Abstract
Estimating regional deformation of the myocardium from Cine MRI has the potential to locate abnormal tissue. Regional deformation of the left ventricle is commonly estimated using either segmentation or 3D + t registration. Segmentation is often performed at each instant separately from the others. It can be tedious and does not guarantee temporal causality. On the other hand, extracting regional parameters through image registration is highly dependent on the initial segmentation chosen to propagate the deformation fields and may not be consistent with the myocardial contours. In this paper, we propose an intermediate approach that couples segmentation and registration in order to improve temporal causality while removing the influence of the chosen initial segmentation. We propose to apply the deformation fields from image registration (sparse Bayesian registration) to every segmentation of the cardiac cycle and combine them for more robust regional measurements. As an illustration, we describe local deformation through the measurement of AHA regional volumes. Maximum regional volume change is extracted and compared across scar and non-scar regions defined from delayed enhancement MRI on 20 ST-elevation myocardial infarction patients. The proposed approach shows (i) more robustness in extracting regional volumes than direct segmentation or standard registration and (ii) better performance in detecting scar.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Local tracking of the myocardium has shown to help determining the local viability of the heart from MRI [10] or echocardiography [4]. Two ways to measure regional deformation are reported in most papers: (i) the sequence of 3D segmentations (named here Segmentation) and (ii) the sequence made of an initial 3D geometry propagated in time using the output of the image registration along the sequence (named here 3D segmentation + registration). Manual or semi-automatic segmentation as offered in commercial software is straightforward, does not rely on any hypothesis from a registration algorithm and is usually considered as ground truth. However, it is a fastidious process leading to variable results between observers, non-consistency between slices or frames and requiring several manual adjustments. To tackle this issue, many research groups have worked on automatic segmentation [11], with some recent methods that include spatio-temporal information to propagate the segmentation [7, 13]. Despite these progresses, routine delineation of the ventricles is still semi-automatic, which offers more confidence and flexibility to the cardiologists. 3D segmentation + registration, on the other hand, gives smooth results in space and time and better consistency between frames. However, full temporal consistency is still not guaranteed and the method can be inconsistent with the myocardial contours (Fig. 1 left). Moreover, standard 3D segmentation + registration heavily depends on the first 3D segmentation used to propagate the deformation fields (Fig. 1 right) leading to high uncertainty on the quantification of the deformation. The dependence on the frame selection and the temporal consistency issue have already been considered in the design of registration algorithms, for instance on 3D echocardiographic data [3, 12], although the segmentation from a single instant is considered.
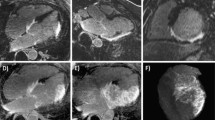
(Left) Segmentation (green contours) compared to registration (purple contours) on short-axis and long-axis view. (Right) Comparison between segmentation (pink contours), the registration propagated from end-diastole (green contours) and the registration propagated from mid-diastole (frame 20, blue contours), for a mid-systole short-axis slice. (Color figure online)
We suggest to combine both the segmentation (of all time frames) and an independent registration algorithm by averaging the propagated mesh from every frame (and not only the first frame) in order to leverage the drawbacks of both while maintaining their assets. This approach is simple, registration-independent, and could be directly translated to clinical practice using already available segmentation software and image registration algorithms. Using this approach reduces the need for a temporally consistent segmentation or registration, since all frames of the cardiac cycle are used to propagate the registration output.
The ultimate goal of registration or segmentation is to extract quantitative parameters in order to understand, estimate, and classify patient’s motion, deformation or shape abnormalities. As an application, we intend to detect abnormal AHA zones from regional volume changes using the standard two approaches and our proposed coupling method. Regional volumes are clinical indices already measured in clinical practice from echocardiography [2] in some cases but rarely measured in MRI partly due to insufficient reliability of the current methods. Previous work also used regional volumes as a way to personalise an electromechanical model as it overcomes the aperture problem of tracking contours using Cine MRI only [9].
2 Methods
2.1 Patient Population and Pre-processing
Image Acquisition. To validate the proposed method against alternatives, 20 patient scans were collected from 3 different clinical studies. The first 10 patients were recruited after ST-elevation myocardial infarction and images were acquired on a Siemens 3 T mMR. The next 10 patients were scanned on a Philips 3T Ingenia after ST-elevation myocardial infarction. Ethical approval and written consent were obtained for all patients. Imaging protocol consisted of 2 chambers, 3 chambers, 4 chambers and short-axis stack Cine images to evaluate the cardiac function as well as short-axis delayed enhancement (LGE) sequences 10 min after injection of 0.4 mmol/kg of Gadolinium. Image resolution varied between 1.32\(\,\times \,\)1.32\(\,\times \,\)9 mm\(^3\) and 1.42\(\,\times \,\)1.42\(\,\times \,\)10 mm\(^3\) and contained 25 to 30 frames per cardiac cycle.
Short-axis image processing. All images were analyzed by 3 experts on SegmentFootnote 1 and then manually corrected after consensus. Image processing of the Cine MRI images consisted of semi-automatic segmentation of the left ventricle endocardium and epicardium on all the short-axis slices and every time frames. Image processing of the LGE MRI required manual segmentation of the left ventricle and semi-automatic segmentation of the scar using Otsu thresholding model [5], as thresholding methods and manual corrections are still the clinical standard despite progress [1, 6] towards automatic infarct delineation as demonstrated in the STACOM’12 challenge.
3D modelling. MR spatial resolution of both the Cine and the LGE sequences is highly heterogeneous with a slice thickness of 9 to 10 mm leading to a staircase effect when creating a mesh directly from the stack of short-axis binary masks. To smooth this effect, the short-axis 2D segmentations were first realigned around the long-axis to prevent from potential artifacts due to to different breath holding positions. For this, the long-axis was defined as the line linking the barycentres of the apical and basal endocardial contours. For each slice, the barycentre of the endocardium was then translated to the long-axis. Second, the polygons formed by each 2D binary masks were linearly interpolated in the z dimension to allow homogeneous resolution. 3D meshes were then created using the CGAL 4.8 libraryFootnote 2. A sequence of 3D segmentations was therefore obtained from Cine MRI. From LGE images, a 3D geometry was created similarly from the endocardium and epicardium delineation. The 2D binary masks of scar delineation were then mapped onto this mesh after iso-resampling. 3D meshes were divided into 17 AHA zones and regional volumes for Cine MRI and scar percentage (number of mesh elements with a scar over the total size of the AHA zone) for LGE MRI, were computed for each AHA zone. Each AHA zone containing at least 1% scarred tissue was labeled as a scar zone. Figure 2 illustrates these two pipelines. The effect of spatial interpolation errors was minimized by averaging the measurements over the AHA zones, larger than the slice resolution.
2.2 Proposed Segmentation and Registration Coupling
Evaluation of local properties such as regional volume change, is usually performed by studying a time sequence of meshes created by segmentation or by propagation of deformation fields on an initial mesh. An intermediate approach that takes the advantages of both approaches is presented here.
Sparse Bayesian Registration. Image registration was based on the sparse Bayesian algorithm presented in [8]. Images were first upsampled to an isotropic resolution using a linear interpolator. Pairs of consecutive images are registered and the estimated transformations are chained along the cycle. Our implementation uses a three-level multiresolution scheme and the parameters described in [8], which were evaluated on the STACOM’11 registration challenge dataset.
Coupling segmentation and registration. In order to soften the heavy influence of the segmentation on the results of a registration, all 3D segmentations of the cardiac cycle are used in our approach as an initial mesh to which the corresponding registration is applied. More precisely, let’s call \(M_t\) the mesh created from the segmentation of the cardiac frame t and \(f_{j \rightarrow j+1}\) the deformation field computed from the registration of frames j to \(j+1\). These deformation fields may be composed and inverted to register any frame into another one: \(f_{i \rightarrow j}=f_{i \rightarrow k}\circ \cdots \circ f_{j-1 \rightarrow j}\). Therefore, if the cardiac cycle is imaged into N frames, there exists N possible meshes for each frame: \(\lbrace M_i^j\rbrace _{i \in [1,N]}\) where \(M_i^j=f_{i \rightarrow j}(M_i)\) is the deformed mesh at time j coming from the segmentation of frame i.
2.3 Regional Volumes
In this paper we decided to focus on regional volume changes as an index for local contraction deficiency caused by the presence of scar tissue (as previously shown using a electromechanical model of the heart in [9]). Regional volumes are defined as the volume formed by the endocardial surface of the AHA zone and the barycentre of the LV (Fig. 2 left). This measure is segmentation-based and can be easily measured from independent meshes. Additionally, this measure is robust to small registration or segmentation errors since it averages the displacements of all points of the selected surface. We compute it via three different ways:
-
(i)
Segmentation: Regional volumes are computed for each mesh created directly from the segmentation and compiled as a time sequence for each AHA zone \(\alpha \):
$$V_{reg_\alpha }^{Seg}(j)=V_{reg_\alpha }(M_j)$$ -
(ii)
3D segmentation + registration: AHA zones are created on the end-diastolic mesh and this mesh is deformed under the registration deformation fields. Consistent regional volumes sequences are then extracted for each zone:
$$ V_{reg_\alpha }^{3DSeg+Reg}(j)=V_{reg_\alpha }(f_{0 \rightarrow j}(M_0)) $$ -
(iii)
Our coupling: segmentation + registration: Regional volumes are computed for every combination of 3D segmentation + registration and the mean value measured for each time point:
$$ V_{reg_\alpha }^{Coupling}(j)=\frac{1}{N}\sum _{i=0}^{N}V_{reg_\alpha }(f_{i \rightarrow j}(M_i)) $$
2.4 Statistical Analysis
We hypothesize that maximum regional volume change enables to detect zones containing scar tissue. Regional volume changes were computed as the relative difference between the regional volume at time t and the regional volume at time 0 (end-diastole). The maximum regional volume change was then measured as the minimum over time of the regional volume change (also called regional ejection fraction). Maximum regional volume change of scar and healthy zones were compared statistically using Student’s t-test and the level of statistical significance was set to a p-value < 0.05. Additionally, in order to evaluate the accuracy of the regional volumes in predicting the position of a scar zone, ROC analysis was performed and the Area Under Curve (AUC) computed. A perfect prediction tool corresponds to an AUC of 1 while a AUC of 0.5 corresponds to a coin toss.
(Top) Examples of regional volume change (between each frame and the first frame, leading to a change of 0% at time 0 for all methods) of 1 patient, for 3 AHA zones: (red) 3D Segmentation + registration, (green) segmentation, (blue) distribution over the set of segmentations where the mean is the selected value for our coupling approach. (Bottom) Corresponding short-axis images where yellow circles highlight the AHA zone to consider. Contours are colored with the same code as the above curves. (Color figure online)
3 Results
3.1 Comparison of Volume Changes Between Methods
Differences in the application of the deformation fields from the end-diastolic frame or any other frame were noticed for every case. Figure 3 (bottom) illustrates three examples on the same patient where the propagation from the first frame (red contours), the segmentation (green contours), and the coupling curve (blue contours), lead to different contraction levels. These differences impact the computation of the regional volumes (Fig. 3 top). The left column illustrates ideal cases where all three methods agree with a small difference. The middle column illustrates examples where the segmentation is unreliable and inconsistent in time leading to noisy regional volumes, probably due to inclusion/exclusion of papillary muscles as shown in the bottom row. For these cases, using a registration algorithm enables to smooth the results and improve the temporal consistency, for both the standard registration or the proposed coupling. For the right column, both the standard approaches are incorrect: the segmentation is noisy but within an acceptable level, however the registration using the first frame leads to overestimated contraction. The standard deviation of volume changes over all the segmentations propagated to a given frame is represented by a blue errorbar. Its average amplitude at end-systole around 20% illustrates the high influence of the initial mesh for the quantification of the deformation.
3.2 Ability to Locate Scar Zones
As shown in Fig. 4, all methods agree that maximum regional volume change is lower for scar zones (in red) than for healthy tissue (in blue) for every AHA zone. This difference is even significant (p < 0.05) for 7 or 6 of the 17 AHA zones depending on the method used to calculate the regional volumes. Note that lateral zones (5, 6, 11, 12) present only 0 to 3 scar regions making statistical significance unreachable. It is also interesting to note that the mean healthy regional volume is highly dependent on the AHA zone. A unique threshold for the full myocardium would therefore be inadequate.
More precisely, segmentation seems to be the least reliable of the three methods and fails to differentiate the scars on zones 1 and 16. Moreover, 3D segmentation + registration fails to separate healthy from scar tissue on zone 11. Our coupling approach, on the other hand always differentiates scar vs non-scar regional volume changes.
Additionally, AUC values are presented Table 1 and examples of ROC curves that led to the measurements of AUC values are shown Fig. 5. The mean accuracy of the coupling method (0.84 ± 0.10) is significantly higher than the standard segmentation method (0.78 ± 0.14) and higher than the standard 3D segmentation + registration approach (0.82 ± 0.10) although not significantly. Additionally, the coupling methods reaches the best detection in 8 zones. In contrast, segmentation is better than our coupling in 5 zones and 3D segmentation + registration in 3 zones. Finally, the proposed approach never shows the worst performance.
4 Discussion and Conclusion
In this paper, we highlighted the lack of consistency between the two standard approaches for deformation estimation and the need for a more robust, intermediate approach. We proposed a coupling method that combines both the output of the registration and the segmentation of all the cardiac frames. We illustrated this method by measuring the regional volumes and studied their ability to detect infarct tissue on 20 patients. Results showed that segmentation, standard registration and our approach can all be accurate in the scar detection. However, the detection was more systematic using the proposed coupling, which gathers the best assets of both methods (ground truth segmentation, spacial and temporal smoothness) without their drawbacks (noisy segmentation, exclusion/inclusion of papillary muscles, influence of the initial frame). Moreover, this method can easily be translated into clinical practice and applied routinely from already available segmentation and registration tools. A larger database would be required to validate these results and allow a more precise localization of the scars from regional volumes. Future work will investigate better fusion algorithms for a more robust coupling approach than the current simple averaging. We will also study the extension of this approach to tagged images for the evaluation of cardiac motion through radial, circumferential and longitudinal strains.
Notes
- 1.
Segment is a freely available software available at http://segment.heiberg.se.
- 2.
The Computational Geometry Algorithms Library is available at www.cgal.org.
References
Albà, X., Figueras i Ventura, R.M., Lekadir, K., Frangi, A.F.: Healthy and scar myocardial tissue classification in DE-MRI. In: Camara, O., Mansi, T., Pop, M., Rhode, K., Sermesant, M., Young, A. (eds.) STACOM 2012. LNCS, vol. 7746, pp. 62–70. Springer, Heidelberg (2013). doi:10.1007/978-3-642-36961-2_8
Auger, D., Ducharme, A., Harel, F., Marcotte, F., Thibault, B., O’Meara, E.: Patient assessment for cardiac resynchronization therapy: past, present and future of imaging techniques. Can. J. Cardiol. 26(1), 27–34 (2010)
De Craene, M., Piella, G., Camara, O., Duchateau, N., Silva, E., Doltra, A., et al.: Temporal diffeomorphic free-form deformation: application to motion and strain estimation from 3D echocardiography. Med. Image Anal. 16(2), 427–450 (2012)
Duchateau, N., De Craene, M., Allain, P., Saloux, E., Sermesant, M.: Infarct localization from myocardial deformation: prediction and uncertainty quantification by regression from a low-dimensional space. IEEE Trans. Med. Imaging 35(10), 2340–2352 (2016)
Heiberg, E., Engblom, H., Engvall, J., Hedström, E., Ugander, M., Arheden, H.: Semi-automatic quantification of myocardial infarction from delayed contrast enhanced magnetic resonance imaging. Scand. Cardiovasc. J. 39(5), 267–275 (2005)
Karim, R., et al.: Infarct segmentation challenge on delayed enhancement MRI of the left ventricle. In: Camara, O., Mansi, T., Pop, M., Rhode, K., Sermesant, M., Young, A. (eds.) STACOM 2012. LNCS, vol. 7746, pp. 97–104. Springer, Heidelberg (2013). doi:10.1007/978-3-642-36961-2_12
Atehortúa Labrador, A.M., Zuluaga, M.A., Ourselin, S., Giraldo, D., Castro, E.R.: Automatic segmentation of 4D cardiac MR images for extraction of ventricular chambers using a spatio-temporal approach. In: SPIE Medical Imaging. International Society for Optics and Photonics (2016)
Folgoc, L., Delingette, H., Criminisi, A., Ayache, N.: Sparse bayesian registration. In: Golland, P., Hata, N., Barillot, C., Hornegger, J., Howe, R. (eds.) MICCAI 2014. LNCS, vol. 8673, pp. 235–242. Springer, Heidelberg (2014). doi:10.1007/978-3-319-10404-1_30
Marchesseau, S., Delingette, H., Sermesant, M., Cabrera-Lozoya, R., Tobon-Gomez, C., Moireau, P., et al.: Personalization of a cardiac electromechanical model using reduced order unscented Kalman filtering from regional volumes. Med. Image Anal. 17(7), 816–829 (2013)
Medrano-Gracia, P., Suinesiaputra, A., Cowan, B., Bluemke, D., Frangi, A., Lee, D., Lima, J., Young, A.: An atlas for cardiac MRI regional wall motion and infarct scoring. In: Camara, O., Mansi, T., Pop, M., Rhode, K., Sermesant, M., Young, A. (eds.) STACOM 2012. LNCS, vol. 7746, pp. 188–197. Springer, Heidelberg (2013). doi:10.1007/978-3-642-36961-2_22
Petitjean, C., Dacher, J.N.: A review of segmentation methods in short axis cardiac MR images. Med. Image Anal. 15(2), 169–184 (2011)
Zhang, Z., Ashraf, M., Sahn, D.J., Song, X.: Temporally diffeomorphic cardiac motion estimation from three-dimensional echocardiography by minimization of intensity consistency error. Med. Phys. 41(5), 052902 (2014)
Zhuang, X., Rhode, K., Razavi, R., Hawkes, D., Ourselin, S.: A registration-based propagation framework for automatic whole heart segmentation of cardiac MRI. IEEE Trans. Med. Imaging 29(9), 1612–1625 (2010)
Acknowledgement
This work has been partially funded by the NMRC NUHS Centre Grant Medical Image Analysis Core (NMRC/CG/013/2013) and by the European Research Council (MedYMA ERC-AdG-2011-291080).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Marchesseau, S., Duchateau, N., Delingette, H. (2017). Segmentation and Registration Coupling from Short-Axis Cine MRI: Application to Infarct Diagnosis. In: Mansi, T., McLeod, K., Pop, M., Rhode, K., Sermesant, M., Young, A. (eds) Statistical Atlases and Computational Models of the Heart. Imaging and Modelling Challenges. STACOM 2016. Lecture Notes in Computer Science(), vol 10124. Springer, Cham. https://doi.org/10.1007/978-3-319-52718-5_6
Download citation
DOI: https://doi.org/10.1007/978-3-319-52718-5_6
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-52717-8
Online ISBN: 978-3-319-52718-5
eBook Packages: Computer ScienceComputer Science (R0)