Abstract
Lipidomics is a lipid-targeted metabolomics approach aiming at comprehensive analysis of lipids in biological systems. Recent technological progresses in mass spectrometry, nuclear magnetic resonance spectroscopy, and chromatography have significantly enhanced the developments and applications of metabolic profiling of lipids in more complex biological samples. As many diseases reveal a notable change in lipid profiles compared with that of healthy people, lipidomics have also been broadly introduced to scientific research on diseases. Exploration of lipid biochemistry by lipidomics approach will not only provide insights into specific roles of lipid molecular species in health and disease, but it will also support the identification of potential biomarkers for establishing preventive or therapeutic approaches for human health. This chapter aims to illustrate how lipidomics can contribute for understanding the biological mechanisms inherent to schizophrenia and why lipids are relevant biomarkers of schizophrenia. The application of lipidomics in clinical studies has the potential to provide new insights into lipid profiling and pathophysiological mechanisms underlying schizophrenia. The future perspectives of lipidomics in mental disorders are also discussed herein.
The original version of this book was revised. An erratum to this chapter can be found at DOI 10.1007/978-3-319-47656-8_14
An erratum to this chapter can be found at http://dx.doi.org/10.1007/978-3-319-47656-8_14
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
- Biomarkers
- Liquid Chromatography
- Lipidomics
- Mass Spectrometry
- Nuclear Magnetic Resonance Spectroscopy
- Metabolomics
- Schizophrenia
1 Introduction
With the progress of “omics,” lipidomics, a branch of metabolomics, was first put forward by Han and Gross [1]. Lipidomics aims to characterize and quantify the range of intact lipid molecules in cells and biological fluids, allowing to correlate the lipid compositions to genomics, proteomics, diet, and diseases. The amount of genomic and proteomic data is greater than that in the lipidomics field, because of the complex nature of lipids and the limitations of tools available for such investigations. The key revolution that has incited advances in lipid analysis in the recent years was the development of new mass spectrometry techniques, particularly the “soft ionization” techniques, as the electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI). Such developments that provided a high sensitivity and specificity, excellent mass, and chromatographic resolutions, which in addition to an increased accessibility to authentic synthetic lipid standards, coupled to the remarkable developments in data and bioinformatics analysis, have facilitated the analysis of a wide diversity of lipids, ranging from phospholipids (PLs) and triacylglycerols (TGs) to sterols and glycolipids [2]. Lipid metabolism may be of particular importance for the central nervous system (CNS), due to its characteristic high concentration of lipids. The complexity of such analysis is highlighted by the recent characterization of over 500 different lipid species in a collective human serum sample conducted by the LIPID MAPS consortium (www.lipidmaps.org) [3].
The critical role of lipids in cell signaling and tissue physiology is demonstrated by the many neurological disorders, including bipolar disorder (BD) and schizophrenia (SCZ), and neurodegenerative diseases such as Alzheimer’s, Parkinson’s, and Niemann-Pick diseases, which present all deregulated lipid metabolism [4]. However, little is known about the molecular mechanisms that are altered in changes of states such as relapse and remission in mental illness patients. Lipidomics can be used in the search for biomarkers for specific diseases. Lipid-based biomarkers offer new prospects for precision medicine by providing sensitive diagnostic tools for disease forecast and monitoring.
In this book chapter, we describe the lipidomics approaches, summarize promising biomarkers reported in SCZ, and conclude with commentaries on the future contribution of the lipidomics approach within the larger biomarker discovery framework currently employed in the field of SCZ.
2 Lipid Classification
Lipids are water-insoluble compounds due to their hydrophobic features. The International Lipid Classification and the Nomenclature Committee, together with the Lipids Metabolites and Pathways Strategy (LIPID MAPS) Consortium, defined eight groups of lipids and divided them into classes and subclasses [2]. They classified the lipids by their chemically functional backbones and biochemical principles in:
-
1.
Fatty acyls (FAs): fatty acids and conjugates, octadecanoids, eicosanoids, docosanoids, and fatty alcohols
-
2.
Glycerolipids (GLs): monoradylglycerols, diradylglycerols, and triradyglycerols
-
3.
Glycerophospholipids (GPs): glycerophosphocholines, glycerophosphoglycerols, glycerophosphoethanolamines, glycerophosphoglycerophosphates, glycerophosphoserines, and glycerophosphoinositols
-
4.
Sphingolipids (SPs): sphingoid bases, ceramides, phosphosphingolipids, neutral glycosphingolipids, and acidic glycosphingolipids
-
5.
Sterol lipids (ST): sterols
-
6.
Prenol lipids (PR): isoprenoids
-
7.
Saccharolipids (SL): acrylamide sugars
-
8.
Polyketides (PK): linear polyketides
Fatty acids may be saturated, monounsaturated, or polyunsaturated. In animals, the residues of predominant fatty acids are the ones with a 16 or 18 carbon atoms chain – the palmitic and the stearic acids, which are saturated; oleic acid (C18Δ9) and linoleic acid (C18Δ9.12), which are unsaturated. The linolenic (C18Δ9.12,15) and linoleic acids form arachidonic, eicosapentaenoic, and docosahexaenoic acids and are essential fatty acids. TGs are the most important way to store energy in the organism, consisting of deposits in the adipose and muscle tissues.
3 The Molecular Biology of Lipids
Lipids are metabolites that play a significant role in different metabolic pathways. They are structural components of the cellular membranes, in which protein complexes, such as ion channels, receptors, and scaffolding complexes, are embedded, whether as a substratum, a product, or as a cofactor of biochemical reactions within a cell. Many of the biologically applicable lipids aggregate into macromolecular assemblies such as micelles or bilayers in aqueous environments of the human body. These aggregates comprise proteins, of course, in addition to lipid (e.g., apolipoproteins). Typically, the polar ends of lipid molecules face the aqueous milieu, whereas the more nonpolar fatty acyl moieties form a hydrophobic core (Fig. 11.1). Such aggregates therefore have unique biophysical properties and surface chemistries, which have important significances for the mechanisms of lipid function.
Monolayers of lipid and protein arrangements: (a) binding of lipid ligands by CD1 protein occurs via the hydrophobic acyl chains of the lipid molecules; (b) protein modules that specifically interact with lipid head groups. Interfacial binding is a significant mode of interaction for many lipid enzymes, as well as effectors of lipids. It is directed by electrostatic interactions at the interfacial region and the characteristics of lipid head groups; (c) the interior portion of a lipid assembly (e.g., the bilayer interior) contributes with interactions that arise from the hydrophobic parts of lipids molecules that have a role in the regulation of membrane channels
The finding that membrane lipids can act as precursors for second messengers has added to a dramatically different view of lipid action however. Phospholipase-mediated hydrolysis of phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) produces a variety of second messengers, such as diacylglycerol (DG), inositol 1,4,5-trisphosphate (IP(1,4,5)P3), and arachidonic acid (AA), which by themselves are precursors of biologically active molecules. DG is rapidly phosphorylated to phosphatidic acid (PA) (glycerophosphatidic acid, GPA), which is an important intermediate in PL biosynthesis and a potent regulator of enzyme function and bilayer structure. IP(1,4,5)P3 is metabolized by complicated enzymatic machinery, which leads to the generation of many different polyphosphorylated inositols. AA is the precursor for eicosanoids, which have an important and well-recognized role in inflammatory processes.
SPs are another example of highly bioactive membrane lipids and, like in the case of PI(4,5)P2, various components of their structures exert different activities (Fig. 11.2). The ceramide backbone is found in many complex glycolipids, and ceramides are potent regulators of cellular growth and death. Sphingosine and its phosphorylated derivative, sphingosine-1-phosphate, control the migration of immune cells and act via binding to specific receptors.
Furthermore, and in addition to their role as precursors, it is now clear that many membrane lipids act as signaling components themselves. Phosphoinositides (PIs; phosphorylated derivatives of phosphatidylinositol, GPIns), of which PI(4,5)P2 is a prominent representative, are an important class of such signaling lipids and contribute to a wide variety of cellular processes, including calcium homeostasis, membrane trafficking, and cytoskeletal dynamics. Indeed, it is becoming increasingly evident that understanding the biology of lipids often relates to understanding the responses that are mediated via lipids.
4 Lipids and the Central Nervous System (CNS)
A large number of diseases and neurological disorders in which lipid metabolism is altered confirm the crucial role of lipids in cell signaling and tissue physiology. Lipid metabolism may be of precise significance for the nervous system, as this organ has the second highest concentration of lipids, only after the adipose tissues. As mentioned, many neurological disorders, including BD and SCZ, and neurodegenerative diseases, such as Alzheimer’s, Parkinson’s, and Niemann-Pick diseases, involve deregulated lipid metabolism [4]. Altered lipid metabolism is also supposed to be an important event that contributes to CNS injury.
SCZ is noticeable by disturbances in thinking, emotional reactions, and social behavior, with delusions and hallucinations. Drugs that block dopamine receptors alleviate symptoms of SCZ, indicative of surplus dopaminergic function, while agents that block glutamate receptors induce some of the symptoms of SCZ in normal persons.
Current concepts on the neurological deficits of SCZ have focused on aberrations in PL metabolism, mainly increased activity of phospholipase A2 (PLA2) enzymes, and reduced activity of the system which integrates polyunsaturated fatty acids (PUFAs) into PLs (a simultaneous increase in PL hydrolysis and decrease in synthesis). Neither aberration alone produces SCZ but the presence of both does. These aberrations lead to changes in membrane structure and thus in the function of membrane-bound proteins, accessibility of cell signaling molecules, and in neurotransmitter systems. This assumption is reinforced by animal studies representing that application of PLA2 into the brain produces alterations in the dopamine system [5]. Furthermore, since PL metabolism has a crucial role in neuronal and synaptic growth and remodeling, it is believable that defects in this system result in failure of normal neurodevelopment in SCZ. There is also proof that SCZ is related with changes in lipid transport proteins and membrane PL composition (increase in phosphatidylserine (PS) and decrease in PC and PE) [6]. Genome studies have found that numerous genes involved in myelination have decreased expression levels in SCZ. Recently, significantly increased myelin basic protein (MBP) expression was observed in first-episode psychosis patients compared to sex and age paired healthy controls [7].
5 Reactive Oxygen Species (ROS) and Lipid Peroxidation (LPO)
ROS, including superoxide anion radical and hydrogen peroxide, are produced by a number of cellular oxidative metabolic processes, including oxidative phosphorylation by the mitochondrial respiratory chain, which includes the xanthine oxidase, NAD(P)H oxidases, monoamine oxidases and metabolism of AA by cyclooxygenases/lipoxygenases (COX/LOX) [8]. Though current literature proposes that COX does not directly produce ROS during AA oxidative metabolism, COX does form free radicals (i.e., carbon-centered radicals on AA). There are numerous reports in the literature on ROS production by COX, and this is possibly due to a secondary ROS generation induced by several eicosanoids. Interruption of mitochondria during COX-2-associated apoptosis is a probable source of ROS production, as it has been established for a number of different cells [4, 9]. ROS then cause oxidative damage to nucleic acids, proteins, carbohydrates, and lipids. Beyond impairment to membranes, lipid peroxides give rise to reactive α,β-unsaturated aldehydes including malondialdehyde, 4-hydroxynonenal (HNE), and acrolein. These aldehydes covalently bind to proteins through reaction with thiol groups and modify their function. Although there are intracellular defenses against ROS, increased production of ROS or loss of antioxidant defenses leads to progressive cell damage and decline in physiological function. The “oxidative stress” results when generation of ROS exceeds the cell’s capacity to detoxify them. The brain is believed to be particularly vulnerable to oxidative stress as it comprises high concentrations of PUFAs that are vulnerable to LPO, consumes relatively large amounts of oxygen for energy production, and has lower antioxidant defenses compared to other organs [8].
6 Systems-Level Approaches for Lipidomics
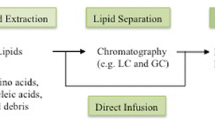
Traditional approaches for lipid analysis typically pre-fractionate lipids into classes using thin-layer chromatography (TLC), normal-phase liquid chromatography (NPLC), or solid-phase extraction (SPE), and then, distinct specific classes of lipids are fractionated into individual molecular species by high-performance liquid chromatography (HPLC) coupled with either ultraviolet (UV) or evaporative light-scattering (ELS) detector. With these traditional approaches, individual molecular species of many lipid classes can be evaluated. However, such “classical” techniques often lack sensitivity or need large sample volumes and multistep procedures for sample preparation; in addition, the resolution is inadequate, i.e., only a limited set of individual molecular species are evaluated. Alternatively, gas chromatography (GC) has been, and is still, frequently used for lipid analysis, although usually involving time-consuming procedures, which consist of hydrolysis and derivatization steps that are essential as most lipids are not GC amenable otherwise. Proper GC-based techniques meet the requirement of lipidomics with regards to the wider distribution of molecular component detection and physical properties and the wider dynamic range coverage of lipid concentrations; very often mass spectrometric (MS) detection is used.
6.1 MS as the First Choice and Successful Technique for Lipidomics
With the introduction of soft ionization techniques such as matrix-assisted laser desorption/ionization (MALDI), electrospray ionization (ESI), and atmospheric pressure chemical ionization (APCI) for MS, being the last two easily coupled to LC, the rapid and sensitive analysis for the majority or of a substantial fraction of lipids in one single experiment was made possible. Consequently, new soft ionization MS-based analytical strategies have been, and are still, emerging in lipidomics research. Strategies currently used in lipidomics include direct infusion ESI-MS and ESI-MS/MS, LC coupled with ESI-MS or MS/MS, and MALDI combined with Fourier transform ion cyclotron resonance MS (MALDI-FTICR-MS) or time-of-flight MS (MALDI-TOF-MS) [2].
Recently, novel multidimensional methodologies and ion mobility-MS (IM-MS) became available for lipidomics. A rapid separation of isomers, conformers, and enantiomers can be achieved by two-dimensional IM-MS, in addition to a resolving power analogous to that obtained with capillary GC. IM-MS has already shown enormous potential in lipid characterization and analysis of complex biological samples [10].
Identification of lipids can be achieved by using MS/MS, recorded in positive or in negative modes, as to get information on the head group (positive ESI), and on the carbon chain length and degree of unsaturation of the fatty acid chains of the lipid (negative ESI) (Table 11.1). Most GPs, such as phosphatidylcholines (PCs), phosphatidylethanolamines (PEs), lysoPCs, ceramides, and cardiolipins, can be identified with both positive and negative ionization modes, whereas TGs and DGs are identified only in positive mode. TGs are mainly distinguished as ammonium and sodium adducts, whereas DGs form sodium adducts and display neutral loss of water caused by in-source fragmentation. The formation of these adducts varies depending on the chain length of the fatty acids. Shorter TG species ionize preferentially as [M + Na]+ adducts, whereas longer chain length leads to predominant [M + NH4]+ species. In-house libraries are commonly created for specific lipids and use retention time, m/z value, and MS/MS data. All these tools that have recently been established facilitate automated lipid identification. However, it is difficult to get information on the double-bond positions, and typically, lipid structures are often stated as a single isomer. With the use of MS/MS, the sn-1 or sn-2 position can be determined; however, for the thorough data on the lipid identity, there is a need for using specific techniques, such as high-energy collision-induced dissociation, specific multistage fragmentation approaches, or ozone-induced dissociation. Another possibility is to gather specific lipid fractions and, after hydrolysis and methylation, to examine the fatty acid composition with GC-electron ionization MS. Fatty acid methyl esters can be separated according to the carbon number and saturation, and positional isomers – that are n3, n6, n9, etc. fatty acid methyl esters – can be determined. In addition, with the use of appropriate stationary phases, cis and trans isomers can also be separated. Thorough structural information is important to comprehend the data in the biochemical context.
6.2 NMR as a Powerful Technique for Lipid Assessment
Nuclear magnetic resonance (NMR) spectroscopy has been used to study the physical properties of membrane components. This technique has been changed to study the properties of lipid mixtures in order to determine their structures and functions. Information from these studies is significant with regard to obtaining structure, composition of lipids in cells, turnover of lipids, and characterization of lipid synthesis/transport and degradation pathways [11].
Because NMR spectroscopy is an analytical technique widely employed for characterization and identification of many classes of substances, it also has emerged as suitable analytical platform for the study of biological systems. There are many attempts for optimization and development of pulse sequences and for methodologies of analysis that allow the identification and the classification of organic compounds in complex samples. A great variety of NMR experiments (e.g., HSQC, HMBC, TOCSY, etc.) are being used to solve biological issues where biofluid samples such as serum, plasma, urine, cerebrospinal fluids (CSF), and others are being investigated. Biofluids are constituted from a mixture of organic compounds as amino acids, carbohydrates, organic acids, lipids, etc. [12]. Beside of this, many of these molecules are present in different concentrations in samples and have different physicochemical properties (mass, mobility, functions, etc.), making very challenging lipid and lipoprotein analyses in metabolomics [13]. Nuclear magnetic resonance comes to facilitate the analysis of hundreds of metabolites in a single sample, with or without any previous treatment [14].
One-dimensional (1D) NMR spectroscopy using solvent suppression is often used for metabolomics analysis. Recently, the new approach is using lipidomics of biological samples from the isolated lipids, being one of the successful strategies for lipid biological system comprehension [15].
There are diverse types of clinical sample preparations for lipid analysis, from simple approaches such as single organic solvent extraction, liquid-liquid extraction, and solid-phase microextraction to advanced techniques such as supercritical fluid extraction, microwave-assisted extraction, and ultrasound-assisted extraction. The common approach is liquid-liquid extraction, which is a method involving the use of two immiscible organic solvents. In order to reach exhaustive extraction of lipid classes, comprehends from phospholipids and glycolipids to fatty acids, DAGs and TAGs, usually for lipids extraction a mixture of chloroform and methanol with water is used [16].
Modified methods aiming to reproduce all properties of the clinical samples, from those classical methods as by Folch et al. [17] (chloroform/methanol/water ratio 8:4:3 v/v/v) and by Bligh and Dyer [18] (chloroform/methanol/water ratio 1:2:0.8 v/v/v), are broadly used nowadays [16–20]. Tukiainen et al. [15] use NaCl 0.15 mol L−1 solution instead of water to extract lipids from plasma. For NMR analysis, samples do not have to pass through processes of derivatization or sequences of dilutions. Also, after lipid extraction, a solubilization using deutered solvent is performed, and this solution is placed into a NMR tube.
However, lipidomics analysis is still very challenging, once lipids show some properties that may bring difficulties in analysis performed by some analytical techniques. The typical difference in the polarity between the lipids and the lack of chromophores in their structures, for example, complicate the separation and identification of the classes [21]. In this context, NMR spectroscopy may help with the simplification in the sample preparation also, physicochemical properties of samples are not changed, and the data on diffusion coefficients though peaks at intensity measurements, using the pulsed field gradients (PFG) applied during the FT NMR experiment [13], can be done.
The use of NMR spectroscopy in lipidomics is not as widespread as in metabolomics because of its low sensitivity compared to MS. However, the application of NMR techniques that help in the discrimination of individual molecules, as well as in complex mixtures analysis, principally with the employment of two-dimensional NMR (DETOCSY, 31P, 1H COSY, among others), which have a detection limit around 4 nmol L−1, is increasing in the lipidomics research based on NMR spectroscopy [14, 22].
The nuclei detected in NMR spectroscopy that commonly are used in the characterization of lipids are 1H, 13C, and 31P, being the 13C NMR spectroscopy very employed for triradylglycerols (GL03) and fatty acid analyses. 31P NMR is more suitable for glycerophospholipid analysis, and 1H NMR allows analysis of all types of lipids [21, 23]. In general, the advantages using NMR for lipid analysis when compared to other techniques are direct measurement; nondestructive techniques, which allow the recovery of the sample after analysis; and structural analysis of compounds. Disadvantages are low sensitivity, spectra dominated by very abundant lipids (cholesterol, PC) in 1H NMR, and line broadening of lipids in aqueous solutions in 31P NMR [14, 23].
Studies on 13C and 31P NMR spectroscopy data, and understanding of relations between the analytical parameters and physicochemical properties of lipids, help to achieve a reliable determination of the composition of phospholipids that constitute the matrix of cell membranes [24, 25].
In the 1H NMR spectra acquisition of a blood serum sample, the resonances referred to lipid moieties show broad signals and high intensities of peaks similar to the signals of lipoproteins thus interfering the analyst in the assignments of the peaks [26, 27]. One method used to solve this problem is the application of diffusion NMR spectroscopy that is based on the diffusion process of molecules or ions, i.e., in the random translational motion also called Brownian motion of these molecules [28].
Liu et al. [13] developed a pulse sequence for spectra edition of biofluid samples based on the combination of relaxation time of spin, molecular diffusion, and water suppression (WATERGATE) [29], which was named diffusion and relaxation editing (DIRE) pulse sequence. In this work, different combinations between times of longitudinal relaxations (T 1 ) and transversal relaxations (T 2 ) and spin-echo values for attenuation of broad peaks from molecules of high molecular mass (or fast relaxing) as the lipoproteins and albumins were studied. These allowed the NMR analysis of samples constituted from molecules with different molecular masses without the need of a pretreatment as the dialysis.
Posteriorly, Liu et al. [30] studied the measurement of diffusion coefficients of individual molecules in blood plasma samples through 1H-1H diffusion-edited total-correlation NMR spectroscopy (DETOCSY) for better understanding of the transport of molecules in biological system. When they applied a low gradient strength, the cross peaks of small molecules were attenuated, while high gradient strength was responsible for attenuation of macromolecules or small molecules bound to them, and so enabled a more accurate measure of the diffusion coefficient through the pairs of cross peaks relating to the separation of molecules.
Lopes et al. [31] studied the lipids in plasma samples of overweight subjects that underwent Roux-en-Y gastric bypass surgery (RYGB), a clinical method for weight loss that helped the glycemic control induced by hormonal changes. The T 2 -edited (Carr-Purcell-Meiboom-Gill pulse sequence) and diffusion-edited 1H NMR spectra (Fig. 11.3) were acquired for monitoring the bariatric surgery and the gastric mixed-meal tolerance test (MMTT). By T 2 -edited 1H NMR spectrum, it was possible to monitor levels of small molecules (β-glucose, alanine, lactate, etc.) and some amino acids, while the diffusion-edited 1H NMR enabled to measure the level changes in HDL, LDL, VLDL, phosphatidylcholine, etc.
(a) T 2 -edited and (b) diffusion-edited 1H NMR spectrum for overweight subjects before Roux-en-Y gastric bypass surgery. BCAA branched-chain amino acids, EDTA ethylenediaminetetraacetic acid, PtoCho phosphatidylcholine, TMSP 2,2,3,3-d 4 -3-(trimethylsilyl)propionic acid (Permission from Omics Journal)
Cai et al. [32] analyzed the lipoproteins in plasma and urine samples of first-episode neuroleptic-naive schizophrenia (FENNS) patients and after the treatment with risperidone drug using the diffusion-edited experiments and bipolar pulse pair longitudinal eddy current delay (BPP-LED) pulse sequence [33–35]. The LED is used to prevent spin relaxation during the diffusion, thus allowing to obtain information that can help in the determination of molecular diffusion coefficients [33, 36]. In their work, Cai et al. [32] observed a reduction in the levels of high-, low-, and very-low-density lipoproteins (HDL, LDL, and VLDL), phosphatidylcholine (PC), lipids, and unsaturated fatty acids (UFA) and an increase in lysophosphatidylcholines (LPC) in FENNS when compared to the control groups.
The 1H NMR spectroscopy gives information about chemical shifts, intensities, and chemical environments regarding all compounds that have hydrogen atoms in their organic functions. The data from the NMR spectra such as chemical shift and intensities (variables) are preprocessed through different methods, as the normalization scaling and the transformation after do the correction of baseline and the standardization of reference peaks. The preprocessing passes the set of spectra in variables that are named of buckets or binning and used in chemometric analysis [37].
7 Lipidomics Data Analysis and Bioinformatics
Data processing, data mining, and identification are critical steps in the lipidomics analytical workflow. Numerous preprocessing tools that incorporate different approaches and possibilities are available for chromatography-MS data, and different approaches are typically used for shotgun data. The core functions achieved by most tools typically comprise of peak detection, filtering and artifact removal, alignment (for LC-MS data), normalization, etc. In addition, “gap filling” is a significant feature, i.e., finding “missing” peaks in the data to avoid zero values that make the statistical analysis difficult. Filtering and peak detection focus on detecting real chromatographic peaks in each data file, and peak alignment focuses on locating and listing detected peaks found in the sample files.
Also, the data processing tools have clear advantages but also limitations. In a recent study, four automated preprocessing tools for GC-MS and LC-MS data were assessed in terms of their ability to ascertain the greatest number of metabolites consistently in a set of samples, as well as the robustness of the methods [38]. The preprocessing tools selected were MetAlign, MZmine, XCMS, and SpectConnect. The results revealed that different tools accomplished better for the GC-MS data than for the LC-MS data and that the qualitative and quantitative performance also displayed perfect variances between the tools. For GC-MS data, MetAlign had the most component recognitions, followed by MZmine, SpectConnect, and XCMS, whereas for the accurate-mass LC-MS data, the order was MetAlign, XCMS, and MZmine. The best presentation was obtained when two methods (e.g., MetAlign with MZmine or MetAlign and XCMS) were combined. However, in terms of quantitative presentation, SpectConnect and MetAlign achieved clearly worse than, for example, XCMS and MZmine for the accurate-mass LC-MS data. Both MZmine and XCMS provided acceptable quantitative results for GC-MS data as well as LC-MS data. All software tools stated a large number of false peaks, and thus manual examination of the chromatographic runs is required to eradicate those peaks. The study also indicated that although preprocessing tools have automated steps that are impractical to perform manually, a significant level of manual input is required in selecting the optimal parameters, processing peak tables, and validation. These outcomes clearly display that further expansion of the data processing algorithms is required.
Numerous software tools have been established for the identification of lipids in shotgun lipidomics, including LipidQA, LIMSA, FAAT, lipID, LipidSearch, LipidView, LipidInspector, LipidXplorer, LipidBlast, and ALEX, both for specific applications/instrumentation and for cross-platform software presenting user-specified instructions interrogating spectral data in an open-source format [38]. Typically, in the top-down approach, the data are investigated for specific molecular fragmentation. For example, in LipidXplorer individual spectra are systematized in a single flat file database that is further questioned by user-defined queries written in the molecular fragmentation query language (MFQL). In each MS spectrum, the lipid class-specific MFQL query checks if plausible precursor masses match the elemental composition likely for the corresponding molecular species. Optional search criteria can also be applied, for example, based on the odd or even number of carbon atoms in the fatty acid residues or the anticipated number of double bonds.
8 Physiological Factors Affecting Lipidomic Analysis
8.1 Lipid Composition in Blood-Based Samples
Blood is composed by (a) cellular components encompassing red and white blood cells and platelets and (b) a liquid carrier, named plasma. In blood, complex lipids are primarily found in the lipoprotein particles, while smaller, more polar lipids (e.g., free fatty acids (FFAs), bile acids, sterols) are found in free form or bound to protein carriers, such as albumin. Among these lipids, free cholesterol is the most abundant and is found primarily in lipoprotein particles. More complex lipids are also found in the red blood cells (RBCs) and platelets. Quehenberger et al. [3] accomplished a comprehensive characterization of lipids in human reference plasma, i.e., in pooled human plasma obtained from healthy individuals after overnight fasting and with gender balance and ethnic distribution that is symbolic of the US population. A summary of the foremost lipid subclasses and their concentrations is given in Table 11.2, with the most abundant lipids in each subclass noticeable. In individual lipoprotein fractions, lipid profiles differ considerably. Specific lipids, such as ceramides, have been distinguished primarily in very low-density lipoprotein (VLDL) and low-density lipoprotein (LDL) fractions, whereas ethanolamine plasmalogens are found primarily in LDL and high-density lipoprotein (HDL) subfraction 2. LysoPCs and ether-linked PCs have been found in all lipoprotein fractions, with the greatest abundances in HDL subfraction 2, HDL subfraction 3, and LDL. Most large-scale studies are based on the extent of the lipids in total serum or plasma, because it is too time-consuming to first separate, for example, individual lipoprotein fractions, followed by lipid analysis. However, the lipid characterization of lipoprotein fractions can give a much more thorough view of the lipid metabolism. For example, the VLDL fraction can give improved perception into lipid metabolism in liver than the overall lipid composition [39].
The most abundant lipids in human blood are GLs, mainly TGs. The TGs are most abundant in chylomicrons and in VLDL and intermediate-density lipoprotein fractions. Most recent methods can detect and recognize about 50 or more TGs in human plasma or serum; however, the definite amount of TGs is substantially higher than that. A large number of different GPs have been recognized in plasma and serum, including PA, PC, PE, PG, PI, and PS. By total class concentration, the overwhelming majority of GPs in human plasma are PCs and PEs (Table 11.2). These two classes also cover substantial amounts of ether-linked lipids. SPs in the blood correspond to a part of the circulating lipoprotein particles (VLDL, LDL, and HDL), they are carried by serum albumin, and they are also present in blood cells and platelets. Over 200 individual SPs were identified in human plasma, and the sphingomyelins (SMs) account for the largest fraction of SPs in plasma [40]. Cholesteryl esters (CEs) are one of the most abundant lipid classes (Table 11.2) in human serum and plasma, with CE (18:2) constituting a major fraction of the CE subclass. Some lipids are usually not identified in blood samples; for example, cardiolipins are present in blood-based samples at very low concentrations and are typically not detected with the current nontargeted profiling methods.
8.2 Effect of Gender, Age, Diet, and Sampling Time on Lipid Levels
Numerous phenotypic and physiological parameters, including medication and specific food supplements (e.g., fish oil, and vitamin D), have a significant influence on circulating lipid levels, and thus the medication should be documented and considered during the study planning and for the interpretation of the outcomes. Although the huge biological variation of lipid levels, the levels of lipids have been shown to have a relatively small difference in the same individual when analyzed under identical sampling conditions [41].
The levels of lipids such as SMs, PCs, and TGs in blood may vary between the genders. Most lipids do not present a strong association with gender, but for different age groups, gender does have an effect on specific lipids. Age also has a prominent effect on lipid concentrations, mainly in females. Numerous TGs along with a few other lipids have been shown to present significantly higher levels in elderly females than in young females in both serum and plasma samples [42]. The same study showed that female-specific age-associated changes of the levels of lysoPCs were detected in plasma but that were markedly less in serum.
Diet also affects the levels of lipids and individual fatty acids in blood, both in the long and short term. In fact, it has been proposed that about 50 % of metabolites measured are dependent on diet [43].
In addition to gender and age, the circadian system controls lipid and carbohydrate homeostasis, thus augmenting energy storage and its utilization during the day. Thus, the time of sample collection also has an effect on lipid profiles. Naturally, the diet and fasting conditions play a critical role, but even in fasting conditions, specific lipids have revealed difference due to the circadian time course. There are still an inadequate number of studies related to circadian time course of lipids in humans; however, the studies reveal so far that specific lipids have a circadian cycle pattern [44].
Guidelines usually recommend that lipid profiles should be attained from individuals in the fasting state. However, it is not always promising to obtain fasting samples, for example, in the case of very young children. Also, fasting is uncomfortable predominantly in individuals with diabetes, due to the fear of either hyperglycemia or hypoglycemia. It is also essential to test the individuals’ capacity and flexibility to respond with typical environmental factors, such as physical activity or eating. However, most studies to date are inadequate to the analysis of samples obtained in a fasted state.
The body mass index (BMI) of individuals also affects the lipid profile. In most studies, lipid profiles have been studied in relation to specific obesity-associated disease, and precise alterations were observed. However, a substantial portion of obese subjects does not present any of the well-known metabolic abnormalities. Thus, it is essential to recognize lipids that are associated with obesity rather than with a specific disease [45].
9 Lipidomics-Based Biomarkers
Since lipids retain a diversity of biological functions in the processes of life such as formation of cellular membranes, energy storage, and cell signaling, they can be anticipated to reflect much of the metabolic status in health and disease. In addition, several studies have demonstrated that lipid metabolic disorders or abnormalities can lead to various human diseases including diabetes, obesity, arteriosclerosis, coronary heart disease, and brain injuries [2, 46]. Therefore, monitoring the changes of lipid metabolites of certain molecular species in biological samples, as influenced by external stimuli or disturbance by disease processes, will be helpful for the discovery of lipid metabolites with potential of being indicative of metabolic disorders or diseases.
Biomarkers could be considered to extend all the way to include our fixed genomic characters. At the level of the subcellular and tissue, the research in this field has employed the transcriptomics, proteomics, metabolomics, lipidomics, immunological, and biological epigenetics [46, 47]. The recent attention in biomarker discovery is encouraged by new molecular biology techniques, which allows the rapid finding of relevant biomarkers, without specific comprehensive insight into the mechanisms of a disease. Lipidomics is the most appropriate approach for the study of pathways and networks of lipids in biological systems. There is no question that clinical requirement and complexity of lipid metabolism will pose a series of novel challenges for researchers devoted to lipidomics of various diseases. Numerous risk factors contributing to disease are suggested to influence lipid metabolism, and thus, they may be reflected in the lipid profile of an individual. One of the most extensively known lipid biomarkers is cholesterol, which, in the form of total blood cholesterol and/or HDL cholesterol, has been used for more than 50 years for the determination of risk for cardiovascular disease [48]. Apart from applications in human diseases, the approach of lipidomics-driven biomarker discovery has also been used in nutrition and health fields, aiming the health promotion and the disease prevention [49].
In spite of the numerous analytical strategies available to identify changes in lipid metabolism related to disorders or diseases, multivariate statistical analysis was almost invariably performed to assist the finding of novel lipid molecular species that could serve as potential biomarkers.
10 Lipidomics in Schizophrenia Research
One of the goals of schizophrenia (SCZ) research is to explain the disorder in clear and simple biological terms and, in doing so, relegate it to the index of mundane – and more amenable – human disorders. The existing pace of advances in large-scale biological data acquisition and data processing might make the task appear eminently achievable. However, even the most avid optimists would have to disclose that, despite this effort, we are still piecing together the edge of the jigsaw puzzle rather than seeing the full picture (in contraposition to the situation of Parkinson’s disease – a cousin by neurotransmitter of SCZ). A cross-disciplinary understanding of environmental impacts, genetic risk, cellular pathology, anatomical pathology, network dysfunction, and outward symptomatology has converged to inform the development of novel therapies. Multiple explanations have been put forward for this qualitative difference in disorder complexity, with main focus on the idea that SCZ is not a unitary disorder but rather a common symptomatic endpoint of a great variety of brain dysfunctions and insults. In support to this, SCZ now belongs to a wide family of disorders that have proven interrelationships at the genetics level. These include BD, depression, intellectual disability, autism spectrum disorders, and, recently, multiple sclerosis [46]. Biomarker identification is one such arena of research that will have to meet these demands. The advancement in development of commercial SCZ biomarker tests depends on economic models, regulation, and perceived healthcare need, just as much as bioinformatics. Hence, there may be a more pragmatic short-term goal for biomarkers: to help the subdivision and classification of SCZ according to its principal molecular etiopathology.
Current technological developments for lipidomics analysis have provided remarkable outcomes in other areas of research, with potential to be useful for biomarker discovery in SCZ. The lipid biomarkers offer a new outlook for achieving better diagnosis either in preclinical experiments using mammal animal models or in human clinical evaluations. Lipidomics has the potential for the discovery of biomarkers for SCZ, not only in brain tissue but also in the blood (serum or plasma) and CSF (Table 11.3; referred from Sethi et al. [2]).
In one study, Kaddurah-Daouk et al. [19] used a specific lipidomics platform and found changes in different lipid classes (PC, PE, TG) in the plasma of SCZ patients after 2–3 weeks of treatment with atypical antipsychotic drugs. A recent study has also proved a significant downregulation of several n3 and n6 PUFAs compositions in PE and in lipid classes in the blood plasma of first-episode SCZ patients [55]. These changes in lipid metabolism could indicate a metabolic vulnerability in patients with SCZ that may occur early in the development and onset of the disease.
Alterations in the peripheral tissue membrane PLs levels have also been detected [61] in RBCs of SCZ patients. Schwarz et al. [51] observed lipid changes in postmortem brain samples from SCZ, and they found significant changes in the levels of FFAs and PCs in the gray and white matter of SCZ patients compared to control samples. Ceramides were significantly increased in white matter of SCZ as compared to control levels. In addition, lipid profiling of RBCs of SCZ accused significantly decreased levels of FFAs and ceramides in drug-naїve first-onset patients. Reductions of PC levels have formerly been reported for different regions of the SCZ brain [50], which was linked to an increase in SM turnover, as PC is the choline donor to SM in neurons and oligodendrocytes.
Lipidomic analyses can be complicated due to the high occurrence of metabolic syndrome in SCZ and its induction/worsening by treatment with antipsychotics. However, in the case of plasmalogen (Pl) analyses, about 20 % decrease in circulating Pls was described in 20 first-episode and 20 recurrent SCZ patients [62], suggesting that this may represent an intrinsic biochemical deficit involved in this developmental disorder. A recent study also revealed significant decrease in choline plasmalogens, ethanolamine plasmalogens, and DHA in the plasma of patients with SCZ. In contrast, increased cellular levels of choline plasmalogens and decreased levels of ethanolamine plasmalogens and DHA were found in platelets of patients with SCZ [58]. Another recent study from the same research group demonstrated elevated level of sulfatides, choline plasmalogens, ethanolamine plasmalogens, and N-acyl-phosphatidylserines (NAPS) in the gray matter of postmortem frontal cortex SCZ subjects [59]. As major components of membranes, Pls are essential for membrane fluidity, lipid raft formation, membrane fusion for neurotransmitter release, ion transport, and regulation of cholesterol efflux. Pls are also essential in brain development, both for white and gray matters.
In conclusion, several studies showed significant changes in prefrontal cortex FA concentrations, particularly within cholesteryl ester (CE) and abnormalities in Pl levels of SCZ patients compared to controls [20, 62]. An increase in CE-FA concentration turnover in SCZ may reflect the excitotoxicity and neuronal loss reported in SCZ patients [63]. This is consistent with findings in postmortem brains from SCZ patients, suggesting a hypoglutamatergic and hyperdopaminergic neurotransmitter signaling, in association with neuronal loss and disease worsening over time [64]. Upcoming studies of Pls are required to explore their role in the pathogenesis of SCZ and to clarify whether restoration of normal Pl levels is linked with the therapeutic effects of antipsychotic drugs.
11 The Future of Clinical Lipidomics
Lipids offer the possibility of discovery novel key biomarkers for many areas and different diseases. They could also fuel diagnostic developments and, thus, support more personalized methodologies of treatment. Lipidomics can also be used for studying numerous experimental disease models, and this could offer an enormous boost for the translational medicine.
During the discovery stage, detailed lipidomic studies currently demand a repertoire of several different analytical platforms. Such a quest is currently time-consuming, and hence, it is appropriate only to a limited amount of samples. However, technology and process improvements are developing and may enable to output thorough lipidome data sets based on large sample sets more rapidly in the near future. Another challenge is the bias of the final output arising from the chosen methodological setups and instrumentations. Although internal standards are typically applied to quantify the lipids of interest, owing to the lack of proper non-endogenous standards, only a subset of lipid species can be measured with acceptable precision. It is notable that the endpoint results depend on the internal standards applied, and, unfortunately, most aberrations among users are found in this aspect. Therefore, due to the lack of standardization, it is difficult to relate or combine lipidomic data from different laboratories. Although rigorous standardization and validation processes are required for conveying lipidomic assays to clinical practice, the benefits are assorted. Once set up, MALDI-MS assays are analytically robust and efficient clinical solutions. The assays can be scaled down from the discovery throughput mode involving long per sample scan times to just 1–2 min analysis time per sample, with no extensive washing or incubation steps in the overall assays. Lipidomic analyses are directed in a 96-well format and in a robot-assisted workflow, and therefore, this platform can attain substantial sample throughputs. More critical for wider assumption remain the issues centering on sample transportation, storage, number of freeze-thaw cycles, preparation, and sample handling during the analytical process.
Given the obtainability of better and more appropriate internal standards and MS analysis methods, the eminence of lipidomic outputs will be higher with more lipid species determined in absolute quantities. Bioinformatics solutions will permit the processing of all data and to put out lipidomic results instantly, in an accessible way. Concurrently, informatics setups can monitor all processes in the lipidomic workflow, identify automatically any failure in the process, and also monitor continuously the sample quality. Placing all these pieces together will guide lipidomic standardization, which will make lipidomic solutions attractive for preclinical and clinical studies and pertinent to the regulatory environment in a cost-effective manner.
12 Concluding Remarks and Perspectives
Abnormal lipid or metabolism dysfunction is considered to be the major influence aspect in many lifestyle-associated diseases and hereditary/genetic conditions. Lipidomics is an emerging approach for a widespread and systematic study of a variety of lipids. The field of lipidomics is under a continuous investigation to further explore the lipidome with the eventual goal to broaden our biological knowledge for different diseases. Currently, lipidomics has an enormous prospective in lipid research, in which different lipid profiling is associated to various diseases, and changes of lipid metabolism or pathway modulation can be identified in human complex diseases. This offers new perceptions into metabolic and inflammatory diseases. The combination of lipid profiles and multivariate statistics can help us in novel biomarker discovery, disease pathology explanation, drug-response monitoring in therapy and toxicity, translational medicine, and in-depth uncovering mechanisms of lipid-mediated disease.
MS and chromatography techniques have significantly encouraged the developments and applications of lipidomics in clinical chemistry. According to different research objectives, different MS and chromatography approaches can be selected, and selected approaches must be appropriate for application to the specific lipid species. Direct infusion ESI-MS has been used to distinguish whole lipid extracts, and it has revealed significant potential in the identification and quantification of PLs and fatty acid species in a quick and robust manner. However, direct infusion ESI-MS is vulnerable to ion suppression; this shortcoming can be overwhelmed to some extent by chromatographic techniques. LC-MS can also be applied to separation lipids from complex samples into individual lipid classes or separate the same lipid class. GC-MS is appropriate for the fatty acids and their derivatives, but it is limited by the necessity of analytes being volatile and due to its dynamic range. Combined analytical approaches can be acquired by overcoming the limitations of individual techniques for a wide range of the lipidome.
Although a number of lipidomic experiments has been carried out on exploring diseases through analyzing biomarkers and metabolic pathways in clinics, clinical lipidomics is still in its infancy compared to proteomics and metabolomics. The current researches on lipidomics focus predominantly on biomarker searching, which is apparently inadequate. There is no question that clinical requirement and complexity of lipid metabolism will pose researchers enthusiastic to lipidomics of various diseases with a series of novel challenges. The combined techniques will help advance our understanding of the physiological functions of lipid species and depict the etiology and pathophysiology of multiple lipid-related diseases, such as cancer, obesity, diabetes, and atherosclerosis. Integrated with other omics strategies, this platform will offer a new outlook for dissecting and improving disease diagnosis and prevention.
Abbreviations
- AA:
-
Arachidonic acid
- APCI:
-
Atmospheric pressure chemical ionization
- BD:
-
Bipolar disorder
- BMI:
-
Body mass index
- CE:
-
Cholesteryl ester
- Cer:
-
Ceramide
- CNS:
-
Central nervous system
- COX:
-
Cyclooxygenase
- DG:
-
Diacylglycerol
- DHA:
-
Docosahexaenoic acid
- ELSD:
-
Evaporative light-scattering detector
- ESI:
-
Electrospray ionization
- FA:
-
Fatty acyl
- FFA:
-
Free fatty acid
- FID:
-
Flame ionization detector
- FTICR:
-
Fourier transform ion cyclotron resonance
- GC:
-
Gas chromatography
- GL:
-
Glycerolipid
- GP:
-
Glycerophospholipid
- GPA:
-
Glycerophosphatidic acid
- HDL:
-
High-density lipoprotein
- hexCer:
-
Monohexosylceramide
- HNE:
-
4-Hydroxynonenal
- HPLC:
-
High-performance liquid chromatography
- IM-MS:
-
Ion mobility-mass spectrometry
- LDL:
-
Low-density lipoprotein
- LOX:
-
Lipoxygenase
- LPC:
-
Lysophosphatidylcholine
- LPE:
-
Lysophosphatidylethanolamine
- LPO:
-
Lipid peroxidation
- MALDI:
-
Matrix-assisted laser desorption/ionization
- MS:
-
Mass spectrometry
- MS:
-
Mass spectrometry
- MS/MS:
-
Tandem mass spectrometry
- NAPS:
-
N-acyl-phosphatidylserine
- NMR:
-
Nuclear magnetic resonance
- NPLC:
-
Normal-phase liquid chromatography
- PA:
-
Phosphatidic acid
- PC:
-
Phosphatidylcholine
- PE:
-
Phosphatidylethanolamine
- PG:
-
Phosphoglycerol
- PI:
-
Phosphatidylinositol
- PK:
-
Polyketide
- Pl:
-
Plasmalogen
- PL:
-
Phospholipid
- PLA2 :
-
Phospholipase A2 PS: Phosphatidylserine
- PR:
-
Prenol lipid
- PS:
-
Phosphatidylserine
- PUFA:
-
Polyunsaturated fatty acid
- Q:
-
Quadrupole
- RBC:
-
Red blood cell
- ROS:
-
Reactive oxygen species
- S1P:
-
Sphingosine-1-phosphate
- SCZ:
-
Schizophrenia
- SL:
-
Saccharolipid
- SM:
-
Sphingomyelin
- SP:
-
Sphingolipid
- SPE:
-
Solid-phase extraction
- ST:
-
Sterol lipid
- TG:
-
Triacylglycerol
- TLC:
-
Thin-layer chromatography
- TOF:
-
Time of flight
- UPLC:
-
Ultra-performance liquid chromatography
- VLDL:
-
Very low-density lipoprotein
References
Han X, Gross RW. Electrospray ionization mass spectroscopic analysis of human erythrocyte plasma membrane phospholipids. Proc Natl Acad Sci U S A. 1994;91:10635–9.
Sethi S, Hayashi MA, Sussulini A, et al. Analytical approaches for lipidomics and its potential applications in neuropsychiatric disorders. World J Biol Psychiatry. 2016:1–15. doi:10.3109/15622975.2015.1117656.
Quehenberger O, Armando AM, Brown AH, et al. Lipidomics reveals a remarkable diversity of lipids in human plasma. J Lipid Res. 2010;51:3299–305.
Adibhatla RM, Hatcher JF. Role of lipids in brain injury and diseases. Future Lipidol. 2007;2:403–22.
Horrobin D. The lipid hypothesis of schizophrenia. In: Skinner ER, editor. Brain lipids and disorders in biological psychiatry, vol. 35. Amsterdam: Elsevier Science; 2002. p. 39–52.
Berger GE, Smesny S, Amminger GP. Bioactive lipids in schizophrenia. Int Rev Psychiatry. 2006;18:85–98.
Ota VK, Noto C, Santoro ML, et al. Increased expression of NDEL1 and MBP genes in the peripheral blood of antipsychotic-naïve patients with first-episode. Eur Neuropsychopharmacol. 2015;25:2416–25.
Maurya PK, Noto C, Rizzo LB, et al. The role of oxidative and nitrosative stress in accelerated aging and major depression disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2016;65:134–44.
Kunz A, Anrather J, Zhou P, et al. Cyclooxygenase-2 does not contribute to postischemic production of reactive oxygen species. J Cereb Blood Flow Metab. 2007;27:545–51.
Paglia G, Kliman M, Claude E, et al. Applications of ion-mobility mass spectrometry for lipid analysis. Anal Bioanal Chem. 2015;407:4995–5007.
Vilella F, Ramirez LB, Simón C. Lipidomics as an emerging tool to predict endometrial receptivity. Fertil Steril. 2013;99:1100–6.
Smolinska A, Blanchet L, Buydens LMC, et al. NMR and pattern recognition methods in metabolomics: from data acquisition to biomarker discovery: a review. Anal Chim Acta. 2012;750:82–97.
Liu M, Nicholson JK, Lindon JC. High-resolution diffusion and relaxation edited one- and two-dimensional 1H NMR spectroscopy of biological fluids. Anal Chem. 1996;68:3370–6.
Rolim AEH, Henrique-Araújo R, Ferraz EG, et al. Lipidomics in the study of lipid metabolism: current perspectives in the omic sciences. Gene. 2015;554:131–9.
Tukiainen T, Tynkkynen T, Mäkinen VP, et al. A multi-metabolite analysis of serum by 1H NMR spectroscopy: early systemic signs of Alzheimer’s disease. Biochem Biophys Res Commun. 2008;375:356–61.
Teo CC, Chong WPK, Tan E, et al. Advances in sample preparation and analytical techniques for lipidomics study of clinical samples. Trends Anal Chem. 2015;66:1–18.
Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem. 1957;226:497–509.
Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–7.
Kaddurah-Daouk R, McEvoy J, Baillie RA, et al. Metabolomic mapping of atypical antipsychotic effects in schizophrenia. Mol Psychiatry. 2007;12:934–45.
Taha AY, Cheon Y, Ma K, et al. Altered fatty acid concentrations in prefrontal cortex of schizophrenic patients. J Psychiatr Res. 2013;47:636–43.
Carrasco-Pancorbo A, Navas-Iglesias N, Cuadros-Rodríguez L. From lipid analysis towards lipidomics, a new challenge for the analytical chemistry of the 21st century. Part I: modern lipid analysis. Trends Anal Chem. 2009;28:263–78.
Li M, Yang L, Bai Y, et al. Analytical methods in lipidomics and their applications. Anal Chem. 2014;81:161–75.
Wenk MR. The emerging field of lipidomics. Nat Rev Drug Discov. 2005;4:594–610.
Lutz NW, Cozzone PJ. Principles of multiparametric optimization for phospholipidomics by 31P NMR spectroscopy. Biophys Rev. 2013;5:295–304.
Leftin A, Mologu TR, Job C. Area per lipid and cholesterol interactions in membranes from separated local-field 13C NMR spectroscopy. Biophys J. 2014;107:2274–86.
Ala-Korpela M. 1H NMR spectroscopy of human blood plasma. Prog Nucl Magn Reson. 1995;27:475–554.
Barrilero R, Llobet E, Mallol R, et al. Design and evaluation of standard lipid prediction models based on 1H-NMR spectroscopy of human serum/plasma samples. Metabolomics. 2015;11:1394–404.
Nicolay K, Braun KPJ, de Graaf RA, et al. Diffusion NMR spectroscopy. NMR Biomed. 2001;14:94–111.
Piotto M, Saudek V, Sklenář V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR. 1992;2:661–5.
Liu M, Nicholson JK, Parkinson JA, et al. Measurement of biomolecular diffusion coefficients in blood plasma using two-dimensional 1H-1H diffusion-edited Total-Correlation NMR Spectroscopy. Anal Chem. 1997;69:1504–9.
Lopes TI, Geloneze B, Pareja JC, et al. “Omics” prospective monitoring of bariatric surgery: roux-en-Y gastric bypass outcomes using mixed-meal tolerance test and time-resolved (1)H NMR-based metabolomics. OMICS. 2016;20:415–23.
Cai HL, Li HD, Yan XZ, et al. Metabolomic analysis of biochemical changes in the plasma and urine of first-episode neuroleptic-naiv̈e schizophrenia patients after treatment with Risperidone. J Proteome Res. 2012;11:4338–50.
Gibbs SJ, Johnson Jr CS. A PFG NMR experiment for accurate diffusion and flow studies in the presence of eddy currents. J Magn Reson. 1991;93:395–402.
Fordham EJ, Gibbs SJ, Hall LD. Partially restricted diffusion in a permeable sandstone: observations by stimulated echo PFG NMR. Magn Reson Imaging. 1994;12:279–84.
Wu D, Chen A, Johnson CS. An improved diffusion-ordered spectroscopy experiment incorporating bipolar-gradient pulses. J Magn Reson A. 1995;115:260–4.
Beckwith-Hall BM, Thompson NA, Nicholson JK, et al. A metabonomic investigation of hepatotoxicity using diffusion-edited 1H NMR spectroscopy of blood serum. Analyst. 2003;128:814–8.
Checa A, Bedia C, Jaumot J. Lipidomic data analysis: tutorial, practical guidelines and applications. Anal Chim Acta. 2015;885:1–16.
Hyötyläinen T, Orešič M. Optimizing the lipidomics workflow for clinical studies-practical considerations. Anal Bioanal Chem. 2015;407:4973–93.
Kotronen A, Velagapudi VR, Yetukuri L, et al. Saturated fatty acids containing triacylglycerols are better markers of insulin resistance than total serum triacylglycerol concentrations. Diabetologia. 2009;52:684–90.
Vieu C, Terce F, Chevy F, et al. Coupled assay of sphingomyelin and ceramide molecular species by gas liquid chromatography. J Lipid Res. 2002;43:510–22.
Breier M, Wahl S, Prehn C, et al. Targeted metabolomics identifies reliable and stable metabolites in human serum and plasma samples. PLoS One. 2014;9:e89728.
Ishikawa M, Maekawa K, Saito K, et al. Plasma and serum lipidomics of healthy white adults shows characteristic profiles by subjects’ gender and age. PLoS One. 2014;9:e91806.
Zivkovic AM, Wiest MM, Nguyen U, et al. Assessing individual metabolic responsiveness to a lipid challenge using a targeted metabolomic approach. Metabolomics. 2009;5:209–18.
Gooley JJ, Chua EC. Diurnal regulation of lipid metabolism and applications of circadian lipidomics. J Genet Genomics. 2014;41:231–50.
Pietilainen KH, Sysi-Aho M, Rissanen A, et al. Acquired obesity is associated with changes in the serum lipidomic profile independent of genetic effects – a monozygotic twin study. PLoS One. 2007;2:e218.
Sethi S, Brietzke E. Omics-based biomarkers: application of metabolomics in neuropsychiatric disorders. Int J Neuropsychopharmacol. 2016;19(3):pyv096. doi:10.1093/ijnp/pyv096.
Sethi S, Chourasia D, Parhar IS. Approaches for targeted proteomics and its potential applications in neuroscience. J Biosci. 2015;40:607–27.
Meikle P, Barlow C, Weir J. Lipidomics and lipid biomarker discovery. Aus Biochemist. 2009;40:12–6.
Draisma HH, Reijmers TH, Bobeldijk-Pastorova I, et al. Similarities and differences in lipidomics profiles among healthy monozygotic twin pairs. OMICS. 2008;12:17–31.
Schmitt A, Wilczek K, Blennow K, et al. Altered thalamic membrane phospholipids in schizophrenia: a postmortem study. Biol Psychiatry. 2004;56:41–5.
Schwarz E, Prabakaran S, Whitfield P, et al. High throughput lipidomic profiling of schizophrenia and bipolar disorder brain tissue reveals alterations of free fatty acids, phosphatidylcholines, and Ceramides. J Proteome Res. 2008;7:4266–77.
Hamazaki K, Choi KH, Kim HY. Phospholipid profile in the postmortem hippocampus of patients with schizophrenia and bipolar disorder: no changes in docosahexaenoic acid species. J Psychiatr Res. 2010;44:688–93.
Orešič M, Tang J, Seppänen-Laakso T, et al. Metabolome in schizophrenia and other psychotic disorders: a general population-based study. Genome Med. 2011;3:19.
Orešič M, Seppänen-Laakso T, Sun D, et al. Phospholipids and insulin resistance in psychosis: a lipidomics study of twin pairs discordant for schizophrenia. Genome Med. 2012;4:1.
McEvoy J, Baillie RA, Zhu H, et al. Lipidomics reveals early metabolic changes in subjects with schizophrenia: effects of atypical antipsychotics. PLoS One. 2013;8:e68717.
Wood PL. Accumulation of N-acylphosphatidylserines and N-acylserines in the frontal cortex in schizophrenia. Neurotransmitter. 2014;1:e263.
Wood PL, Filiou MD, Otte DM, et al. Lipidomics reveals dysfunctional glycosynapses in schizophrenia and the G72/G30 transgenic mouse. Schizophr Res. 2014;159:365–9.
Wood PL, Unfried G, Whitehead W, et al. Dysfunctional plasmalogen dynamics in the plasma and platelets of patients with schizophrenia. Schizophr Res. 2015;161:506–10.
Wood PL, Holderman NR. Dysfunctional glycosynapses in schizophrenia: disease and regional specificity. Schizophr Res. 2015;166:235–7.
Weng R, Shen S, Burton C, et al. Lipidomic profiling of tryptophan hydroxylase 2 knockout mice reveals novel lipid biomarkers associated with serotonin deficiency. Anal Bioanal Chem. 2016;408:2963–73.
Ponizovsky AM, Modai I, Nechamkin Y, et al. Phospholipid patterns of erythrocytes in schizophrenia: relationships to symptomatology. Schizophr Res. 2001;52:121–6.
Kaddurah-Daouk R, McEvoy J, Baillie R, et al. Impaired plasmalogens in patients with schizophrenia. Psychiatry Res. 2012;198:347–52.
Rao JS, Kellom M, Reese EA, et al. Dysregulated glutamate and dopamine transporters in postmortem frontal cortex from bipolar and schizophrenic patients. J Affect Disord. 2012;136:63–71.
Moghaddam B, Javitt D. From revolution to evolution: the glutamate hypothesis of schizophrenia and its implication for treatment. Neuropsychopharmacology. 2012;37:4–15.
Acknowledgments
We thank the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brasília, Brazil) and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, São Paulo, Brazil) for their financial support and fellowship. SS received a Young Talent Scholarship from the CNPq. BSB received a scholarship from the FAPESP (2013/14707-9), and JGMP received a scholarship from the CNPq.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this chapter
Cite this chapter
Sethi, S., Hayashi, M.A.F., Barbosa, B.S., Pontes, J.G.M., Tasic, L., Brietzke, E. (2017). Lipidomics, Biomarkers, and Schizophrenia: A Current Perspective. In: Sussulini, A. (eds) Metabolomics: From Fundamentals to Clinical Applications. Advances in Experimental Medicine and Biology(), vol 965. Springer, Cham. https://doi.org/10.1007/978-3-319-47656-8_11
Download citation
DOI: https://doi.org/10.1007/978-3-319-47656-8_11
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-47655-1
Online ISBN: 978-3-319-47656-8
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)