Abstract
Segmentation of the heart ventricles from short axis Cine MRI is an active area of research. However, most of the solutions offered to radiologists are still semi-automatic. Several commercial software require from the users to input the centres of the ventricles for every image to segment which is fastidious and time-consuming. The automatic detection of these centres is challenging, especially, in the case of the right ventricle (RV). The variability in image quality, heart shape, thickness and motion, have led researchers to make assumptions not always valid regarding its position, blood pool intensity or shape. We aim in this work to offer a fast automatic, robust and accurate solution to this issue. By using the motion, and the pixel intensity, we are able to localize, recognize and select centres for both ventricles. First, our approach focuses on performing a coarse segmentation of each ventricle at the basal slice at the end-diastolic frame. The coarse segmentation of the left ventricle (LV) is then propagated to the following frames and below slices to reduce the region of interest. The greater reliability of the LV centre detection allows its use to define an area of search for the RV. We tested our method on 32 patients from the MICCAI 2012 RVSC Test1 and Test2 datasets and 10 volunteers, totalling 7485 images. We achieved a 99.3% success detection rate in the case of the LV, and 89.8% for the RV. We also show how the LV centre detection can be applied to define the LV central axis, and used to detect and correct misaligned slices.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
The segmentation of the ventricles of the heart remains an active area of medical image analysis. It is of key interest to radiologists who use the results to evaluate the cardiac function. Many fully automatic solutions have been presented in the literature for the left ventricle (LV) [5], and the right ventricle (RV) [6]. However, most of the tools available to clinicians are semi-automatic and require manual input. This manual input is usually required for every image and, therefore, represents a tedious task prone to observer variability.
In this paper, we focus on the automatic detection of the centres of the ventricles. Localizing the centres of the ventricles will remove the need for manual input in commercial software such as Segment MedvisoFootnote 1, CMR tools from Cardiovascular Imaging SolutionsFootnote 2 or Circle Cardiovascular ImagingFootnote 3. Additionally, the centres are necessary to define the LV central axis. From the LV central axis, 3D reconstruction can be performed after realignment of the short axis slices. Moreover, LV central axis and RV centres allow the definition of the 17 AHA zones for regional measurements.
The automatic detection of these centres is challenging, especially, in the case of the RV. Despite the variability in image quality, heart shape, thickness as illustrated in Fig. 1, current methods make assumptions such as [7] on the respective position, or [2] on the intensity profile, which remains difficult to predict because of the blood flow, the fat, and the trabeculations. We tackle this centre detection without these assumptions by a coarse-to-fine approach to detect the centre of each ventricle in every single image of a 4D sequence.
The proposed method was tested on 32 patients from the MICCAI 2012 RVSC Test1 and Test2 datasets and 10 volunteers. We also present an application of the LV centre detection algorithm to realign misaligned short-axis slices.
2 Methods
The proposed method uses motion and pixel intensity to detect, recognize, and select centres for both ventricles. First, the heart is located and cropped by estimating the motion throughout the frames at the basal slice. Then, a coarse segmentation of each ventricles is performed at the basal slice at the end-diastolic (ED) frame. The segmented LV is then propagated through the slices and frames to reduce the area of search. The LV centres are used to define a region of interest for the RV. The pipeline is illustrated in Fig. 2.
Pipeline for 4D automatic centre detection. First, a cropping is performed on the 4D image. Then at the basal slice, both ventricles are detected, recognized and centres are selected. The result of this detection is then used to search in the 4D volume the centres of the LV. Finally, the LV centres are used to define elliptic areas of search around the RV centres.
2.1 Cropping
The first step of the proposed coarse-to-fine approach consists in cropping the image to delimit the heart. This will help in the detection by preventing possible false positives, and also reduce the overall computational time. The cropping method is first used on the basal slice. The basal slice is selected manually, as the first slice where the ventricles are the biggest and form a full ring (for instance Fig. 3a). However, automatic selection will be considered in the future using recently published method [4].
This cropping method is quite similar to the heart localization approach presented in [2], as we look for the region presenting most of the motion in the image. To observe the motion, we compute the sum of the absolute pixel-wise difference between 2 images of the cardiac cycle:
where \(I_t\) is the 2D image representing the basal slice at time t.
The effect of the background noise is reduced by summing the absolute difference between the first frame and the following frames. Moreover, the small motions are attenuated by smoothing \(I_\varDelta \) with a Gaussian kernel (choosing a large \(\sigma \) to reduce the amount of noise). Otsu’s thresholding algorithm is then applied to approximate the shape of the heart (Fig. 3), which may contain surrounding objects. The bounding box is defined as the smallest window W containing every pixel of the mask. In order to make up for possible errors W’s size is increased by 5 pixels in every directions. The bounding box defined at the basal slice is then used for all the slices.
2.2 Centre Detection at Basal Slice
The ventricles are the most filled with blood in the ED frame, where they appear as big structures with high intensity. At that specific time, the ventricles can easily be discriminated from the background by using a thresholding method. In the literature, Otsu’s model has been used for the same purpose [7].
The result obtained is an approximation of the blood pool of the ventricles, on which is applied a morphological reconstruction [3] to fill the holes present in all connected components and remove small papillary muscles.
The previously computed \(I_\varDelta \) is then used to reduce the number of potential ventricles. Only the 3 largest connected components sharing pixels with \(I_\varDelta \) are kept as illustrated in Fig. 4. We identify the LV as being the object with the highest circularity measure C [1] defined as \( C = \dfrac{4\pi A}{P^2} \) where P is the perimeter, and A the area. The centre of the LV is chosen as the barycentre of the object and denoted \(LV_c\).
The RV is then estimated between the two other objects \(\varOmega _1\), \(\varOmega _2\), by optimising the following function
(a) Original image. (b) The LV extracted at the previous slice (in green) is intersected with the result of Otsu on the original image. (c) The result of the intersection. In yellow, the centre of the previous slice, and in green the LV segmentation chosen for this slice. (d) The result of the detection in green. (Color figure online)
The largest and closest object to the LV with the highest circularity measure is selected. The centre of the RV is chosen as the maximum of the distance map (Fig. 4d and e) to account for the potential presence of fat and the non-circular shape of the RV, that will misplace the barycentre.
2.3 Left Ventricle Centre Detection on the 4D Sequence
To detect the centre of the other slices and frames, the area of search is reduced to allow faster computation, to prevent false positive due to fat tissue around the heart and to take into account the muscle narrowing toward the apex. Both the coarse segmentation of the LV and the detected centre (Fig. 4c) from the basal slice at ED are used to detect the LV centre of first the other slices and then the other frames of the cardiac cycle. First, for each new slice, Otsu’s binarization is performed and intersected with the coarse mask from the previous slice, as described Fig. 5b. This intersection may contain several disconnected objects (Fig. 5c) from which the LV is detected as the object containing, or closest to, the LV centre from the previous slice. The centre of this LV is then defined as the barycentre of the selected coarse object (Fig. 5d).
To detect the centre of the LV for the other time frames, a similar process is applied using the coarse LV masks and the LV centres detected at ED. However, instead of performing Otsu on the whole image, it is only applied on the intersection with the coarse mask in order to give an apriori localization of the blood pool at end-systole (ES). Using the same ED inputs for all time frames allows parallel computing.
2.4 Right Ventricle Centre Detection on the 4D Sequence
Even more importantly for the RV, the area of search needs to be reduced to prevent the risk of detecting fat tissue which presents brighter intensity and sharper contrast than the very thin RV muscle. Similarly to the LV, the centre detection is first performed on all slices at ED, then on the other time frames of the cardiac cycle. Due to the reliability of the LV detection, the LV centre can be used as a landmark to define the area of search in which the RV blood pool is assumed to be found. First, an ellipse is defined around the RV centre obtained from the previous slice at ED. Based on geometrical observations, the main axis size is set to a fourth of the distance between the current LV centre and the previous RV centre, and the minor axis size is set to a sixth of this distance (Fig. 6a). The ellipse is assumed to follow the narrowing of the RV towards the apex, and its motion during contraction. Otsu’s algorithm is once again used to differentiate the blood pool from the background within the ellipse (Fig. 6b). Similarly to LV detection, the RV is chosen as the closest object to the centre of the ellipse and its barycentre defined as the centre of the RV for the current slice. The ellipses defined at ED are used to detect the RV centre in the other frames of the cardiac cycle, which allows parallel computing.
3 Results
We evaluated our algorithm on 32 patient short-axis volumes from the Test1 and Test2 datasets of the MICCAI 2012 RVS challenge [6] acquired on a Siemens Symphony Tim 1.5T MRI, and 10 volunteer scans acquired on a Siemens 3T Prisma. In total, the algorithm has been tested on 7485 images. The cropping is done on average in 65 ms, the detection on the basal slice (all frames) in 250 ms, and the detection on a slice (all frames) in 110 ms. The overall time for a 4D detection of 1 subject is less than 2 s. The tests were done on a Intel Xeon CPU ES-1650 3.20 GHz (12 cores). The detection is considered as successful if the centre is detected in the middle for the LV, and in the blood pool for the RV. Examples of successful detection are shown in the first row of the Fig. 7, for 2 apical and 2 basal slices.
The algorithm performs extremely well for the LV with a 99.3% success rate for all the patients, and a mean of 99.4% per patient. The failed detection happens only when the blood pool is not visible anymore due to the contraction. As for the RV, we achieved a satisfying 89.8% success rate over the 7485 images. There are several reasons to be considered for the 10% failures, Fig. 7 second row illustrates some of them.
In some cases, the presence of fat or fluid around the wall may be detected as part of the blood pool, especially, close to the apex at ES (Fig. 7e). Similarly to the LV, it might fail when the ventricle is completely closed by the contraction which usually happens around the apex at ES as shown in Fig. 7f. Overall, most of the errors are close to the apical slice, the algorithm fails to follow the sudden narrowing of the heart ventricles (Fig. 7g and h). Future improvements will consider a linear interpolation of the distance between the RV and LV centres to redefine the area of search towards the apex, and hopefully decrease the error rate.
3.1 LV Central Axis and Alignment
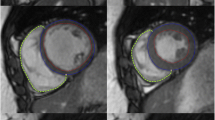
Misaligned slices are a frequent issue in short axis Cine MRI due to different breath-hold positions. In order to detect and correct the misaligned slices, we propose to use the LV central axis. The LV central axis is defined as the line joining the LV centre of the basal slice and the apex. To estimate the misalignment, the distance between the LV centre of each intermediates slices and the axis is computed. Then, Tukey’s boxplot method [8] is used to detect the outliers among the set of distances. The outliers are considered as misaligned slices, and the translations needed to align them is calculated and applied. In our example, two iterations of the LV centre detection and alignment algorithm was needed to perfectly align the slices as shown in Fig. 8. This alignment method can only be efficient for a small number of misaligned slices. Aligning short-axis slices allows to perform 3D reconstruction of the LV geometry for further analysis such as motion tracking.
First row: results of LV detector on misaligned slices, the middle image is clearly misaligned. Second row: results of the alignment algorithm, the middle image is perfectly aligned with the other slices. Last column illustrates on top a LV misaligned, and in the bottom a LV after alignment, the blue line represents the central axis. (Color figure online)
4 Discussion and Conclusion
The segmentation of ventricles in cine MRI is a challenging task due to the large variability in shape, motion, and image quality. Especially, in the case of the RV where, to the best of our knowledge, no robust automatic method has been made available to clinicians. We aimed through this work at presenting a robust technique for ventricle centre detection in 4D. Our goal is to simplify the traditional usage of segmentation tools by replacing the manual input required from clinicians. Our solution was tested on a database of 7485 images, presenting a large variability, in heart sizes, image quality, presence of artefact, and motion.
The LV centre detector showed close to 100% success rate, which makes it extremely reliable as an add-on to any semi-automatic tool directed to clinicians. Moreover, its precision allows us to use it to define the LV central axis. This central axis can be useful to detect and correct misalignment but also for 3D LV models and AHA regional segments definition. The results of RV detector were also satisfying as they reached the 90% threshold and represent, therefore, a usable solution to initialize a centre-based automatic segmentation method.
References
Jiang, L., Ling, S., Li, Q.: Fully automated segmentation of left ventricle using dual dynamic programming in cardiac cine MR images. In: SPIE Medical Imaging. International Society for Optics and Photonics (2016)
Atehortúa Labrador, A.M., Zuluaga, M.A., Ourselin, S., Giraldo, D., Romero Castro, E.: Automatic segmentation of 4D cardiac MR images for extraction of ventricular chambers using a spatio-temporal approach. In: SPIE Medical Imaging. International Society for Optics and Photonics (2016)
Lehmann, G.: Label object representation and manipulation with ITK. Insight J. 8 (2007)
Paknezhad, M., Marchesseau, S., Brown, M.S.: Automatic basal slice detection for cardiac analysis. In: SPIE Medical Imaging. International Society for Optics and Photonics (2016)
Petitjean, C., Dacher, J.-N.: A review of segmentation methods in short axis cardiac MR images. Med. Image Anal. 15(2), 169–184 (2011)
Petitjean, C., Zuluaga, M.A., Bai, W., Dacher, J.-N., Grosgeorge, D., Jérôme Caudron, S., Ruan, I.B., Ayed, M.J., Cardoso, H.-C.C., et al.: Right ventricle segmentation from cardiac MRI: a collation study. Med. Image Anal. 19(1), 187–202 (2015)
Ringenberg, J., Deo, M., Devabhaktuni, V., Berenfeld, O., Boyers, P., Gold, J.: Fast, accurate, and fully automatic segmentation of the right ventricle in short-axis cardiac MRI. Comput. Med. Imaging Graph. 38(3), 190–201 (2014)
Tukey, J.W.: Box-and-whisker plots. In: Exploratory Data Analysis, pp. 39–43 (1977)
Acknowledgement
This work has been partially funded by the NMRC NUHS Centre Grant Medical Image Analysis Core (NMRC/CG/013/2013).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Fadil, H., Totman, J.J., Marchesseau, S. (2017). 4D Automatic Centre Detection of the Right and Left Ventricles from Cine Short-Axis MRI. In: Mansi, T., McLeod, K., Pop, M., Rhode, K., Sermesant, M., Young, A. (eds) Statistical Atlases and Computational Models of the Heart. Imaging and Modelling Challenges. STACOM 2016. Lecture Notes in Computer Science(), vol 10124. Springer, Cham. https://doi.org/10.1007/978-3-319-52718-5_16
Download citation
DOI: https://doi.org/10.1007/978-3-319-52718-5_16
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-52717-8
Online ISBN: 978-3-319-52718-5
eBook Packages: Computer ScienceComputer Science (R0)