Abstract
Missing values are a common issue in many real-world datasets, and therefore coping with such datasets is an essential requirement of classification since inadequate treatment of missing values often leads to large classification errors. One of the most popular ways to address incomplete data is to use imputation methods to fill missing fields with plausible values. Multiple imputation, which fills each missing field with a set of plausible values, is a powerful approach to dealing with incomplete data, but is mainly used for statistical analysis. Ensemble learning which constructs a set of classifiers instead of one classifier has proven capable of improving classification accuracy, but has been mainly applied to complete data. This paper proposes a combination of multiple imputation and ensemble learning to build an ensemble of classifiers for incomplete data classification tasks. A multiple imputation method is used to generate a set of diverse imputed datasets which is then used to build a set of diverse classifiers. Experiments on ten benchmark datasets use a decision tree as classification algorithm and compare the proposed approach with two other popular approaches to dealing with incomplete data. The results show that, in almost all cases, the proposed method achieves significantly better classification accuracy than the other methods.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Classification is one of the main tasks in data mining and machine learning. Classification has been successfully applied to many scientific areas such as computer science, engineering, statistic, medicine, biology, etc [4]. In spite of receiving great attention over many decades, there are still open issues in classification; one of these issues is incomplete data [10].
An incomplete dataset is a dataset containing some fields which are missing values. Missing values are a unavoidable problem in many real-world datasets [15, 18]. For instance, 45 % of the datasets in the UCI repository [1], which is one of the most popular data repositories for machine leaning, have the issue of missing values [10]. The reasons for missing values are various. For example, in a social survey, respondents often ignore to answer some questions; some results collected from industrial experiments may be missing values due to mechanical failures while collecting data; medical datasets are often incomplete because not all tests can be run on every patient [9].
Missing values lead to severe issues for classification. One of the most severe issues is non-applicability of many classification algorithms. Although some classification algorithms are able to deal with incomplete data, many others require complete data. Therefore, these classification algorithms cannot directly work with incomplete data. Even for algorithms that can cope with incomplete data, missing values often result in large classification errors [10, 21].
One approach to handling classification with incomplete data is to use imputation methods to replace missing fields with plausible values before using classification algorithms. For example, mean imputation replaces each missing field with the average of the complete values of the same feature. Imputation methods provide complete data that can be then used by any classifier. Consequently, imputation methods are one of the most popular approaches to addressing classification with incomplete data [10].
Multiple imputation is an approach to tackling incomplete data by creating multiple imputed datasets to reflect better the uncertainty in incomplete data. In statistical fields, multiple imputation has become increasingly popular because of its convenience and flexibility [15, 18, 20]. Multiple imputation also has been a powerful technique for addressing classification with incomplete data [9, 19, 23]. However, when multiple imputation is used for classification with incomplete data, multiple imputed datasets are simply averaged to generate a single imputed dataset which is then used by classification algorithms [9, 23]. The disadvantage of this approach is that it ignores the ability of multiple imputation to reflect the uncertainty of incomplete data. How to exploit this ability of multiple imputation in classification with incomplete data is still an open issue.
Ensemble learning algorithms can build a set of classifiers for classification task instead of a single classifier. After that, a new instance is classified by taking a vote of their predictions. Both theoretical development and empirical research have showed that an ensemble can help to improve classification accuracy [8, 16]. However, ensemble methods are mainly applied to complete data. Therefore, how to use ensemble methods for improving classification with incomplete data should be further investigated.
1.1 Research Goals
The goal of this paper is to propose a combination of multiple imputation with ensemble learning for improving classification with incomplete data. The proposed method is compared with two other popular approaches to dealing with missing values. One approach is to use single imputation to generate a single imputed dataset. Another approach is to use multiple imputation to generate a single imputed dataset by averaging multiple imputed datasets. Results from experiments are used to address the following objectives:
-
1.
Whether the combination of multiple imputation with ensemble learning can achieve better classification than using single imputation; and
-
2.
Whether the combination of multiple imputation with ensemble learning can achieve better classification than using multiple imputation to generate a single imputed dataset by averaging multiple imputed datasets.
1.2 Organisation
The rest of the paper is organised as follows. Section 2 discusses related work. Section 3 outlines the proposed method. Section 4 presents experiment design. Section 5 shows results and analysis. Section 6 draws conclusions and presents future work.
2 Related Work
This section discusses related work including classification with missing data, imputation methods and ensemble learning.
2.1 Classification with Missing Data
There are four major approaches to addressing classification with incomplete data including the removal approach, the imputation approach, the model-based approach and the machine learning approach [10].
The removal approach eliminates all instances containing missing values before using classifiers. The main benefit of this approach is to provide complete data that can be then classified by any classifiers. Nevertheless, incomplete instances are not classified by the classifier. Therefore, this approach is only able to be applied to the training process and when a dataset includes a small number of incomplete instances [9].
The imputation approach uses imputation methods to replace missing values with suitable values before using classifiers. For instance, mean imputation fills all missing fields in a feature with the average of complete values in the feature. The main benefit of this approach is to provide complete data which can be used by any classification algorithm. By using imputation methods, both complete and incomplete instances are attended in the classification process. Furthermore, most imputation methods can enhance classification accuracy compared to the corresponding methods without using imputation. Therefore, the imputation approach is a main way to address classification with incomplete datasets [9].
The model-based approach generates a data distribution model from input data. Thereafter, a combination of the data distribution model and Bayesian decision theory [3] is used to classify both complete and incomplete instances. Although this approach can classify both complete and incomplete instances, it requires to make assumptions about the joint distribution of all features in the model [10].
The machine learning approach makes classifiers that are able to directly classify incomplete datasets without using nay imputation methods. For instance, C4.5 [17] can tack with missing values in both training data and test data by using a probabilistic approach.
2.2 Imputation Methods
The goal of imputation methods is to fill missing fields with plausible values [15]. Imputation methods can be categorized into single imputation and multiple imputation [9]. While single imputation methods search one value for each missing value, multiple imputation methods search multiple values for each missing value.
2.2.1 Single Imputation
Each missing field is filled by one value in single imputation methods. This paper uses three single imputation methods: mean imputation, hot deck imputation and K nearest neighbours-based imputation.
Mean imputation replaces all missing fields in each feature with the average of the complete values in the feature. The advantage of this method is that it maintains the mean of each feature, but it under-represents the variability in the data since all missing fields in each feature have the same value [10].
In hot deck imputation, for each incomplete instance, the most similar instance with the incomplete instance is found, and missing fields are replaced with complete values from the most similar instance. The main merit of hot deck imputation is that it fills missing fields by real values from the data. Nonetheless, this method only utilises the information of one instance; thus, it ignores all global properties of the data [15].
KNN-based imputation is based on K-nearest neighbors algorithm for classification. For each incomplete instance, firstly, it finds the K most similar instances with the incomplete instance, and then fills missing fields of the incomplete instance with the average of values in the K most similar instances. KNN-based imputation often performs better than mean imputation and hot deck imputation [2]. However, this method is often computationally intensive owing to having to search through all instances to find the K most similar instances for each incomplete instance [10].
2.2.2 Multiple Imputation
Multiple imputation has three main steps. Firstly, incomplete data is put N times (N > 1) into an imputation model incorporating random variation to build N different imputed datasets. After that, each imputed dataset is separately analysed by standard procedures for complete data. The second step provides N analysis results. Finally, the N analysis results are combined to provide a final result [15, 18].
Multiple imputation has become more and more popular because of several reasons. Firstly, multiple imputation often reflects better uncertainty related to a particular model used for imputation, though it is computationally more expensive than single imputation [9]. Moreover, many recent software developments have based on the multiple imputation framework [12].
One of the most convenient and powerful multiple imputation methods is multivariate imputation by chained equations (MICE) [22]. The first step to generate multiple imputed datasets in MICE is multiple imputation by chained equations. MICE utilises a set of regression methods such as classification and regression trees (CART) [5] and Random forest [14]. Initially, each missing field is replaced by a complete value randomly chosen from the same feature. Afterwards, each incomplete feature is regressed on all other features to compute a better estimate for the feature. The process is repeated several times for all incomplete features to generate a single imputed dataset. The whole procedure is repeated N times to generate N imputed datasets which are then used to calculate the final imputed dataset [22]. MICE software [6] makes it easy to use this method.
2.3 Ensemble Learning
Ensemble learning is the process that builds a set of classifiers for classification. Thereafter, a new instance is classified by voting the decision of the individual classifiers. Ensemble learning has been proved capable of achieving better classification accuracy than any single classifier [8, 16].
An ensemble of classifiers is good if the individual classifiers in the ensemble is accurate and diverse. Bagging and Boosting are two popular approaches to building accurate ensembles [16]. Both Bagging and Boosting use “resampling” techniques to manipulate the training data. Bagging manipulates the original training dataset of N instances by randomly drawing with replacement instances. Therefore, in the resulting training dataset, some of the original instances may appear multiple times while others might disappear. Bagging is often effective on “unstable” learning algorithms such as neural networks and decision trees where small changes in the training dataset lead to major changes in predictions. Experimental results show that Bagging ensemble almost always performs better than a single classifier. Boosting manipulates the original dataset for each individual classifier by using the performance of the previous classifier(s). In Boosting, instances which are incorrectly classified by previous classifiers are selected more often than instances which are correctly classified. Therefore, Boosting tries to build new classifiers that are better to classify instances for which the current ensemble’s performance is poor. Empirical results show that with little or no classification noise, Boosting ensemble also almost always performs better than a single classifier, and it is sometimes more accurate than Bagging ensemble. However, in situations with substantial classification noise, Boosting ensemble is often less accurate than a single classifier because Boosting often overfits noisy datasets [16].
An ensemble of classifiers trained with random subsets of features is presented in [13] to classify with incomplete data. In this approach, each base classifier is trained with a randomly selected subset of features. In [7], a combination of data analysis and ensemble learning is proposed to deal with classification with incomplete data. Firstly, the incomplete data is analysed and grouped into complete data subsets, and then each data subset is used to train one classifier. In the both approaches, when an incomplete instance needs be classified, only those classifiers trained with those features that are available in the instance are used to classify the instance. Although, the two methods are able to cope with incomplete data in some degree, they cannot guarantee to classify all incomplete instances, especially when data contains many missing values. Moreover, combining ensemble learning and multiple imputation has not been investigated. Therefore, using ensemble learning for classification with incomplete data should be more investigated.
3 Multiple Imputation and Ensemble Learning for Classification with Missing Data
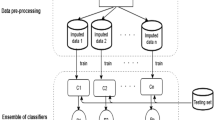
The proposed algorithm has two phases: the training process and the application process. The training process uses a multiple imputation method combined with ensemble learning to build a set of classifiers. After that, the application process uses the multiple imputation method and the set of classifiers to classify a new incomplete instance (Fig. 1).
In the training process, a training incomplete dataset is put into a multiple imputation method to build a set of imputed datasets. Afterwards, each imputed dataset is used as a training data by a classification algorithm to train a classifier. As a result, a set of classifiers are generated from the set of imputed datasets.
In the application process, if an instance which needs to be classified is incomplete, the incomplete instance is put into the multiple imputation method (along with the training data) to generate a set of imputed instances. After that, each classifier is applied to each imputed instance to generate a large set of predicted classes. The final predicted class will be the most frequent class of all the predictions. If an instance which needs to be classified is complete, the complete instance does not need the imputation method. Rather, they are classified directly by each of the classifiers and the most frequent class is chosen.
A key requirement of ensemble methods is that the set of classifiers should be diverse. The key idea of the proposed algorithm is that it exploits the ability of the multiple imputation method to build a set of diverse imputed datasets from which diverse classifiers can be constructed. This is in contrast to the usual use of multiple imputation for classification which averages the imputed datasets into a single dataset. From one incomplete dataset, multiple imputation is able to generate a set of diverse imputed datasets because the initial step of the multiple imputation is to fill each missing field with a randomly chosen complete value. Therefore, the initial step generates different temporary imputed datasets. Although the same regression method is then used to improve the temporary imputed datasets, the multiple imputation method is able to generate a set of diverse imputed datasets, especially when the training dataset contains many missing fields. As a result, a classifier ensemble which is then built by using the set of imputed datasets is hopefully diverse.
4 Experiment Design
This section shows detailed experiment design including the method, datasets, imputation methods and classification algorithms.
4.1 Comparison Method
This study is designed to empirically evaluate the proposed method for classification with incomplete datasets. In order to achieve this objective, the proposed method is compared to two popular benchmark methods for tacking with classification with incomplete datasets. The first popular benchmark method for classification with incomplete datasets is to use multiple imputation to generate multiple imputed datasets. After that the multiple imputed datasets are averaged to generate a single imputed dataset which is used to build a classifier. The second popular benchmark method for classification with incomplete datasets is to use a single imputation method to generate a single imputed dataset which is then used to build a classifier.
In the first benchmark method for classification with incomplete data, a training incomplete dataset is put into a multiple imputation method to generate a set of imputed datasets. After that, the set of imputed datasets is averaged to generate a single imputed dataset which is then used to learn a classifier. In the application process, each incomplete instance is combined with the training dataset, and then is put into the multiple imputation method to generate a set of imputed instances. Subsequently, the set of imputed instances is averaged to generate a single imputed instance which is then classified by the classifier.
In the second benchmark method for classification with incomplete data, a training incomplete dataset is put into a single imputation method to generate a single imputed dataset. Thereafter, the imputed dataset is used to learn a classifier. In the application process, each incomplete instance is combined with the training dataset, and then is put into the single imputation method to generate a single imputed instance. Afterwards, the single imputed instance is classified by the classier.
4.2 Datasets
Ten datasets, summarised in Table 1, are used in the experiments. These are taken from the UCI Repository of Machine Learning Databases [1]. Each dataset is presented in one row in Table 1 including the number of instances, the number of features, the number of classes, the proportion of instances containing at least one missing field and the proportion of missing values.
The first five datasets suffer from missing values in a “natural” way. In the datasets, we do not know any information related to the randomness of missing values, so we make assumption that missing values in the datasets are distributed in a missing at random (MAR) way [15].
In order to test the performance of the proposed feature selection method with datasets containing different levels of missing values, the missing completely at random (MCAR) mechanism [15] was utilised to introduce missing values into the last five complete datasets. Three different levels of missing values: 10 %, 30 % and 50 % were used to introduce missing values into the datasets. With each dataset in the last five datasets and each level of missing values in the three levels, repeat 30 times: introduce randomly the level of missing values in all features. Hence, from one dataset and one level of missing values, 30 artificial datasets containing missing values were generated. Therefore, from one complete dataset, 90 (\({=}{30\,\times \,3}\)) artificial datasets containing missing values were generated and a total of 450 (\({=}{90\,\times \,5}\)) artificial datasets containing missing values were used in the experiments.
None of the datasets in the experiments comes with a specific test set. Moreover, in some datasets, the number of instances is relatively small. Therefore, the ten-fold cross-validation method was used to measure the performance of the learned classifiers. With the first five incomplete datasets, the ten-fold cross-validation method was performed 30 times. With the last five complete datasets, with each dataset and each level of missing values, the ten-fold cross-validation method was performed on the 30 incomplete datasets. Consequently, for each incomplete dataset in the first five datasets and each level of missing values on one dataset in the last five datasets, 300 pairs of training and testing sets were generated.
4.3 Imputation Algorithms
The experiments used multiple imputation MICE [6] with the random forest as a regression method. In the multiple imputation method, each incomplete feature was repeatedly regressed on other features 10 times. With each incomplete dataset, the multiple imputation method was performed 20 times to procedure 20 imputed datasets.
Three single imputation methods including mean imputation, hot deck imputation and KNN-based imputation were used in the experiment. The three single imputations were in-house implementations. With KNN-based imputation, the number of neighbors K were set five.
4.4 Classification Algorithms
The experiment used C4.5 [17] to classify data. For the classifiers, WEKA’s implementation [11] was used and all parameters were set to WEKA’s defaults. The number of classifiers in an ensemble is equal to the number of imputed datasets generated by multiple imputation; therefore, the number of classifier in an ensemble is set 20.
5 Results and Analysis
This section presents the comparison between the proposed method with other methods on classification accuracy, and further analysis.
5.1 Results
Table 2 shows the average of classification accuracy and standard deviation using C4.5. In the tables, and in the following ones, MIEL column presents results by using the proposed method, AvgMI column presents results by using the first benchmark method; Mean, HDI and KNNI columns present results from the second benchmark method by using mean imputation, hot deck-based imputation and KNN-based imputation, respectively. With each dataset in the first five datasets, the classification accuracy is the average of accuracies of the 30 times performing ten-fold cross-validation (30 \(\times \) 10 \(=\) 300 experiments).
Table 3 shows the average of classification accuracy and standard deviation using C4.5 with three levels of missing values. With each dataset and each missing level in the last five datasets, the classification accuracy is the average of accuracies of the 30 generated incomplete datasets at each missing level and ten-fold cross-validation (30 \(\times \) 10 \(=\) 300 experiments).
To compare the performance of MIEL with the other methods, the Wilcoxon signed-ranks tests at 95 % confidence interval is used to compare the classification accuracy achieved by MIEL with the other methods. “T” columns in Tables 2 and 3 show significant test of the columns before them against MIEL, where “\(+\)”, “\(=\)” and “−” mean MIEL is significantly more accurate, not significantly different and significantly less accurate, respectively.
Table 2 shows that MIEL can achieve significantly better classification accuracy than the other methods in almost all cases with the datasets containing natural missing values. MIEL achieves similar classification accuracy to the other methods on Housevotes dataset and significantly better classification accuracy than the other methods on the other four datasets.
Table 3 shows that MIEL also can achieve significantly better classification accuracy than the other methods in almost all cases with the datasets containing artificial missing values. MIEL achieves significantly better classification accuracy than the other methods on all fifteen cases.
It is clear from the results that AvgMI is generally better than single imputation methods showing that multiple imputation generates a more reliable imputed dataset. Furthermore, a combination of multiple imputation and ensemble learning is significantly better than using multiple imputation to generate a single imputed data by averaging imputed datasets.
In summary, the proposed method combining multiple imputation combined with ensemble learning is able to enhance classification accuracy of a classifier not only with natural incomplete datasets, but also with artificial incomplete datasets.
6 Conclusions and Future Work
This paper proposed a new combination of multiple imputation and ensemble learning for classification with incomplete data. Firstly, multiple imputation is used to generate a set of imputed datasets from one incomplete dataset. After that, the set of imputed datasets is used to build an ensemble classifier. The proposed approach was compared with two other popular approaches to dealing with incomplete data: one using multiple imputation to generate one single imputed dataset and the other using single imputation to generate a single imputed dataset. The experiments on ten datasets used C4.5 as classification algorithms. The experimental results showed that the proposed method can achieve better classification accuracy than the two other methods. The experimental results also showed that it is advantageous to exploit the natural diversity generated by multiple imputation, rather than averaging the diverse imputed datasets. Even if the averaged imputed datasets is reliable, using the diversity of imputed datasets in an ensemble method leads to a more effective classifier.
The experiments in the paper used random forest as a regression method in MICE. There are some other regression methods in MICE such as linear regression and CART [5]. Further work could perform this investigation with linear regression and CART. Furthermore, the proposed method uses the majority vote. Therefore, another future work could develop a more powerful vote method to improve the proposed method.
References
Asuncion, A., Newman, D.: UCI machine learning repository (2007)
Batista, G.E., Monard, M.C.: A study of k-nearest neighbour as an imputation method. In: Hybrid Intelligent Systems - HIS. pp. 251–260 (2002)
Berger, J.O.: Statistical decision theory and Bayesian analysis. Springer Science & Business Media (2013)
Bishop, C.M.: Pattern Recognition and Machine Learning. Springer-Verlag New York, Inc. (2006)
Breiman, L., Friedman, J., Stone, C.J., Olshen, R.A.: Classification and regression trees. CRC Press (1984)
Buuren, S., Groothuis-Oudshoorn, K.: MICE: Multivariate imputation by chained equations in R. Journal of statistical software 45, 1–67 (2011)
Chen, H., Du, Y., Jiang, K.: Classification of incomplete data using classifier ensembles. In: Systems and Informatics (ICSAI), 2012 International Conference on. pp. 2229–2232 (2012)
Dietterich, T.G.: Ensemble methods in machine learning. In: International workshop on multiple classifier systems. pp. 1–15 (2000)
Farhangfar, A., Kurgan, L.A., Pedrycz, W.: A novel framework for imputation of missing values in databases. Systems, Man and Cybernetics, Part A: Systems and Humans, IEEE Transactions on 37, 692–709 (2007)
García-Laencina, P.J., Sancho-Gómez, J.L., Figueiras-Vidal, A.R.: Pattern classification with missing data: a review. Neural Computing and Applications 19, 263–282 (2010)
Hall, M., Frank, E., Holmes, G., Pfahringer, B., Reutemann, P., Witten, I.H.: The WEKA data mining software: an update. ACM SIGKDD explorations newsletter 11, 10–18 (2009)
Harel, O., Zhou, X.H.: Multiple imputation: review of theory, implementation and software. Statistics in medicine 26, 3057–3077 (2007)
Krause, S., Polikar, R.: An ensemble of classifiers approach for the missing feature problem. In: Neural Networks, 2003. Proceedings of the International Joint Conference on. vol. 1, pp. 553–558 (2003)
Liaw, A., Wiener, M.: Classification and regression by randomforest. R news 2, 18–22 (2002)
Little, R.J., Rubin, D.B.: Statistical analysis with missing data. John Wiley & Sons (2014)
Opitz, D., Maclin, R.: Popular ensemble methods: An empirical study. Journal of Artificial Intelligence Research 11, 169–198 (1999)
Quinlan, J.R.: C4. 5: programs for machine learning. Elsevier (2014)
Schafer, J.L., Graham, J.W.: Missing data: our view of the state of the art. Psychological methods 7, 147 (2002)
Tran, C.T., Andreae, P., Zhang, M.: Impact of imputation of missing values on genetic programming based multiple feature construction for classification. In: 2015 IEEE Congress on Evolutionary Computation (CEC). pp. 2398–2405 (2015)
Tran, C.T., Zhang, M., Andreae, P.: Multiple imputation for missing data using genetic programming. In: Proceedings of the 2015 Annual Conference on Genetic and Evolutionary Computation. pp. 583–590 (2015)
Tran, C.T., Zhang, M., Andreae, P.: A genetic programming-based imputation method for classification with missing data. In: European Conference on Genetic Programming. pp. 149–163 (2016)
White, I.R., Royston, P., Wood, A.M.: Multiple imputation using chained equations: issues and guidance for practice. Statistics in medicine 30, 377–399 (2011)
Williams, D., Liao, X., Xue, Y., Carin, L., Krishnapuram, B.: On classification with incomplete data. IEEE Transactions on Pattern Analysis and Machine Intelligence 29(3), 427–436 (2007)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Tran, C.T., Zhang, M., Andreae, P., Xue, B., Bui, L.T. (2017). Multiple Imputation and Ensemble Learning for Classification with Incomplete Data. In: Leu, G., Singh, H., Elsayed, S. (eds) Intelligent and Evolutionary Systems. Proceedings in Adaptation, Learning and Optimization, vol 8. Springer, Cham. https://doi.org/10.1007/978-3-319-49049-6_29
Download citation
DOI: https://doi.org/10.1007/978-3-319-49049-6_29
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-49048-9
Online ISBN: 978-3-319-49049-6
eBook Packages: EngineeringEngineering (R0)