) and optimising E, which is done with a Gauss-Newton iteration and backtracking line search. This continues until either convergence is reached or 10 iterations have passed. At the final stiffness level we optimize until convergence. Convergence however is not guaranteed because of the point-to-plane distance function in \(E_{point}\). Specifically, the energy may increase after \(\mathcal {P}\) is re-computed. We handle this by using the point-to-point distance at the final level, because this ensures E will decrease at each iteration.
2.4 The Tracking Stage
Having solved the initial registration we initiate the tracking stage, which updates the initial registration in real-time using live images streamed from the laparoscope. We solve this with an existing feature-based method [3]. This works by first extracting 2D features in the keyframe images, then matching them with RANSAC-based rigid registration to each new image. The advantages of [3] are it is robust to occlusions from e.g. surgical tools, handles partial views and viewpoint changes. Unlike SLAM-based tracking methods it does not use frame-to-frame tracking. Instead it performs tracking-by-detection. This allows it to register over long durations and can trivially recover when the organ is not visible for certain periods, such as when the surgeon removes and then reinserts the laparoscope or cleans the lens. In cases when the organ is assumed to be fixed relative to background structures, we can track using features from both the organ and background structures. This improves stability if the organ’s texture is weak, and we do this in the ex-vivo user study.
2.5 AR Guidance with Tool Access Visualisation
Having registered, the final task is AR visualisation. We briefly describe Transparent Blending (TB) visualisation, which is the previous approach used with monocular laparoscopes. It works by first rendering the tumours on the laparoscope’s image plane, then a composite image is made by blending the render with the real image to give the impression the organ is transparent. An example from [4] is shown in Fig. 2(a) where two myomas are visualised with TB. TB however has a serious limitation which has not been previously addressed, and we find it can actually mis-guide the surgeon. The problem is illustrated in Fig. 3(a) and is as follows. When a surgeon actually uses TB to resect a tumour they usually assume it indicates where they should cut to access the tumour. This however is incorrect. It just shows the position of the tumour from the viewpoint of the laparoscope. Often they assume the tumour’s centre would be reached by cutting into the organ from the rendered tumour’s centre \(\mathbf {c}\in \mathbb {R}^2\). This is not the case as shown in Fig. 3(a). In our user study we found this is a significant problem with smaller and/or deeper tumours, and can cause them to be missed.
(a) AR with Transparent Blending (TB) visualisation taken from [4]. (b) Our AR visualisation combining Transparent Blending with Tool Access Visualisation. (c) Our AR system in live operation during the ex-vivo user study.
What the surgeon actually wants is to be shown how to reach the tumour using the incision tool. Furthermore, surgeons typically want to also see the tumour’s safe tissue margin. We provide both information with what we call Tool Access Visualisation, which is shown in Fig. 2(b). Its associated geometry is shown in Fig. 3(b). Tool Access Visualisation works by showing the tumour’s safe tissue margin projected onto the organ’s surface as a ring, which we call the tumour guidance ring. The idea is that if the surgeon were to cut into the organ along the guidance ring, they would segment the tumour with a minimal margin of \(w\,\)mm. At present we do not visualise uncertainty in the margin’s location, which is important for real clinical use, and leave this to future work.
We achieve Tool Access Visualisation with two projections. The first is a perspective projection of the margin’s surface onto the organ’s surface, using a centre-of-projection located at the incision tool’s port centre \(\mathbf {p}\in \mathbb {R}^3\). The second is a perspective projection of the projected margin’s perimeter onto the laparoscope’s optical image (shown as rings in Fig. 2(b)). To achieve this we need to know \(\mathbf {p}\). Recall that the organ has been registered in laparoscope coordinates, therefore we need \(\mathbf {p}\) in laparoscope coordinates. It may be possible to estimate \(\mathbf {p}\) automatically using external and/or internal tool tracking, however this is left to future work. Here we assume \(\mathbf {p}\) is given a priori. In our user study, where the ports are located on a pelvic trainer, this is simple and can be done offline by taking physical measurements. We complete the visualisation by combining Tool Access Visualisation with TB visualisation (Fig. 2(b)) to show tumours (solid fill), organ (wireframe) and safe tissue margins (wireframe).
3 Evaluation
This section is divided into two parts. The first is the main part describing our ex-vivo user study. The second part shows our system doing live in-vivo registration of a porcine kidney during a laparoscopic training exercise. All the procedures were performed in the operating room of the International Centre of Endoscopic Surgery (CICE), France, approval number C63 18 113.
3.1 Ex-vivo User Study Evaluation
We used 29 porcine kidneys recovered from pigs operated after resident training. For each kidney pseudo-tumours were created by injecting alginate, a hardening hydrocolloid, of between 4 mm and 10 mm in diameter. In total 59 pseudo-tumours were injected at arbitrary sub-surface positions, with an average of 2.5 per kidney. We used safe tissue margins of 5 mm. Kidney models were made as described in Sect. 2.1 from 3T MRI images (0.4 mm resolution and slice thickness 1.5 mm). The interventional equipment is shown in Fig. 2(c) and consisted of a Karl Storz 10 mm laparoscope column with CLARA image enhancement, a surgical grasper, an incision tool, a laparoscopic pelvic trainer and an instrument with a surgical marker pen attached at the tip (referred to as the marker instrument). The AR software ran on a mid-range Intel i7 desktop workstation with an NVidia 980 Ti GPU, with visualisations shown on a 26 inch monitor. Laparosurgery was performed by a skilled final-year resident. The resident spent time training before evaluation to familiarise the task, the guidance software and to provide feedback to improve visualisation. In total 28 pseudo-tumours were resected during this time.
3.2 Interventional Protocol and Equipment
Laparosurgery was performed using the pelvic trainer, with the kidney inserted on a ground surface and the laparoscope and instruments inserted through three ports. The same port configuration was used in all cases. The surgeon was tasked to remove each tumour by cutting out a conic tissue section which included the tumour and its safe tissue margin. The kidneys were divided into two groups (non-randomised): the AR group and the Non-AR group, with 13 kidneys in the AR group with 29 tumours, and 19 kidneys in the Non-AR group with 33 tumours. Kidneys in the AR group were operated with the AR guidance system activated. Recall that the guidance system is not designed to handle significant deformation or topology change after the initial registration, which occurs when a tumour is resected. This was dealt with in the protocol by having the surgeon first mark dots along the tumour guidance ring using the marker instrument, guided by the AR visualisation. Once completed they used the marks to guide the resection with AR disactivated. For the Non-AR group, the surgeon first consulted the MRI using interactive slice-based visualisation [14]. The task was then performed without AR guidance using the same safe tissue margin of 5 mm.
3.3 Results
We present results with the negative margin rate. A negative margin occurs when the tumour is contained entirely within the resected tissue. A positive margin occurs when either the tumour is completely absent from the resected tissue (a complete miss), or if it is partially contained (a contact). For three tumours the protocol was not completed properly (the conic section did not cut fully through the kidney) and were excluded. There were 13 negative margins in the Non-AR group (41.9%), with 4 complete misses and 14 contacts. There were 23 negative margins in the AR group (85.2%), with 0 complete misses and 4 contacts. Statistical significance was measured with Fisher’s exact two-tailed test with \(p=0.0010\). Therefore the user study indicates a very significant benefit for using the AR guidance system.
3.4 Live In-vivo Registration of a Porcine Kidney
We finish by showing our system in live use for registering in-vivo a porcine kidney during a laparoscopic training exercise (Fig. 4). The kidney did not contain a tumour and no ground truth information was available, so the results are merely to demonstrate that the registration system works live and in-vivo. The biomechanical kidney model was build as described in Sect. 2.1. The same Storz laparoscope column was used as the user study. An exploratory video was captured lasting approximately 30 s. We show sample keyframes in Fig. 4(b–d) with contour fragments overlaid. Note that only the kidney’s upper silhouette contours are visible, with the lower ones being occluded by intestine and peritoneum, which is why there are no contour fragments on the lower half. In Fig. 4(e) we overlay the registered model’s surface onto one of the keyframes, showing the upper contours aligning well with the image. In Fig. 4(f,g) we show frames from the live tracking stage, which shows robustness to tool occlusions and mild deformations.
Live in-vivo registration of a porcine kidney. (a) the organ model’s surface mesh. (b–d) contour fragments in three keyframes. (e) the surface mesh overlaid on a keyframe after the initial registration with its silhouette contours in red. (f–g) Snapshots of the surface mesh during live tracking, with ground-truth silhouette contours shown in green. Note that the kidney’s lower portion is occluded by intestine and peritoneum. Best viewed in colour. (Color figure online)
4 Conclusion
We have presented a complete system for AR guided laparoscopic tumour resection, and a quantitative ex-vivo user study to measure its benefit in live use, which is the first of its kind. The system has been based on [4] with several major improvements. These include supporting general biomechanical models as inputs, less manual processing and a new visualisation method, called Tool Access Visualisation (TAV), which shows the surgeon how to access a tumour and its safe tissue margin with an incision tool. In future work we aim to conduct a similar user study in-vivo, and to test with stereo laparoscopic images, where the point cloud reconstruction would come from stereo triangulation. We also aim to extend Tool Access visualisation to handle non-straight incision tools, such as articulated ones used in robotic laparosurgery.
References
Agisoft photoscan. http://www.agisoft.com. Accessed 07 Feb 2016
Bay, H., Ess, A., Tuytelaars, T., Van Gool, L.: Speeded-up robust features (SURF). Comput. Vis. Image Underst. 110(3), 346–359 (2008)
Collins, T., Pizarro, D., Bartoli, A., Canis, M., Bourdel, N.: Realtime wide-baseline registration of the uterus in laparoscopic videos using multiple texture maps. In: Liao, H., Linte, C.A., Masamune, K., Peters, T.M., Zheng, G. (eds.) AE-CAI/MIAR -2013. LNCS, vol. 8090, pp. 162–171. Springer, Heidelberg (2013). doi:10.1007/978-3-642-40843-4_18
Collins, T., Pizarro, D., Bartoli, A., Canis, M., Bourdel, N.: Computer-assisted laparoscopic myomectomy by augmenting the uterus with pre-operative MRI data. In: ISMAR (2014)
Egorov, V., Tsyuryupa, S., Kanilo, S., Kogit, M., Sarvazyan, A.: Soft tissue elastometer. Med. Eng. Phys. 30, 206–212 (2008)
Haouchine, N., Dequidt, J., Berger, M.-O., Cotin, S.: Monocular 3D reconstruction and augmentation of elastic surfaces with self-occlusion handling. Trans. Vis. Comput. Graph., 14 (2015)
Haouchine, N., Dequidt, J., Peterlik, I., Kerrien, E., Berger, M.-O., Cotin, S.: Image-guided simulation of heterogeneous tissue deformation for augmented reality during hepatic surgery. In: ISMAR (2013)
Mortensen, E.N., Barrett, W.A.: Intelligent scissors for image composition. In: SIGGRAPH, pp. 191–198 (1995)
Nosrati, M.S., Peyrat, J.-M., Abinahed, J., Al-Alao, O., Al-Ansari, A., Abugharbieh, R., Hamarneh, G.: Simultaneous multi-structure segmentation and 3D non-rigid pose estimation in image guided robotic surgery. Trans. Med. Imaging 35(1), 1–12 (2016)
Pearsall, G., Roberts, V.: Passive mechanical properties of uterine muscle (myometrium) tested in vitro. J. Biomech. 4(11), 167–176 (1978)
Plantefève, R., Peterlik, I., Haouchine, N., Cotin, S.: Patient-specific biomechanical modeling for guidance during minimally-invasive hepatic surgery. Ann. Biomed. Eng. (2015)
Puerto-Souza, G., Cadeddu, J.A., Mariottini, G.: Toward long-term and accurate AR for monocular endoscopic videos. Biomed. Eng. (2014)
Su, L.-M., Vagvolgyi, B.P., Agarwal, R., Reiley, C.E., Taylor, R.H., Hager, G.D.: Augmented reality during robot-assisted laparoscopic partial nephrectomy: toward real-time 3D-CT to stereoscopic video registration. Urology 73, 896–900 (2009)
Wolf, I., Vetter, M., Wegner, I., Nolden, M., Böttger, T., Hastenteufel, M., Schöbinger, M., Kunert, T., Meinzer, H.-P.: The medical imaging interaction toolkit (MITK). http://www.mitk.org/
Acknowledgements
This research was funded by the EU FP7 ERC research grant 307483 FLEXABLE and Almerys Corporation.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Collins, T. et al. (2017). A System for Augmented Reality Guided Laparoscopic Tumour Resection with Quantitative Ex-vivo User Evaluation. In: Peters, T., et al. Computer-Assisted and Robotic Endoscopy. CARE 2016. Lecture Notes in Computer Science(), vol 10170. Springer, Cham. https://doi.org/10.1007/978-3-319-54057-3_11
Download citation
DOI: https://doi.org/10.1007/978-3-319-54057-3_11
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-54056-6
Online ISBN: 978-3-319-54057-3
eBook Packages: Computer ScienceComputer Science (R0)