Abstract
Fe is an essential micronutrient for Brucella strains. Meeting their physiologic need for this metal is especially challenging for these bacteria because they live in close association with their mammalian hosts, and Fe-sequestration is a well-documented host defense against microbial pathogens. The following chapter will describe what is presently known about Fe homeostasis in Brucella strains, and how the individual cellular components involved in this process contribute to virulence.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
2.1 Fe as a Micronutrient for Brucella Strains
Iron (Fe) is a widely used prosthetic group in proteins due to its exceptional reactivity under physiologic conditions (Crichton 2009). Its capacity to undergo redox reactions, for instance, makes it an important component of the proteins that comprise electron transport chains as well as those that participate in maintaining the redox balance of cells. The cationic nature of Fe also allows this metal to stabilize substrates and reaction intermediates in the active sites of enzymes. Ferrous (Fe2+) and ferric (Fe3+) iron are the main oxidation states found in biological systems, and the redox potential of the Fe2+/Fe3+ couple can vary greatly based on the ligands to which this cation is bound (Kosman 2013). This makes Fe extremely versatile in biochemical reactions. Fe can be found in proteins as either the free metal or as a component of heme or Fe-S centers. Because of its versatility, Fe is an essential co-factor for many cellular enzymes, including those involved in DNA synthesis, central carbon metabolism, cellular respiration and signal transduction. With the notable exception of bacteria in the genus Lactobacillus (Archibald 1983) and Borrelia (Posey and Gherardini 2000), all living organisms that have been studied require Fe as a micronutrient. For instance, early studies showed that Brucella strains require at least 0.5 μM Fe for growth and grow best when supplied with Fe concentrations >1.8 μM (Waring et al. 1953), which is typical of Fe requirements that have been reported for other bacteria (Lengeler et al. 1999).
2.2 The Mammalian Host Is an Fe-Deprived Environment
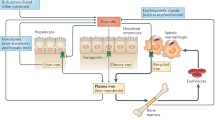
Acquiring sufficient Fe to meet their physiologic needs is a particular challenge for Brucella strains because they reside predominately in close association with their mammalian hosts (Roop et al. 2009). One of the reasons for this is that the vast majority of the Fe in mammals is incorporated into proteins as heme, Fe-S clusters or mononuclear or dinuclear Fe centers, and is thus not readily accessible (Hood and Skaar 2012). Another consideration is that the soluble and biologically active form of Fe, Fe2+, has the capacity to react with reactive oxygen species and generate toxic hydroxyl radicals (OH−), consequently the Fe homeostasis systems of mammals maintain the levels of ‘free’ Fe in their tissues at concentrations <10−23 M in the extracellular environment and within the range of 10−8–10−6 M in the intracellular environment (Hider and Kong 2013) to avoid Fe toxicity. Some of the major components of these Fe homeostasis systems include the Fe transport protein transferrin (Tf) which binds Fe3+ with high affinity in the blood and serum, the exporter ferroportin (Fp) which actively transports Fe2+ out of cells, and the Fe storage protein ferritin (Ft), which converts excess intracellular soluble and reactive Fe2+ into insoluble and inert (e.g. less toxic) Fe3+ complexes (Fig. 2.1). As will be discussed later in this chapter, Brucella strains also have the capacity to utilize heme (Paulley et al. 2007; Ojeda 2012) as an Fe source, and thus another relevant mechanism by which mammalian hosts limit the availability of Fe to these bacteria in the extracellular environment is through the activity of the heme-binding protein hemopexin and the hemoglobin-binding protein haptoglobin, which scavenge these molecules from the serum (e.g., after leakage from damaged cells) and target them for degradation in macrophages.
The innate and acquired immune responses also have significant impacts on the availability of Fe to invading microbes, and in fact is it well documented that Fe deprivation is an important component of protective immunity in mammals (Nairz et al. 2014). During the inflammatory response, for instance, neutrophils release the Fe-binding proteins lactoferrin (Vogel 2012) and calprotectin (Nakashige et al. 2015) at the site of infection. These phagocytes also release the protein lipocalin-2, also known as siderocalin, which inhibits the activity of catechol-based siderophores, small molecular weight chelators that microbes secrete into their environment to capture and transport Fe (Goetz et al. 2002; Sia et al. 2013). In addition, the liver produces the peptide hormone hepcidin (Hp), which induces the degradation of ferroportin preventing the release of Fe from the liver and spleen into circulation (Michels et al. 2015). The combined activities of lactoferrin, calprotectin and Hp thus further reduce the already low levels of free Fe available in the extracellular environment of mammals (Fig. 2.2).
The manner in which macrophages are ‘activated’ also has an impact on the availability of ‘intracellular’ Fe to microbes that reside within these phagocytes. This is an especially important consideration for Brucella strains for two reasons. First of all, the capacity of these bacteria to survive and replicate in macrophages is a critical component of their virulence (Roop et al. 2009). Secondly, it has recently been demonstrated that the activation state of host macrophages influences how well these phagocytes support the intracellular persistence of the brucellae (Xavier et al. 2013). Macrophages activated by the Th1 cytokine interferon-γ (IFN-γ), e.g. classically-activated macrophages (CAMs), for instance, have reduced levels of Tf receptors on their surface (Byrd and Horwitz 1989), which reduces their capacity to import Fe (Fig. 2.3). The divalent cation transporter Nramp1 also pumps Fe2+ out of phagosomes in CAMs (Cellier et al. 2007). In addition, although hepcidin blocks the capacity of macrophages in the spleen and liver to release Fe through inhibition of Fp activity as shown in Fig. 2.2, experimental evidence indicates that CAMs have the capacity to overproduce Fp to bypass this effect and thus utilize Fp-mediated Fe release as a further mechanism for depriving intracellular pathogens of this micronutrient (Nairz et al. 2013) (Fig. 2.3). This combination of so-called ‘Fe-withdrawal defenses’ has been shown to be a critical component of the capacity of CAMs to control the intracellular replication of several intracellular pathogens including Salmonella typhimurium (Nairz et al. 2008) and Legionella pneumophila (Byrd and Horwitz 1989). It is well known that IFN-γ plays a crucial role in protective immunity against Brucella infections (Zhan and Cheers 1993; Baldwin and Goenka 2006) and that CAMs kill intracellular brucellae much better than resting macrophages (Xavier et al. 2013). But the degree to which the Fe-withdrawal defenses contribute to the brucellacidal activity of CAMs is currently unknown. Macrophages activated by the Th2 cytokines interleukin-1 and interleukin 13 (IL-1 and IL-13) (alternatively-activated macrophages [AAMs]) are much less restrictive for the intracellular growth of Brucella strains than CAMs, but more importantly, recent studies have shown that AAMs provide a favorable host cell for maintaining chronic infection in experimentally infected mice (Xavier et al. 2013). Because of their role in scavenging damaged cells and tissue components, AAMs take up a lot of Fe-and heme-containing proteins (Cairo et al. 2011). Despite the resulting increased Fe flux through AAMs, however, less of this Fe is stored in ferritin in AAMs than in CAMs, and AAMs have a larger intracellular labile Fe pools than CAMs, although increased Fp activity in AAMs prevents intracellular Fe levels from reaching toxic levels (Fig. 2.3). Whether or not the increased labile Fe pool in AAMs contributes to these phagocytes being a preferential niche for Brucella strains during chronic infections remains to be determined.
Three other points that warrant consideration in terms of potential Fe sources that are available to Brucella strains in their mammalian hosts are-(a) the recycling of Fe from erythrocytes in mammals; (b) the enhanced uptake of heme-containing proteins by AAMs; and (c) the intracellular trafficking of Brucella strains in host cells. The vast majority of the Fe in mammals is actively recycled by the ingestion and degradation of aged erythrocytes by macrophages in the spleen and liver (Korolnek and Hamza 2014), which are preferred organs for colonization by the brucellae in non-pregnant hosts. AAMs also ingest a considerable amount of heme-containing proteins during their role in tissue repair and remodeling (Cairo et al. 2011). The heme that is released by the degradation of hemoglobin in lysosomes during both of these processes is transported into the cytoplasm and ultimately to the endoplasmic reticulum for degradation by heme oxygenase. The Fe liberated in this fashion is incorporated into the cellular labile iron pool, stored in Ft or released from these cells by Fp for transport and redistribution to other cells (Fig. 2.3). When Brucella strains are ingested by host cells, effector proteins secreted by the Type IV secretion system interact with host cell proteins, which results in the Brucella-containing vacuoles maintaining interactions with components of the host cell endoplasmic reticulum (Celli 2015) (Fig. 2.3). As was alluded to before and will be discussed in more detail later, heme can be readily used as an Fe source by Brucella strains (Paulley et al. 2007; Ojeda 2012). Thus, it is intriguing that these bacteria proactively direct their intracellular trafficking in host cells toward a compartment where there is a continuous flux of a relevant Fe source.
2.3 Fe Acquisition by Brucella
2.3.1 Siderophore Production
Siderophores are low molecular weight chelators that bacteria and other microbes secrete into their environments under Fe-deprived conditions to capture Fe3+ and import it into the microbial cell using energy-dependent transport systems (Raymond and Dertz 2004). Fe3+ is very insoluble in aqueous environments under aerobic conditions at neutral pH due its propensity to form Fe(OH)3, which has a solubility of 10−18 M. Thus, Fe3+ is only readily available under these conditions when it is bound to ligands that prevent its hydrolysis (Kosman 2013). Consequently, in order for siderophores to be effective at scavenging Fe3+, these chelators must have affinities for this cation that are higher than the ligands to which it is bound in the surrounding environment. A variety of different chemical classes of siderophores have been described in the literature based on the structure of their metal binding components. Siderophores containing catechol, hydroxamate, and/or hydroxycarboxylate Fe-binding domains are found in many bacteria, but siderophores with other Fe-binding domains have also been described (Raymond and Dertz 2004). The most highly efficient siderophores contain multiple metal binding domains linked together by amino acids, polyamines or other small molecules into flexible ‘scaffolds’ that allow them to serve as effective Fe3+ chelators (Walsh and Marshall 2004).
The first siderophore described in Brucella was the monocatechol 2,3-dihydroxybenzoic acid (2,3-DHBA) (Fig. 2.4). López-Goñi et al. (1992) employed Fe-affinity chromatography to isolate this catechol from the supernatants of Fe-deprived cultures of B. abortus 2308. They also showed that 2,3-DHBA could facilitate the import of 55Fe3+ into this bacterium by an energy-dependent process, and rescue this strain from Fe deprivation in bioassays. No other siderophores were identified in the supernatants from Fe-deprived cultures of 2308 or other B. abortus strains using this classical approach, and no known siderophores other than 2,3-DBHA or the simple monocatechols 2,3-DHBA-serine or 2,3-DHBA-glycine were found to be able to support Fe acquisition by B. abortus 2308 in in vitro assays. These experimental findings suggested that 2,3-DHBA might be the only siderophore produced and used by Brucella strains, and subsequent work by this research group provided further evidence to support this proposition (López-Goñi and Moriyón 1995).
a Genetic organization of the Brucella genes involved in brucebactin biosynthesis, b roles of the dhbC, B and A gene products in the conversion of chorismate to 2,3-dihydroxybenzoic acid (2,3-DHBA), and c proposed roles of the dhbB, dhbE, entD and vibH gene products in the biosynthesis of brucebactin from 2,3-DHBA and spermidine. Gene designations are those used in the B. abortus 2308 genome sequence in GenBank
Despite these compelling findings, and the fact that others have reported that 2,3-DHBA can serve as a siderophore in other bacteria (Hancock et al. 1977; Smith et al. 1990; Persmark et al. 1992), the ability of this simple monocatechol to serve as an ‘effective’ siderophore when produced endogenously by a bacterium has been called into question from a theoretical perspective (Chipperfield and Ratledge 2000) based on its ‘low’ Fe3+ binding capacity. The stability of Fe3+-siderophore complexes serves as a measure of a siderophore’s affinity for Fe, and is often expressed as its pFe value (Crumbliss and Harrington 2009). This value represents the negative log of the concentration of uncomplexed Fe3+ remaining after an equilibrium reaction between 10 μM siderophore and 1 μM Fe3+ at pH 7.4. The larger the pFe value, the more stable the complex formed between Fe and the siderophore under standard laboratory conditions. The reported pFe value for 2,3-DHBA is 15 (Ollinger et al. 2006), but most microbial siderophores have pFe values ranging from 18.2 to 34.3 (Crumbliss and Harrington 2009), which means they have affinities for Fe3+ that are orders of magnitude greater than that of 2,3-DHBA.
A possible answer to this question was provided by two subsequent studies aimed at identifying the genes involved in siderophore biosynthesis in B. abortus 2308. In one of these studies, a transposon mutant derived from B. abortus 2308 was identified that displayed increased sensitivity to the Fe-‘specific’ chelator diethylenediamine di(o-hydroxyphenylacetic acid) (EDDHA) compared to the parental strain, and supernatants from this mutant were found to lack a second Fe-binding catechol in addition to 2,3-DHBA that was present in 2308 (González-Carreró et al. 2003). It was also noted that the Fe acquisition defect of the B. abortus Tn mutant could be rescued by supplementation of cultures with the second Fe-finding catechol, but not by supplementation with 2,3-DHBA. The mTn5-disrupted gene in this mutant resides next to the genes required for 2,3-DHBA synthesis (Bellaire et al. 1999) (Fig. 2.4) and is predicted to encode a homolog of VibH, the amidase that links 2,3-DHBA to the polyamine norspermidine during vibriobactin synthesis in V. cholerae (Keating et al. 2002). These findings, coupled with the observation that the B. abortus Tn mutant produced more 2,3-DHBA than the parental strain when faced with Fe deprivation, led the authors to propose that the Brucella VibH homolog is involved in the conversion of 2,3-DHBA into a more complex catechol-based siderophore that has a higher affinity for Fe3+ than 2,3-DHBA. They proposed the name ‘brucebactin’ for this siderophore, but were unable to evaluate its chemical or structural features due to its reported instability. When the genes responsible for 2,3-DHBA biosynthesis in B. abortus 2308 were cloned and characterized by another group, it was likewise noted that these genes reside in a locus with genes homologous to those involved in the condensation of 2,3-DHBA with norspermidine during vibriobactin production (Fig. 2.4), but that the Brucella 2,3-DHBA/brucebactin locus notably lacks a critical gene required for the assembly of 2,3-DHBA-norspermidine complexes into vibriobactin (Bellaire et al. 2003a). This led the latter group to postulate that brucebactin is likely to be a monocatechol siderophore consisting of 2,3-DHBA linked to a small molecule such as a polyamine or an amino acid.
Based on the experimental evidence provided by the studies described in the previous paragraph, and what is known about the biosynthesis pathways typically used to assemble catechol-based siderophores in other bacteria (Walsh and Marshall 2004), it is possible to propose a biosynthetic pathway for the production of brucebactin (Fig. 2.4). The products of the dhbC, B and A genes convert chorismate into 2,3-DHBA in a pathway used by many bacteria to produce 2,3-DHBA as a precursor for the assembly of more complex catechol based siderophores such as the tricatechols enterobactin (Young et al. 1971; Luke and Gibson 1971) and bacillibactin (May et al. 2001). The products of the dhbB, dhbE, entD and vibH genes are then predicted to link one or two 2,3-DHBA molecules to the polyamine spermidine to produce brucebactin.
Two important theoretical considerations were taken into account in formulating the brucebactin biosynthetic pathway. The first is that Brucella strains do not have a gene encoding an ‘EntF-type’ modular non-ribosomal peptide synthetase (NRPS). This large enzyme contains adenylation, peptidyl carrier, condensation and thioesterase domains, and is required for both the activation of an amino acid and its condensation with 2,3-DHBA, and the assembly of the multiple 2,3-DHBA-small molecule complexes into complex tricatechol siderophores such as enterobactin (Walsh and Marshall 2004). Although a Brucella ‘entF’ gene has been described previously in the literature (González-Carreró et al. 2002; Jain et al. 2011), the gene being referred to in those citations is in fact the Brucella vibH homolog. The second consideration has to do with the specificity of VibH itself. In Vibrio, this enzyme links 2,3-DHBA to norspermidine (Keating et al. 2002), but this particular polyamine is only found in a few bacteria outside of this genus. Thus, it is likely that the Brucella VibH homolog links 2,3-DHBA to another structurally similar polyamine such as spermidine (L. Quadri, personal communication).
A recent chemical and structural characterization of brucebactin performed in the laboratory of one of the authors of this chapter (E.S. Anderson) has provided evidence that this siderophore is indeed a dicatechol comprised of two 2,3-DHBA molecules linked by a spermidine as shown in Fig. 2.4 (Atkinson 2015). Further characterization of brucebactin will be required to determine its Fe3+-binding properties, but one would predict that they are comparable to the value of 23.3 reported for the dicatechol azotochelin, which is comprised of two 2,3-DHBA molecules linked by a lysine (Cornish and Page 2000). If this is correct, it would give brucebactin an affinity for Fe3+ that is 8 logs higher than that of 2,3-DHBA. Perhaps more importantly, it would give the Brucella siderophore a pFe value in the range (e.g., >20) that has been proposed to be required for bacterial siderophores to be able to effectively compete with transferrin for Fe3+ in mammalian tissues (Evans et al. 2012).
2.3.2 Fe3+-Siderophore Transport Systems in Brucella
Due to their size, Fe3+-siderophore complexes require energy for their transport into the cytoplasm of bacteria. In Gram-negative bacteria, transport across the outer membrane is typically facilitated by ‘gated’ porins which obtain the energy they need to drive this transport from the ExbBD-TonB system (Noinaj et al. 2010). These Fe3+-siderophore complexes are then bound by specific periplasmic binding proteins which direct them to cytoplasmic ABC-type permeases that mediate their passage across the cytoplasmic membrane. Once in the cytoplasm, Fe3+ is released from the siderophore by its reduction to Fe2+and/or degradation of the siderophore (Cooper et al. 1978; Brickman and McIntosh 1992; Harrington and Crumbliss 2009).
Two genetic loci involved in Fe3+-siderophore transport have been identified in Brucella—fatBDCE and exbBD-tonB (Fig. 2.5). The former is predicted to encode a transporter homologous to the periplasmic-binding-protein dependent ABC transporter that imports Fe3+—anguibactin complexes across the cytoplasmic membrane in Vibrio anguillarum (Köster et al. 1991). As described in the previous paragraph, the latter encodes the energy transduction system required for Fe3+-siderophore transport across the outer membrane (Postle and Kadner 2003). Published studies have shown that B. abortus fatB and B. melitensis fatC and exbB mutants cannot use brucebactin and 2,3-DHBA, respectively, as Fe sources in vitro (González-Carreró et al. 2002; Danese et al. 2004). In contrast, the identity of the genes that encode the TonB-dependent OM protein that transports Fe3+-brucebactin across the outer membrane is presently unclear. The two best candidates for performing this function are the Brucella fiu (Fig. 2.5) and cir (Fig. 2.6) homologs (Roop et al. 2012). Fiu and Cir are TonB-dependent outer membrane proteins that transport Fe3+-2,3-DHBA and Fe3+-2,3-DHBA-serine complexes into E. coli (Hantke 1990; Nikaido and Rosenberg 1990). However, the Brucella cir homolog is annotated in most in most Brucella genome sequences as being part of an operon encoding a BtuBCDE-type TonB-dependent cobalamin (vitamin B12) transporter (Kadner 1990), and bioinformatics studies suggest that these genes are regulated by a cobalamin-dependent riboswitch (Rodionov et al. 2003). A more detailed experimental examination will be required to determine the contributions that Fiu and Cir make to Fe3+-brucebactin transport in Brucella.
2.3.3 Siderophore Production and Virulence of Brucella abortus in Pregnant Ruminants
A B. abortus dhbC mutant, which cannot produce 2,3-DHBA or brucebactin, is extremely attenuated in pregnant goats (Bellaire et al. 2000) and cattle (Bellaire et al. 2003a). This mutant does not, however, display significant attenuation in BALB/c (Bellaire et al. 1999) or C57BL/6 mice (Parent et al. 2002). One explanation for these findings is the different experimental readouts used to measure virulence in these animal models. Abortion and fetal pathology, for instance, are the parameters used to measure the virulence of Brucella strains during experimental infections in pregnant ruminants (Bellaire et al. 2003a). Rapid and extensive proliferation of the brucellae in the placental trophoblasts surrounding the developing fetus leads to these clinical manifestations (Enright 1990), and this represents the so-called ‘acute’ stage of brucellosis in natural ruminant hosts (Roop et al. 2004). In contrast, the mouse model assesses the capacity of Brucella strains to establish and maintain ‘chronic’ spleen infection for weeks or months after experimental challenge (Baldwin and Winter 1994; Grilló et al. 2012), and the key host cell in these infections is the macrophage. Thus, it is tempting to speculate that siderophore production plays a more important role in Fe acquisition when Brucella strains are replicating in placental trophoblasts than it does when these bacteria are replicating in macrophages. The fact that B. abortus dhbC and vibH mutants exhibit little or no attenuation in cultured murine macrophages (Bellaire et al. 1999; Parent et al. 2002; González-Carreró et al. 2002; Jain et al. 2011), and a dhbC mutant is not attenuated in the human monocytic cell line THP-1 (B. Bellaire, personal communication), would also seem to support this proposition. But a preliminary study has also shown that a B. abortus dhbC mutant is more susceptible to killing by cultured bovine macrophages than the parental 2308 strain (Bellaire 2001), suggesting that ruminant macrophages may represent a more Fe-restrictive host cell than their murine or human counterparts. Still another possibility is that siderophore production is important for Fe acquisition by the brucellae in the extracellular, rather than the intracellular, environment of the ruminant reproductive tract during pregnancy.
The documented link between siderophore production and erythritol catabolism (Bellaire et al. 2003b; Jain et al. 2011) may also explain the differential virulence properties of the B. abortus dhbC mutant in mice and ruminants. Ruminant placental trophoblasts produce large amounts of this 4 carbon sugar alcohol (Smith et al. 1962), while mice do not have substantial levels of this compound in their tissues. Erythritol is the preferred carbon source for B. abortus strains in vitro (Anderson and Smith 1965), and it has been proposed that the capacity of these bacteria to catabolize erythritol is an important virulence determinant in pregnant ruminants (Smith et al. 1962). Studies employing both B. abortus dhbC and vibH mutants have shown that B. abortus 2308 has a greater physiologic need for Fe when growing in the presence of erythritol than it does when growing in the presence of other carbon and energy sources, and that siderophore biosynthesis is required to meet this increased need for Fe (Bellaire et al. 2003b; Jain et al. 2011). Thus, it is conceivable that brucebactin serves as a virulence determinant for B. abortus strains in pregnant ruminants because its production allows the bacteria to acquire the levels of Fe they need to efficiently catabolize erythritol and support their extensive replication in the gravid ruminant reproductive tract. Clearly, there are many questions that need to be answered before we will have an accurate determination of the role that brucebactin plays in the virulence of Brucella strains in their natural hosts.
2.3.4 Heme as an Fe Source for Brucella Strains
By showing that exogenous heme could rescue the heme auxotrophy of a B. abortus hemH mutant, Almirón et al. (2001) also demonstrated that the parental 2308 strain has the capacity to transport the intact heme molecule. This was an important finding because as noted previously, the intracellular trafficking pattern of the brucellae in host macrophages places these bacteria in an environment where heme is conceivably a relevant Fe source. Subsequent studies showed that heme transport in B. abortus 2308 is mediated by the TonB-dependent outer membrane transporter BhuA (Paulley et al. 2007) and the periplasmic binding protein-dependent ABC transporter BhuTUV (Ojeda 2012) (Fig. 2.7). These latter studies also confirmed that Brucella strains can use heme as an Fe source. Moreover, the significant attenuation displayed by a B. abortus bhuA mutant in cultured murine macrophages and experimentally infected mice supports the proposition that heme is indeed an important Fe source for the brucellae during intracellular replication in their mammalian hosts (Paulley et al. 2007)
a Genetic organization of the Brucella bhuA, bhuTUV, bhuQ, omp22, omp25, omp25c omp25d, and omp31-1 loci, and b proposed roles of the corresponding gene products in heme transport and degradation. Gene designations are those used in the B. abortus 2308 genome sequence in GenBank. OM–outer membrane; CM–cytoplasmic membrane
.
The genetic organization of the Brucella bhuA, T, U and V genes is interesting. Bacterial heme transport genes typically reside in operons, which allows their coordinated expression in response to Fe and heme availability. But in Brucella strains, the gene encoding the outer membrane transporter (bhuA) resides >660 kb distant from the genes encoding the rest of the transport system (bhuT, U and V) (Fig. 2.7). The bhuA gene in B. abortus 2308 is also much more responsive to Fe-deprivation at the transcriptional level than the bhuTUV locus (Ojeda 2012). These data suggest that the bhuA and bhuTV may respond differently to relevant environmental stimuli, e.g., cellular Fe levels and the availability of heme in the external environment, but these relationships require further study. It is also not clear what biological benefit differential regulation of the bhuA and bhuTUV would provide to the brucellae.
Heme oxygenases degrade the protoporphyrin backbone of heme and release Fe2+ (Frankenberg-Dinkel 2004). By catalyzing this reaction, these enzymes play an important role in allowing bacteria to use internalized heme as an Fe source. But excess heme in the cytoplasm can also be toxic due of its hydrophobicity and potential to participate in the production of ROS, so heme oxygenases also play an important role in preventing heme toxicity in bacteria (Skaar et al. 2006). To date, a single heme oxygenase, BhuQ, has been identified in Brucella (Ojeda et al. 2012). While phenotypic analysis of a B. abortus bhuQ mutant clearly shows that the corresponding gene product plays a role in the capacity of these bacteria to use heme as an Fe source, these studies have also shown that the brucellae contain other, as yet unidentified, heme oxygenases that also contribute to this process (Fig. 2.7).
Another set of proteins that may contribute to the ability of Brucella strains to capture heme from the external environment are the Omp25/Omp31 family of proteins (Vizcaíno and Cloeckaert 2012) (Fig. 2.7). These proteins are major constituents of the outer membranes of these bacteria, and although their precise functions are unknown, experimental evidence suggests that they play important roles in the interactions of the brucellae with their mammalian hosts. One of the interesting features of the Omp25/31 proteins is that they are homologs of the so-called ‘heme-binding proteins’ (Hbps) that have been described in Bartonella (Minnick et al. 2003). In fact, the Brucella melitensis Omp31 protein binds heme in vitro (Delpino et al. 2006), but whether or not these proteins actually play a role in heme acquisition by Brucella remains to be determined.
2.3.5 Ferrous Iron Transport by Brucella Strains
The brucellae are intracellular pathogens in their mammalian hosts, and their capacity to survive and replicate efficiently in macrophages and placental trophoblasts is critical for their virulence (Roop et al. 2009). During the initial stages of their intracellular life cycle in mammalian cells these bacteria reside in acidified, membrane-bound compartments known as ‘endosomal Brucella-containing vacuoles’, or eBCVs (Starr et al. 2012). The acidic pH of the eBCVs serves as a signal that stimulates expression of the genes encoding the Type IV secretion machinery (Boschiroli et al. 2002), which translocates effector molecules into the cytoplasm of the host cell and directs the intracellular trafficking of the BCVs (de Jong et al. 2008; de Barsy et al. 2011; Marchesini et al. 2011; Salcedo et al. 2013; Myeni et al. 2013). The interactions of the eBCVs with lysosomes potentially provides the brucellae with access to Fe released into the host cell by transferrin-mediated Fe import (Anderson and Vulpe 2009), and the low pH of these intracellular compartments favors the solubility of the ferrous form of Fe (Fe2+) (Kosman 2013). Recent studies have also shown that the majority of the Fe present in the so-called ‘labile Fe pool’ of host cells is in the form of Fe2+ (Hider and Kong 2013). For these reasons, it has been postulated that Fe2+ is a relevant Fe source for the brucellae during their intracellular replication in their mammalian hosts (Roop et al. 2012).
Brucella strains produce a single high affinity Fe2+ transporter (Elhassanny et al. 2013), which is predicted to be structurally and functionally similar to the FtrABCD Fe2+ transporter described in Bordetella spp. (Brickman and Armstrong 2012). Both of these transporters are specific for Fe2+, and a B. abortus ftrA mutant is highly attenuated in experimentally infected mice (Elhassanny et al. 2013), which supports the contention that Fe2+ is a relevant Fe source for Brucella strains during infection. The Brucella FtrABCD transporter and its Bordetella counterpart are proposed to transport Fe2+ by the mechanism shown in Fig. 2.8. This model is based on the predicted functions of their individual components and the phenotypes of defined mutants (Brickman and Armstrong 2012; Elhassanny et al. 2013). FtrA is a member of the P19 family of periplasmic Fe-binding proteins which have well-documented roles in bacterial Fe transport (Chan et al. 2010; Koch et al. 2011). The periplasmic protein FtrB is thought to have ferroxidase activity (e.g. Fe2+ → Fe3+ activity), based on the presence of putative ‘cupredoxin-like’ metal binding domains (Rajasekaran et al. 2010), but this function has not been experimentally validated. FtrC is a homolog of the Fe permease Ftr1p, which has been well-studied in eukaryotic microbes (Kosman 2003). One of the distinctive features of Ftr1p is that it forms a complex with the ferroxidase Fet3p, and Ftr1p transports Fe2+, but only after it has been oxidized to Fe3+ in a step that occurs concomitant with transport (de Silva et al. 1995; Kwok et al. 2006). FtrD shares significant amino acid homology with the ferredoxin NapH, which resides in the cytoplasmic membrane of the bacterium Wolinella succinogenes and is an integral component of an electron transport chain (Kern and Simon 2008). Hence, FtrD has been proposed to serve as a sink for the electrons transferred to FtrB during the oxidation of Fe2+, thereby resetting the redox balance of the transporter.
It is important to note that the necessity for the coupled Fe2+ oxidation/permeation step depicted in Fig. 2.8 has not verified experimentally for either the Brucella or Bordetella FtrABCDs. And if it is required, it is not clear what benefit this mode of transport would provide, if any, compared to ‘direct’ transport of Fe2+. One possibility is that converting Fe2+ to Fe3+ provides a mechanism for ‘trapping’ the insoluble Fe3+ at the site of transport. This proposition is consistent with the observation that this mechanism of Fe2+ transport is kinetically superior to direct Fe2+ transport by at least an order of magnitude in yeast (Kosman 2010). There is also evidence suggesting that this coupled oxidation/permeation step provides an indirect form of oxidative defense by converting the strong oxidant Fe2+ to the less reactive Fe3+ (Shi et al. 2003; Kosman 2010). This latter possibility is particularly intriguing when one considers that both Brucella and Bordetella rely upon a respiratory type of metabolism (Corbel and Brinley-Morgan 1984; Pittman 1984) and would be expected to be transporting Fe2+ in host tissues where O2 is present.
2.3.6 Siderophore-Independent Transport of Fe3+ in Brucella
Fe3+-siderophore complexes generally require energy for transport across the outer membrane of Gram-negative bacteria (Noinaj et al. 2010), but Fe3+ bound to low molecular weight chelators such as citrate can diffuse across the outer membrane via porins, where specialized Fe3+-specific periplasmic protein-dependent ABC transporters can capture this Fe3+ and transport it across the cytoplasmic membrane. Such transporters include the Sfu (Angerer et al. 1992), Afu (Chin et al. 1996) and Yfu (Gong et al. 2001) systems that have been described in Serratia, Actinobacillus and Yersinia, respectively. Two sets of genes predicted to encode Sfu-type Fe3+ transporters have been described in Brucella (Jenner et al. 2009; Roop et al. 2012) (Fig. 2.9), but the roles that the corresponding gene products play in Fe transport and/or virulence are unknown.
2.4 Detoxification of Excess Intracellular Fe by Brucella Strains
Bacteria must not only be able to import enough Fe to meet their physiological needs, but they must also have a means of maintaining cellular levels of unincorporated Fe below ‘toxic’ levels. One ‘indirect’ mechanism that they use is to tightly regulate the genes encoding their Fe import systems so that they only import Fe when cellular levels fall below a certain threshold. How Brucella strains employ this strategy to prevent Fe toxicity will be discussed in detail in a subsequent section. The other two more ‘direct’ mechanisms that bacteria employ to prevent Fe toxicity are to (a) convert excess intracellular Fe2+ into a ‘non-toxic’ form (e.g. Fe3+) for storage in proteins such as bacterioferritin and Dps; and (b) to export excess intracellular Fe2+ from the cell.
2.4.1 Bacterioferritin and Dps
Bacterioferritins (Bfrs) are large, 24 subunit proteins that with 12 heme groups form hollow spheres in bacterial cells (Andrews 2010). These proteins have distinctive ferroxidase centers that convert soluble Fe2+ to insoluble Fe3+ which is then stored as 2Fe(O)OH in the interior of these spherical proteins. Each individual Bfr can store up to 4500 atoms of Fe3+, and this Fe3+ can be converted to Fe2+ and released back into the bacterial cytoplasm as needed to replenish depleted Fe levels. By converting the highly reactive Fe2+ to the less reactive Fe3+ and sequestering it away from the other components of the cytoplasm, Bfr serves not only as a depot for excess Fe but also prevents this excess Fe from reaching toxic levels (Fig. 2.10).
Dps is another spherical protein found in bacteria that oxidizes Fe2+ to Fe3+ and stores the insoluble Fe3+ in its interior (Andrews 2010) (Fig. 2.10). Although Dps, like Bfr, is considered a member of the ferritin-like superfamily of proteins, it has three important structural and functional differences when compared to Bfr. First, Dps is made up of 12 subunits, and is thus smaller than Bfr, and consequently can store less Fe (e.g., 500 atoms) than Bfr. The ferroxidase site in Dps is also different from that of Bfr, and Dps uses H2O2 instead of O2 to catalyze the oxidation of Fe2+. In addition, many, but not all bacterial Dps proteins, bind non-specifically to DNA. In fact, the name of this protein, Dps (DNA-binding protein from starved cells), was derived from the observation that this protein is a major nucleoid-associated protein in E. coli cells after they enter into stationary phase (Almirón et al. 1992). Based on these properties and the phenotypic analysis of bacterial dps mutants, it has been proposed that the major physiologic function of Dps is as a cellular defense against oxidative stress rather than Fe storage (Chiancone and Ceci 2010).
Brucella strains produce both Bfr (Denoel et al. 1997; Almirón and Ugalde 2010) and Dps (Kim et al. 2014) (Fig. 2.10). Phenotypic analysis of a defined mutant indicates that Bfr plays a role in Fe metabolism in B. abortus 2308, e.g., a bfr mutant is more sensitive to Fe deprivation and has reduced levels of intracellular Fe compared to the parent strain (Almirón and Ugalde 2010). The biological function of Dps, on the other hand, is presently unclear. Although the dps gene is strongly regulated by the general stress response sigma factor RpoE1 in B. abortus 2308 (Kim et al. 2014), a B. abortus dps mutant exhibits wild-type resistance to H2O2 in in vitro assays. Considering the possibility that Dps could conceivably play a compensatory role to Bfr in terms of Fe storage, it will be important to evaluate the Fe storage capabilities and virulence properties of Brucella bfr dps double mutants to adequately evaluate their respective biological functions.
2.4.2 MbfA
Another protein that has been linked to Fe detoxification in the α-proteobacteria is MbfA (Rodionov et al. 2006). This protein was initially proposed to be a membrane-bound ferritin based on the presence of conserved ‘Fe-binding domains’ in its N-terminus (Andrews 2010). Another distinctive feature of the MbfA proteins is that they have ‘vacuolar iron transporter (VIT) domains’ in their C-termini. Agrobacterium tumefaciens and Bradyrhizobium japonicum mbfA mutants are defective in Fe export and more susceptible to Fe-mediated H2O2 toxicity than their parental strains (Ruangkiattikul et al. 2012; Bhubhanil et al. 2014; Sankari and O’Brian 2014), which led to the proposition that MbfA serves as an Fe2+ exporter in these bacteria (Fig. 2.10). Consistent with their proposed role in Fe export, the A. tumefaciens and B. japonicum mbfA genes have both been shown to display increased expression in response to increasing levels of cellular Fe and decreased expression in response to Fe deprivation, and both genes are regulated by the iron response regulator Irr (Rudolph et al. 2006; Sangwan et al. 2008; Ruangkiattikul et al. 2012). Homologs of mbfA have also been shown to be targets of Irr repression in Rhizobium leguminosarum (Todd et al. 2006) and Rhodobacter sphaeroides (Peuser et al. 2012), but to the authors’ knowledge the role of the corresponding genes products in Fe metabolism in these latter bacteria has not been examined.
The Brucella mbfA gene shows the same pattern of Fe-responsive expression as its Agrobacterium and Bradyrhizobium counterparts (e.g. low Fe repression, high Fe induction), and is a direct target of Irr repression in B. abortus 2308 (Martinson 2014). While these properties are consistent with MbfA playing a role in Fe export in Brucella as shown in Fig. 2.11, this function has not yet been experimentally verified. But it would certainly be important to know if Fe detoxification by Bfr and MbfA, possibility in concert with Dps, plays an important role in the virulence of Brucella strains in their mammalian hosts. Neither Brucella bfr (Denoel et al. 1997; Almirón and Ugalde 2010) nor dps (Kim et al. 2014) mutants have been shown to be attenuated in mice or cultured mammalian cells. But it is easy to envision how the activity of MbfA could compensate for the loss of Bfr or Dps activity, if this protein is indeed an Fe2+ exporter.
2.5 Regulation of Fe Homeostasis in Brucella
Bacteria tightly regulate the expression of genes involved in Fe import, storage and export, as well as those encoding cellular proteins that require Fe for their activity. This strategy maintains Fe homeostasis, avoids Fe toxicity and sustains cellular metabolism in the face of fluctuating levels of Fe availability in their external environments. Three genetic regulators which directly control Brucella Fe metabolism genes, Irr, DhbR and BsrH, and one which is likely to perform this function based on its activity in other α-proteobacteria, RirA, have been described.
2.5.1 Irr
The Brucella iron response regulator, Irr, was originally identified by Martínez et al. (2005). Subsequent studies by this and other groups have shown that Irr is the predominant regulator of the Fe metabolism genes in these bacteria, and suggest that this regulator functions in much the same manner as the Bradyrhizobium japonicum Irr (Yang et al. 2006; O’Brian 2015), which is the prototype for this family of regulators. Specifically, the Brucella Irr is stable under conditions of Fe limitation, but is rapidly degraded under Fe-replete conditions (Martínez et al. 2005; Anderson et al. 2011). The fact that Irr can bind heme (Martínez et al. 2005) and that the Fe-responsive degradation of this protein depends upon a functional heme biosynthetic pathway (Martinson 2014) also suggests the regulatory activity of this protein is being modulated ‘indirectly’ by cellular Fe levels (e.g. sensing whether or not the cell has sufficient Fe to support heme biosynthesis) rather than directly sensing Fe as is the case with other bacterial Fe-responsive regulators such as Fur (Fillat 2014). Like its B. japonicum counterpart, the Brucella Irr can serve as either a transcriptional activator (Martínez et al. 2006; Anderson et al. 2011; Elhassanny et al. 2013) or repressor (Martínez et al. 2005; Ojeda et al. 2012, Martinson 2014) depending upon where it binds in the promoter region of a gene, and it activates the expression of Fe import genes and represses genes involved in Fe export and storage (Fig. 2.12).
Irr plays a critical role in the ability of B. abortus 2308 to maintain chronic spleen infections in experimentally infected mice (Anderson et al. 2011). This should not be too surprising since this activator is required for optimal expression of the genes encoding all three of the Fe transport systems that have been linked to virulence in Brucella strains (e.g., Fe3+-siderophore, heme and Fe2+ transport) (Martínez et al. 2006; Anderson et al. 2011; Elhassanny et al. 2013). But Irr also represses genes involved in Fe export and storage, and it would be interesting to know if down regulation of these latter genes also plays a role in this regulator’s contribution to virulence.
a Intracellular Fe levels and the status of heme biosynthesis control the regulatory activity of the Brucella iron response regulator (Irr) by affecting its stability. b Fe homeostasis genes regulated by Irr in B. abortus 2308. The solid lines in (b) indicate direct interactions between Irr and the promoters of the genes depicted, the dashed lines indicate that it is currently unknown whether the regulatory link between Irr and these genes is direct or indirect
2.5.2 RirA
Many of the α-proteobacteria, with the notable exception of B. japonicum, produce another Fe-responsive regulator known as RirA (Rodionov et al. 2006). This protein was originally identified in Rhizobium leguminosarum where it serves as a global repressor of Fe uptake genes during growth under Fe-replete conditions, hence its designation, rhizobial iron regulator A (Todd et al. 2002). RirA is a member of the Rrf2 family of transcriptional regulators. These regulators typically rely upon Fe-S clusters for their activity, and hence it has been proposed that like Irr, RirA does not sense Fe levels in the cell directly, but rather monitors the status of Fe-S assembly as an indirect means of sensing cellular Fe levels (Johnston et al. 2007) (Fig. 2.13).
Brucella strains produce a RirA homolog, and the corresponding gene resides in an operon with the gene encoding the heme oxygenase BhuQ (Ojeda et al. 2012). Transcription of this operon is repressed by Irr under low Fe conditions, and it lies in close proximity to the gene encoding Bfr. This genetic context and regulatory pattern is consistent with the possibility that RirA serves as an Fe-responsive regulator in Brucella. Microarray analysis also indicates that Fe and heme transport genes display elevated expression in a B. abortus rirA mutant during growth under Fe-replete conditions (Martinson 2014). Based on these observations, it is tempting to speculate that the Brucella Irr and RirA work together to form an integrated regulatory circuit like that proposed for the related α-proteobacterium Agrobacterium tumefaciens (Hibbing and Fuqua 2011), where Irr activates Fe acquisition genes and represses Fe storage, transport and utilization genes under low Fe iron conditions, and RirA represses Fe uptake genes under high Fe conditions (Fig. 2.13). But confirming this will require further experimental analysis.
2.5.3 DhbR and Other Fe Source-Specific Regulators
In addition to being up regulated in response to Fe deprivation, the genes that encode many bacterial Fe transporters are subject to a second level of regulation that ensures that they are only expressed at maximum levels if the corresponding Fe source is readily available in the external environment. For instance, the genes responsible for both biosynthesis and transport of the siderophore alcaligin in Bordetella are ‘de-repressed’ in response to Fe deprivation, but their maximum expression is dependent upon the transcriptional activator AlcR, which recognizes Fe3+-alcaligin complexes imported into the cell (Brickman et al. 2001). This ‘two-step’ mode of regulation allows bacteria to conserve energy and prioritize the expression of their Fe acquisition genes to best fit the often changing environmental niches they inhabit (Brickman and Armstrong 2009). Similarly, the AlcR homolog DhbR is required for maximum expression of the siderophore biosynthesis genes in B. abortus 2308 in response to Fe deprivation (Anderson et al. 2008). Electrophoretic mobility shift assays have shown that DhbR binds directly to the dhbC promoter region, and genetic analysis suggests that brucebactin serves as the co-activator of DhbR (Fig. 2.14). It is presently unknown, however, if DhbR also plays a role in the regulation of the siderophore transport genes in Brucella, or what role, if any, this transcriptional regulator has in the virulence of these strains in their natural ruminant hosts.
The AraC-type transcriptional activator DhbR is required for maximum expression of the brucebactin biosynthesis genes in B. abortus 2308 in response to Fe deprivation. Experimental suggests that Fe3+-brucebactin likely serves as a co-activator for DhbR (Anderson et al. 2008)
Experimental evidence likewise suggests that ‘Fe source-specific’ regulators play important roles in prioritizing the expression of the Brucella Fe2+ and heme transport genes. For instance, the genes encoding the Fe2+ transporter FtrABCD display maximum expression in B. abortus 2308 when this strain is subjected both Fe deprivation and exposure to acid pH (Elhassanny 2016). Irr is responsible for the low Fe-responsive activation of these genes (Elhassanny et al. 2013), but the transcriptional regulator that controls their acid-responsive induction has yet to be identified. This dual stimulus enhancement of ftrABCD expression may be especially beneficial to the brucellae in the endosomal BCVs in host macrophages, where they would be expected to be exposed to both Fe deprivation and acidic pH, and Fe2+ is likely to be a readily available Fe source (Elhassanny et al. 2013). Preliminary evidence also suggests that the genes encoding the Brucella heme transporters are induced in response to exposure to heme (Paulley 2007; Ojeda 2012), but the nature of this regulation has not been well-characterized.
2.5.4 BsrH
Small, untranslated regulatory RNAs (sRNAs) play critical roles in controlling gene expression in prokaryotes (Waters and Storz 2009). By facilitating or inhibiting the translation of their target mRNAs, and/or modulating the stability of these transcripts, sRNAs provide a quick mode of genetic regulation that complements and amplifies that provided by transcriptional regulators. sRNAs encoded by Fe-responsive genes are essential components of the Fe homeostasis systems of many bacteria (Massé et al. 2007). In E. coli, for instance, the sRNA RyhB inhibits the expression of genes encoding certain metabolic enzymes that require Fe for their activity such as the TCA cycle enzymes succinate dehydrogenase, aconitase, and fumarase A, and the Fe-cofactored superoxide dismutase SodB when the cell is growing under Fe-deprived conditions (Massé and Gottesman 2002). The ryhB gene is repressed by the Fe-responsive repressor Fur, and when cellular Fe levels reach a certain threshold, ryhB expression is repressed, and the sdhDCAB, acnB, fumA and sodB loci are efficiently expressed.
The recent discovery of the 104 nt sRNA BsrH (Peng et al. 2015) suggests that these post-transcriptional regulators may also contribute to the maintenance of Fe homeostasis in Brucella. The bsrH gene partially overlaps the gene encoding the heme biosynthesis enzyme ferrochelatase (hemH), but bsrH and hemH are transcribed in opposite directions. This genetic organization results in a 46 nt region of complementarity between BsrH and the hemH transcript, which led to the proposition that the latter gene is a target for BsrH repression. Overexpression of bsrH leads to reduced hemH expression in an E. coli surrogate, and produces a growth defect when B. abortus 2308 is grown under Fe-deprived conditions. While these findings are certainly consistent with the authors’ proposition that BsrH plays a role in Fe metabolism in Brucella strains (Peng et al. 2015), there are several important questions that need to be addressed to determine its exact function. For instance, bsrH expression is reduced in response to Fe limitation in B. abortus 2308, and it would seem counterintuitive to relieve BsrH repression of hemH expression in the face of reduced cellular levels of Fe. The mechanism by which BsrH is proposed to repress hemH expression (e.g., as a cis-acting sRNA) is also atypical in comparison with known sRNAs that regulate Fe metabolism genes in other bacteria, which are trans-acting sRNAs that regulate genes that lie distant from the genes that encode them. Regardless, it will be important to determine precisely how sRNAs such as BsrH contribute to the regulation of Fe metabolism in Brucella.
2.6 Conclusions
Hopefully, this chapter will provide readers with an appreciation of how well-adapted Brucella strains are to defend themselves against the Fe-withdrawal defenses of their mammalian hosts. But the authors also hope that they will recognize that there is a lot more to be learned about Fe metabolism in Brucella. One particularly important area that needs to be addressed is how changes in Fe trafficking in the host caused by immune and other physiologic responses influences the availability of this micronutrient to Brucella strains in specific host cells and tissues.
References
Almirón M, Link AJ, Furlong D, Kolter R (1992) A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Gens Dev 6:2646–2654
Almirón M, Martínez M, Sanjuan N, Ugalde RA (2001) Ferrochelatase is present in Brucella abortus and is critical for its intracellular survival and virulence. Infect Immun 69:6225–6230
Almirón MA, Ugalde RA (2010) Iron homeostasis in Brucella abortus: the role of bacterioferritin. J Microbiol 48:668–673
Anderson GJ, Vulpe CD (2009) Mammalian iron transport. Cell Mol Life Sci 66:3241–3261
Anderson JD, Smith H (1965) The metabolism of erythritol by Brucella abortus. J Gen Microbiol 38:109–124
Anderson ES, Paulley JT, Roop RM II (2008) The AraC-like transcriptional regulator DhbR is required for maximum expression of the 2,3-dihydroxybenzoic acid biosynthesis genes in Brucella abortus 2308 in response to iron deprivation. J Bacteriol 190:1838–1842
Anderson ES, Paulley JT, Martinson DA, Gaines JM, Steele KH, Roop RM II (2011) The iron-responsive regulator Irr is required for wild-type expression of the gene encoding the heme transporter BhuA in Brucella abortus 2308. J Bacteriol 193:5359–5364
Andrews SC (2010) The ferritin-like superfamily: evolution of the biological storeman from a rubrerythrin-like ancestor. Biochim Biophys Acta 1800:691–705
Angerer A, Klupp B, Braun V (1992) Iron transport systems of Serratia marcescens. J Bacteriol 174:1378–1387
Archibald F (1983) Lactobacillus plantarum, an organism not requiring iron. FEMS Microbiol Lett 19:29–32
Atkinson XJ (2015) Determining the chemical structure of brucebactin, the sole complex siderophore utilized by the pathogenic bacterium Brucella abortus. Masters thesis. East Carolina University
Baldwin CL, Goenka R (2006) Host immune responses to the intracellular bacterium Brucella: does the bacterium instruct the host to facilitate chronic infection? Crit Rev Immunol 26:407–442
Baldwin CL, Winter AJ (1994) Macrophages and Brucella. In: Zwilling BS, Eisenstein TK (eds) Macrophage-pathogen interactions. Marcel Dekker, New York, NY, pp 363–380
Bellaire BH, Elzer PH, Baldwin CL, Roop RM II (1999) The siderophore 2,3-dihydoxybenzoic acid is not required for virulence of Brucella abortus in BALB/c mice. Infect Immun 67:2615–2618
Bellaire BH (2001) Production of the siderophore 2,3-dihydroxybenzoic acid by Brucella abortus is regulated independent of Fur and is required for virulence in cattle. Doctoral dissertation. Louisiana State University Health Sciences Center-Shreveport
Bellaire BH, Baldwin CL, Elzer PH, Roop RM II (2000) The siderophore 2,3-dihydroxybenzoic acid contributes to the virulence of Brucella abortus in ruminants. Abstr 100th Gen Meet Amer Soc Microbiol, Abstr B-17, p 44
Bellaire BH, Elzer PH, Hagius S, Walker J, Baldwin CL, Roop RM II (2003a) Genetic organization and iron-responsive regulation of the Brucella abortus 2,3-dihydroxybenzoic acid biosynthesis operon, a cluster of genes required for wild-type virulence in pregnant cattle. Infect Immun 71:1794–1803
Bellaire BH, Elzer PH, Baldwin CL, Roop RM II (2003b) Production of the siderophore 2,3-dihydroxybenzoic acid is required for wild-type growth of Brucella abortus in the presence of erythritol under low-iron conditions in vitro. Infect Immun 71:2927–2932
Bhubhanil S, Chamsing J, Sittipo P, Chaoprasid P, Sukchawalit R, Mongkolsuk S (2014) Roles of Agrobacterium tumefaciens membrane-bound ferritin (MbfA) in iron transport and resistance to iron under acidic conditions. Microbiology 160:863–871
Boschiroli ML, Ouahrani-Bettache S, Foulongne V, Michaux-Charachon S, Bourg G, Allardet-Servent A, Cazevieille C, Liautard JP, Ramuz M, O’Callaghan D (2002) The Brucella suis virB operon is induced intracellularly in macrophages. Proc Natl Acad Sci USA 99:1544–1549
Brickman TJ, Armstrong SK (2009) Temporal signaling and differential expression of Bordetella iron transport systems: the role of ferrimones and positive regulators. Biometals 22:33–41
Brickman TJ, Armstrong SK (2012) Iron and pH-responsive FtrABCD ferrous iron utilization system of Bordetella species. Mol Microbiol 86:580–593
Brickman TJ, McIntosh MA (1992) Overexpression and purification of ferric enterobactin esterase from Escherichia coli. Demonstration of enzymatic hydrolysis of enterobactin and its iron complex. J Biol Chem 267:12350–12355
Brickman TJ, Kang HY, Armstrong SK (2001) Transcriptional activation of Bordetella alcaligin siderophore genes requires the AlcR regulator with alcaligin as inducer. J Bacteriol 183:483–489
Byrd TF, Horwitz MA (1989) Interferon gamma-activated human monocytes downregulate transferrin receptors and inhibit the intracellular multiplication of Legionella pneumophila by limiting the availability of iron. J Exp Med 83:1457–1465
Cairo G, Recalcati S, Mantovani A, Locati M (2011) Iron trafficking and metabolism in macrophages: contribution of the polarized phenotype. Trends Immunol 32:241–247
Celli J (2015) The changing nature of the Brucella-containing vacuole. Cell Microbiol 17:951–958
Cellier MF, Courville P, Campion C (2007) Nramp1 phagocyte intracellular metal withdrawal defense. Microbes Infect 9:1662–1670
Chan ACK, Doukov TI, Scofield M, Tom-Yew SAL, Ramin AB, MacKichan JK, Gaynor EC, Murphy MEP (2010) Structure and function of P19, a high-affinity iron transporter of the human pathogen Campylobacter jejuni. J Mol Biol 401:590–604
Chiancone E, Ceci P (2010) The multifaceted capacity of Dps proteins to combat bacterial stress conditions: detoxification of iron and hydrogen peroxide and DNA binding. Biochim Biophys Acta 1800:798–805
Chin N, Frey J, Chang CF, Chang YF (1996) Identification of a locus involved in the utilization of iron by Actinobacillus pleuropneumoniae. FEMS Microbiol Lett 143:1–6
Chipperfield JR, Ratledge C (2000) Salicylic acid is not a bacterial siderophore: a theoretical study. Biometals 13:165–168
Cooper SR, McArdle JV, Raymond KN (1978) Siderophore electrochemistry: relation to intracellular iron release mechanism. Proc Natl Acad Sci USA 75:3551–3554
Corbel MJ, Brinley-Morgan WJ (1984) Genus Brucella. In: Krieg NR, Holt JG (eds) Bergey’s manual of systematic bacteriology, vol 1. Williams & Wilkins, Baltimore, MD, pp 377–388
Cornish AS, Page WJ (2000) Role of molybdate and other transition metals in the accumulation of protochelin by Azotobacter vinelandii. Appl Environ Microbiol 66:1580–1586
Crichton R (2009) Iron metabolism—from molecular mechanisms to clinical consequences, 3rd edn. Wiley, West Sussex, UK
Crumbliss AL, Harrington JM (2009) Iron sequestration by small molecules: thermodynamic and kinetic studies of natural siderophores and synthetic model compounds. Adv Inorg Chem 61:179–251
Danese I, Haine V, Delrue R-M, Tibor A, Lestrate P, Stevaux O, Mertens P, Paquet J-Y, Godfroid J, De Bolle X, Letesson JJ (2004) The Ton system, an ABC transporter, and a universally conserved GTPase are involved in iron utilization by Brucella melitensis 16M. Infect Immun 72:5783–5790
de Barsy M, Jamet A, Filipon D, Nicolas C, Laloux G, Rual JF, Muller A, Twizere JC, Nkengfac B, Vandenhaute J, Hill DE, Salcedo SP, Gorvel JP, Letesson JJ, De Bolle X (2011) Identification of a Brucella spp. secreted effector specifically interacting with human small GTPase Rab2. Cell Microbiol 13:1044–1058
de Jong MF, Sun YH, den Hartigh AB, van Dijl JM, Tsolis RM (2008) Identification of VceA and VceC, two members of the VjbR regulon that are translocated into macrophages by the Brucella type IV secretion system. Mol Microbiol 70:1378–1396
de Silva DM, Askwith CC, Eide D, Kaplan J (1995) The FET3 gene product required for high affinity iron transport in yeast is a cell surface ferroxidase. J Biol Chem 270:1098–1101
Delpino MV, Cassataro J, Fossati CA, Goldbaum FA, Baldi PC (2006) Brucella outer membrane protein Omp31 is a haemin-binding protein. Microbes Infect 8:1203–1208
Denoel PA, Crawford RM, Zygmunt MS, Tibor A, Weynants AE, Godfroid F, Hoover DL, Letesson JJ (1997) Survival of a bacterioferritin deletion mutant of Brucella melitensis 16M in human monocyte-derived macrophages. Infect Immun 65:4337–4340
Elhassanny AEM (2016) Characterization of FtrABCD: a ferrous iron-specific transporter that is required for the virulence of Brucella abortus 2308 in mice. Doctoral dissertation. East Carolina University
Elhassanny AEM, Anderson ES, Menscher EA, Roop RM II (2013) The ferrous iron transporter FtrABCD is required for the virulence of Brucella abortus 2308 in mice. Mol Microbiol 88:1070–1082
Enright FM (1990) The pathogenesis and pathobiology of Brucella infections in domestic animals. In: Nielsen KH, Duncan JR (eds) Animal brucellosis. CRC Press, Boca Raton, FL, pp 201–320
Evans RW, Kong X, Hider RC (2012) Iron mobilization from transferrin by therapeutic iron chelating agents. Biochim Biophys Acta 1820:282–290
Fillat MF (2014) The FUR (ferric uptake regulator) superfamily: diversity and versatility of key transcriptional regulators. Arch Biochem Biophys 546:41–52
Frankenberg-Dinkel N (2004) Bacterial heme oxygenases. Antioxid Redox Signal 6:825–834
Goetz DH, Holmes MA, Borregaard N, Bluhm ME, Raymond KN, Strong RK (2002) The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol Cell 10:1033–1043
Gong S, Bearden SW, Geoffroy VA, Fetherston JD, Perry RD (2001) Characterization of the Yersinia pestis Yfu ABC inorganic iron transport system. Infect Immun 67:2829–2837
González-Carreró MI, Sangari FJ, Agüero J, García-Lobo JM (2002) Brucella abortus 2308 produces brucebactin, a highly efficient catecholic siderophore. Microbiology 148:353–360
Grilló MJ, Blasco JM, Gorvel JP, Moriyón I, Moreno E (2012) What have we learned from brucellosis in the mouse model? Vet Res 43:e29
Hancock REW, Hantke K, Braun V (1977) Iron transport in Escherichia coli K-12—2,3-dihydroxybenzoate-promoted iron uptake. Arch Microbiol 114:231–239
Hantke K (1990) Dihydroxybenzoylserine—a siderophore for E. coli. FEMS Microbiol Lett 67:5–8
Harrington JM, Crumbliss AL (2009) The redox hypothesis in siderophore-mediated iron uptake. Biometals 22:679–689
Hibbing ME, Fuqua C (2011) Antiparallel and interlinked control of cellular iron levels by the Irr and RirA regulators of Agrobacterium tumefaciens. J Bacteriol 193:3461–3472
Hider RC, Kong X (2013) Iron speciation in the cytosol: an overview. Dalton Trans 42:3220–3229
Hood MI, Skaar EP (2012) Nutritional immunity: transition metals at the pathogen-host interface. Nature Rev Microbiol 10:525–537
Jain N, Rodriguez AC, Kimsawatde G, Seleem MN, Boyle SM, Sriranganathan N (2011) Effect of entF deletion on iron acquisition and erythritol metabolism by Brucella abortus 2308. FEMS Microbiol Lett 316:1–6
Jenner DC, Dassa E, Whatmore AM, Atkins HS (2009) ABC-binding cassette systems of Brucella. Comp Funct Genom 2009:e354649
Johnston AWB, Todd JD, Curson AR, Lei S, Nikolaidou-Katsaridou N, Gelfand MS, Rodionov DA (2007) Living without Fur: the sublety and complexity of iron-responsive gene regulation in the symbiotic bacterium Rhizobium and other α-proteobacteria. Biometals 20:501–511
Kadner RJ (1990) Vitamin B12 transport in Escherichia coli: energy coupling between membranes. Mol Microbiol 4:2027–2033
Keating TA, Marshall CG, Walsh CT, Keating AE (2002) The structure of VibH represents nonribosomal peptide synthetase condensation, cyclization and epimerization domains. Nat Struct Biol 9:522–526
Kern J, Simon J (2008) Characterization of the NapGH quinol dehydrogenase complex involved in Wolinella succinogens nitrate respiration. Mol Microbiol 69:1137–1152
Kim H-S, Willett JW, Jain-Gupta N, Fiebig A, Crosson S (2014) The Brucella abortus virulence regulator, LovhK, is a sensor kinase in the general stress response signalling pathway. Mol Microbiol 94:913–925
Koch D, Chan ACK, Murphy MEP, Lilie H, Grass G, Nies DH (2011) Characterization of a dipartite iron uptake system from uropathogenic Escherichia coli strain F11. J Biol Chem 286:25317–25330
Korolnek T, Hamza I (2014) Like iron in the blood of the people: the requirement for heme trafficking in iron metabolism. Front Pharmacol 5:e126
Kosman DJ (2003) Molecular mechanisms of iron uptake in fungi. Mol Microbiol 47:1185–1197
Kosman DJ (2010) Redox cycling in iron uptake, efflux and trafficking. J Biol Chem 285:26729–26735
Kosman DJ (2013) Iron metabolism in aerobes: managing ferric iron hydrolysis and ferrous iron autoxidation. Coord Chem Rev 257:210–217
Köster WL, Actis LA, Waldbeser LS, Tolmasky ME, Crosa JH (1991) Molecular characterization of the iron transport system mediated by the pJM1 plasmid in Vibrio anguillarum 775. J Biol Chem 266:23829–23833
Kwok EY, Severance S, Kosman DJ (2006) Evidence for iron channeling in the Fet3p-Ftr1p high-affinity iron uptake complex in the yeast plasma membrane. Biochemistry 45:6317–6327
Lengeler JW, Drews G, Schlegel HG (1999) Biology of the prokaryotes. Georg Thieme Verlag, Stuttgart, Germany
López-Goñi I, Moriyón I (1995) Production of 2,3-dihydroxybenzoic acid by Brucella species. Curr Microbiol 31:291–293
López-Goñi I, Moriyón I, Neilands JB (1992) Identification of 2,3-dihydroxybenzoic acid as a Brucella abortus siderophore. Infect Immun 60:4496–4503
Luke RKJ, Gibson F (1971) Location of three genes concerned with the conversion of 2,3-dihydroxybenzoate into enterochelin in Escherichia coli K-12. J Bacteriol 107:557–562
Marchesini MI, Herrmann CK, Salcedo SP, Gorvel JP, Comerci DJ (2011) In search of Brucella abortus type IV secretion substrates: screening and identification of four proteins translocated into host cells through VirB system. Cell Microbiol 13:1261–1274
Martínez M, Ugalde RA, Almirón M (2005) Dimeric Brucella abortus Irr protein controls its own expression and binds haem. Microbiology 151:3427–3433
Martínez M, Ugalde RA, Almirón M (2006) Irr regulates brucebactin and 2,3-dihydroxybenzoic acid biosynthesis, and is implicated in the oxidative stress resistance and intracellular survival of Brucella abortus. Microbiology 152:2591–2598
Martinson DA (2014) The iron response regulator controls iron homeostasis in Brucella. Doctoral dissertation. East Carolina University
Massé E, Gottesman S (2002) A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci USA 99:4620–4625
Massé E, Salvail H, Desnoyers G, Arguin M (2007) Small RNAs controlling iron metabolism. Curr Opin Microbiol 10:140–145
May JJ, Wendrich MT, Marahiel MA (2001) The dhb operon of Bacillus subtilis encodes the biosynthetic template for the catecholic siderophore 2,3-dihydroxybenzoate-glycine-threonine trimeric ester bacillibactin. J Biol Chem 276:7209–7217
Michels K, Nemeth E, Ganz T, Mehrad B (2015) Hepcidin and host defense against infectious diseases. PLoS Pathog 11:e1004998
Minnick MF, Sappington KN, Smitherman LS, Andersson SG, Karlberg O, Carroll JA (2003) Five-member gene family of Bartonella quintana. Infect Immun 71:814–821
Myeni S, Child R, Ng TW, Kupko JJ III, Wehrly TD, Porcella SF, Knodler LA, Celli J (2013) Brucella modulates secretory trafficking via multiple Type IV secretion effector proteins. PLoS Pathog 9:e1003556
Nairz M, Fritsche G, Brunner P, Talasz H, Hantke K, Weiss G (2008) Interferon-γ limits the availability of iron for intramacrophage Salmonella typhimurium. Eur J Immunol 38:1923–1936
Nairz M, Schleicher U, Schroll A, Sonnweber T, Theurl I, Ludwiczek S, Talasz H, Brandacher G, Moser PL, Muckenthaler MU, Fang FC, Bogdan C, Weiss G (2013) Nitric oxide-mediated regulation of ferroportin-1 controls macrophage iron homeostasis and immune function in Salmonella infection. J Exp Med 210:855–873
Nairz M, Haschka D, Demetz E, Weiss G (2014) Iron at the interface of immunity and infection. Front Pharmacol 5:e152
Nakashige TG, Zhang B, Krebs C, Nolan EM (2015) Human calprotectin is an iron-sequestering host-defense protein. Nature Chem Biol 11:765–771
Nikaido H, Rosenberg EY (1990) Cir and Fiu proteins in the outer membrane of Escherichia coli catalyze transport of monomeric catechols: study with & β-lactam antibiotics containing catechol and analogous groups. J Bacteriol 172:1361–1367
Noinaj N, Guillier M, Barnard TJ, Buchanan SK (2010) TonB-dependent transporters: regulation, structure and function. Ann Rev Microbiol 64:43–60
O’Brian MR (2015) Perception and homeostatic control of iron in the rhizobia and related bacteria. Annu Rev Microbiol 69:229–245
Ojeda JF (2012) The bhuTUV and bhuO genes play vital roles in the ability of Brucella abortus to use heme as an iron source and are regulated in an iron-responsive manner by RirA and Irr. Doctoral dissertation. East Carolina University
Ojeda JF, Martinson DA, Menscher EA, Roop RM II (2012) The bhuQ gene encodes a heme oxygenase that contributes to the ability of Brucella abortus 2308 to use heme as an iron source and is regulated by Irr. J Bacteriol 194:4052–4058
Ollinger J, Song KB, Antelmann H, Hecker M, Helmann JD (2006) Role of the Fur regulon in iron transport in Bacillus subtilis. J Bacteriol 188:3664–3673
Parent MA, Bellaire BH, Murphy EA, Roop RM II, Elzer PH, Baldwin CL (2002) Brucella abortus siderophore 2,3-dihydroxybenzoic acid (DHBA) facilitates intracellular survival of the bacteria. Microb Pathogen 32:239–248
Paulley JT (2007) Production of BhuA by B. abortus is required for hemin utilization and virulence and is dependent on the transcriptional regulators RirA and ChrA. Doctoral dissertation. East Carolina University
Paulley JT, Anderson ES, Roop RM II (2007) Brucella abortus requires the heme transporter BhuA for maintenance of chronic infection in BALB/c mice. Infect Immun 75:5248–5254
Peng X, Dong H, Wu Q (2015) A new cis-encoded sRNA, BsrH, regulating the expression of hemH gene in Brucella abortus 2308. FEMS Microbiol Lett 362:1–7
Persmark MD, Expert D, Neilands JB (1992) Ferric iron uptake in Erwinia chrysanthemi mediated by chrysobactin and related catechol-type compounds. J Bacteriol 174:4783–4789
Peuser V, Remes B, Klug G (2012) Role of the Irr protein in the regulation of iron metabolism in Rhodobacter sphaeroides. PLoS One 7:e42231
Pittman M (1984). Genus Bordetella. In: Krieg NR, Holt JG (eds) Bergey’s manual of systematic bacteriology, vol 1. Williams & Wilkins, Baltimore, MD, pp 388–393
Posey JE, Gherardini FC (2000) Lack of a role for iron in the Lyme disease pathogen. Science 288:1651–1653
Postle K, Kadner RJ (2003) Touch and go: tying TonB to transport. Mol Microbiol 49:869–882
Rajasekaran MB, Nilapwar S, Andrews SC, Watson KA (2010) EfeO-cupredoxins: major new members of the cupredoxin superfamily with roles in bacterial iron transport. Biometals 23:1–17
Raymond KN, Dertz EA (2004) Biochemical and physical properties of siderophores. In: Crosa JH, Mey AR, Payne SM (eds) Iron transport in bacteria. ASM Press, Washington, DC, pp 3–17
Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS (2003) Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J Biol Chem 278:41148–41159
Rodionov DA, Gelfand MS, Todd JD, Curson ARJ, Johnston AWB (2006) Computational reconstruction of iron—and manganese-responsive transcriptional networks in α-proteobacteria. PLOS Comput Biol 2:e163
Roop RM II, Bellaire BH, Valderas MW, Cardelli JA (2004) Adaptation of the brucellae to their intracellular niche. Mol Microbiol 52:621–630
Roop RM II, Gaines JM, Anderson ES, Caswell CC, Martin DW (2009) Survival of the fittest: how Brucella strains adapt to their intracellular niche in the host. Med Microbiol Immunol 198:221–238
Roop RM II, Anderson E, Ojeda J, Martinson D, Menscher E, Martin DW (2012) Metal acquisition by Brucella strains. In: López-Goñi I, O’Callaghan D (eds) Brucella—molecular microbiology and genomics. Caister Academic Press, Norfolk, UK, pp 179–199
Ruangkiattikul N, Bhubhanil S, Chamsing J, Niamyim P, Sukchawalit R, Mongkolsuk S (2012) Agrobacterium tumefaciens membrane-bound ferritin plays a role in protection against hydrogen peroxide toxicity and is negatively regulated by the iron response regulator. FEMS Microbiol Lett 329:87–92
Rudolph G, Semini G, Hauser F, Lindemann A, Friberg M, Hennecke H, Fischer HM (2006) The iron control element acting in positive and negative control of iron-regulated Bradyrhizobium japonicum genes, is a target of the Irr protein. J Bacteriol 188:733–744
Salcedo SP, Marchesini MI, Degos C, Terwagne M, von Bargen K, Lepidi H, Herrmann CK, Santos Lacerda TL, Imbert PRC, Pierre P, Alexopoulo L, Letesson JJ, Comerci DJ, Gorvel JP (2013) BtpB, a novel TIR-containing effector protein with immune modulatory functions. Front Cell Infect Microbiol 3:e28
Sangwan I, Small SK, O’Brian MR (2008) The Bradyrhizobium japonicum Irr protein is a transcriptional repressor with high-affinity DNA-binding activity. J Bacteriol 190:5172–5177
Sankari S, O’Brian MR (2014) A bacterial iron exporter for maintenance of iron homeostasis. J Biol Chem 289:16498–16507
Shi X, Stoj C, Romeo A, Kosman DJ, Zhu Z (2003) Fre1p Cu2+ reduction and Fet3p Cu1+ oxidation modulate copper toxicity in Saccharomyces cerevisiae. J Biol Chem 278:50309–50315
Sia AK, Allred BE, Raymond KN (2013) Siderocalins: siderophore binding proteins evolved for primary pathogen host defense. Curr Opin Chem Biol 17:150–157
Skaar EP, Gaspar AH, Schneewind O (2006) Bacillus anthracis IsdG, a heme-degrading monooxygenase. J Bacteriol 188:1071–1080
Smith AW, Freeman S, Minnett WG, Lambert PA (1990) Characterisation of a siderophore from Acinetobacter calcoaceticus. FEMS Microbiol Lett 70:29–32
Smith H, Williams AE, Pearce JH, Keppie J, Harris-Smith PW, Fitz-George RB, Witt K (1962) Foetal erythritol: a cause of the localization of Brucella abortus in bovine contagious abortion. Nature 193:47–49
Starr T, Child R, Wehrly TD, Hansen B, Hwang S, López-Otin C, Virgin HW, Celli J (2012) Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle. Cell Host Microbe 11:33–45
Todd JD, Wexler M, Sawers G, Yeoman KH, Poole PS, Johnston AWB (2002) RirA, an iron-responsive regulator in the symbiotic bacterium Rhizobium leguminosarum. Microbiology 148:4059–4071
Todd JD, Sawers G, Rodionov DA, Johnston AWB (2006) The Rhizobium leguminosarum regulator IrrA affects the transcription of a wide range of genes in response to Fe availability. Mol Gen Genomics 275:564–577
Vizcaíno N, Cloeckaert A (2012) Biology and genetics of the Brucella outer membrane. In: López-Goñi I, O’Callaghan D (eds) Brucella—molecular microbiology and genomics. Caister Academic Press, Norfolk, UK, pp 133–161
Vogel HJ (2012) Lactoferrin, a bird’s eye view. Biochem Cell Biol 90:233–244
Walsh CT, Marshall CG (2004) Siderophore biosynthesis in bacteria. In: Crosa JH, Mey AR, Payne SM (eds) Iron transport in bacteria. ASM Press, Washington, DC, pp 18–37
Waring WS, Elberg SS, Schneider P, Green W (1953) The role of iron in the biology of Brucella suis I. Growth and nutrition. J Bacteriol 66:82–91
Waters LS, Storz G (2009) Regulatory RNAs in bacteria. Cell 136:615–628
Xavier MN, Winter MG, Spees AM, den Hartigh AB, Nguyen K, Roux CM, Silva TMA, Atluri VL, Kerrinnes T, Keestra AM, Monack DM, Luciw PA, Eigenheer RA, Bäumler AJ, Santos RL, Tsolis RM (2013) PPARγ-mediated increase in glucose availability sustains chronic Brucella abortus infection in alternatively activated macrophages. Cell Host Microbe 14:159–170
Yang J, Sangwan I, Lindemann A, Hauser F, Hennecke H, Fischer HM, O’Brian MR (2006) Bradyrhizobium japonicum senses iron through the status of haem to regulate iron homeostasis and metabolism. Mol Microbiol 60:427–437
Young IG, Langman L, Luke RKJ, Gibson F (1971) Biosynthesis of the iron-transport compound enterochelin: mutants of Escherichia coli unable to synthesize 2,3-dihydroxybenzoate. J Bacteriol 106:51–57
Zhan Y, Cheers C (1993) Endogenous gamma interferon mediates resistance to Brucella abortus infection. Infect Immun 61:4899–4901
Acknowledgements
The authors thank present and former members of the Roop and Almirón laboratories for their contributions to the body of work that provided the basis for preparation of this chapter. We also thank Claire Parker Siburt for preparing the siderophore biosynthesis pathways shown in Fig. 2.4. Work on Brucella Fe metabolism in the Roop lab has been supported by grants from the National Institute of Allergy and Infectious Disease (AI-63516) and the United States Department of Agriculture Competitive Research Grants Program (95-01995; 98-02620 and 35204-12218). Work in the Almiron lab has been supported by the Agencia Nacional de Promoción Científica y Tecnológica de la República Argentina (01-6580 and 06-00651) and the Consejo Nacional de Investigaciones Científicas y Tecnológicas de la Argentina-CONICET (PIP 5463).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this chapter
Cite this chapter
Roop II, R.M., Elhassanny, A.E., Almirón, M.A., Anderson, E.S., Atkinson, X.J. (2017). Iron. In: Roop II, R., Caswell, C. (eds) Metals and the Biology and Virulence of Brucella. Springer, Cham. https://doi.org/10.1007/978-3-319-53622-4_2
Download citation
DOI: https://doi.org/10.1007/978-3-319-53622-4_2
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-53621-7
Online ISBN: 978-3-319-53622-4
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)