Abstract
White matter hyperintensities (WMH) seen on FLAIR images are established as a key indicator of Vascular Dementia (VD) and other pathologies. We propose a novel modality transformation technique to generate a subject-specific pathology-free synthetic FLAIR image from a T\(_1\) -weighted image. WMH are then accurately segmented by comparing this synthesized FLAIR image to the actually acquired FLAIR image. We term this method Pseudo-Healthy Image Synthesis (PHI-Syn). The method is evaluated on data from 42 stroke patients where we compare its performance to two commonly used methods from the Lesion Segmentation Toolbox. We show that the proposed method achieves superior performance for a number of metrics. Finally, we show that the features extracted from the WMH segmentations can be used to predict a Fazekas lesion score that supports the identification of VD in a dataset of 468 dementia patients. In this application the automatically calculated features perform comparably to clinically derived Fazekas scores.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
White matter hyperintensities (WMH) are commonly found in brain fluid attenuated inversion recovery (FLAIR) magnetic resonance imaging (MRI). Their aetiology is diverse but they are known to be associated with an increased risk of stroke, dementia and death [1]. WMH are usually clearly visible as hyperintense regions in FLAIR images, and potentially appear as hypointense regions in T\(_1\) -weighted images (Fig. 1).
The accurate annotation of WMH from FLAIR images is a laborious task that requires a high level of expertise and is subject to both inter- and intra-rater variability. To enable effective image analysis in large scale studies or the reproducible quantification of lesion load in the clinic without expert knowledge (e.g. in the context of a comprehensive decision support system) an accurate and fully automatic method for lesion segmentation is desirable.
In this paper, we present a novel method of segmenting WMH from FLAIR images using modality transformation. Modality transformation is the task of generating a synthetic image with the appearance characteristics of a specific imaging modality (or protocol) by using information from images acquired from one or more other modalities. The accurate generation of these images can be critical in the context of, for example, non-linear multi-modality registration [2] where the problem can be reduced to a mono-modality problem when one modality is synthesised from the other. Additionally, many segmentation or classification algorithms require an input image from a certain modality. The ability to synthesise these modalities from another modality could substantially expand the applicability of these algorithms [3].
This paper investigates the principle of synthesising an image with healthy appearance in order to identify pathology in a real scan. Similar to previous work [4, 5], we aim to produce a “pseudo-healthy” version of a particular modality without any signs of pathology. The synthetic image is then compared with the potentially pathological real image and the differences are identified.
Existing modality transformation algorithms can be divided into model and data driven approaches. In the former, intrinsic physical properties of the tissue being imaged are estimated from the available modalities [6]. Once known, a new modality can be synthesised by simulating the image acquisition protocol. However, accurate estimation of these tissue properties requires particular acquisition protocols, which are not routinely carried out. The more commonly used algorithms therefore rely on a data driven approach where the synthesised image is derived directly from the intensities of the source image(s). Most state of the art algorithms employ a patch based, dictionary learning approach [2, 3, 5]. A dictionary of source-target patch pairs is stored with synthesis being performed by using the target patch with the corresponding source patch which most closely matches a given patch in source image. Approaches using a restricted nearest neighbour search [5], compressed sensing [3] and sparse coding [2] are among those proposed for searching and combining patches from the dictionary. Recently, deep learning approaches have also received attention [7] with good results. Another data driven approach, to which our proposed method is more closely related, uses local joint histograms to find the target image intensity with which a given source image intensity most commonly co-occurs [8].
The problem when employing these existing methods for the synthesis of pseudo-healthy images is that WMH are often synthesised. This is because the relationship between WMH intensities in T\(_1\) -weighted and FLAIR images can be similar to that of gray matter (GM) [9]. Existing methods will learn this WMH-GM similarity and synthesise WMH as hyperintense. Whilst this ability has been exploited for better T\(_1\) -weighted image segmentations [10], it is not desirable for the production of pseudo-healthy images.
In this paper we present a novel modality transformation method, which can be used effectively to generate pseudo-healthy images. The proposed approach exploits only information from small neighbourhoods around a given voxel to predict a synthetic intensity, and will therefore not be influenced by the WMH-GM relationship described above, which would be learnt in other regions of the brain. We employ this method to address the problem of WMH segmentation with results that compare favourably with two established reference methods from the Lesion Segmentation Toolbox (LST). Finally, we demonstrate the clinical potential of the proposed automatic lesion segmentation method when applied to the identification of VD in a clinical dataset, and show performance comparable to identification using manually assessed Fazeka scores, a clinical measure of WMH.
2 Method
In the following, we describe the two essential components of the proposed PHI-Syn method. A pseudo-healthy FLAIR image is first synthesised from a patient’s T\(_1\) -weighted image. The estimated FLAIR image is then compared to the real FLAIR image of the patient and abnormally hyperintense regions are identified.
2.1 Image Synthesis
To synthesise a subject’s FLAIR image that does not exhibit WMH (if present in the T1 weighted image), we propose a method that relies on voxel-wise kernel regression to learn local relationships between intensities in T\(_1\) -weighted and FLAIR image pairs of healthy subjects. The regression model is then used to synthesise pseudo-healthy FLAIR images from T\(_1\) -weighted images. There are three factors that enable the synthesis of a pseudo-healthy image: (a) the pathology is in general not prominent in T\(_1\) -weighted images; (b) the model is trained on image pairs of healthy subjects without WMH and does therefore not learn how to synthesise pathology; (c) the method uses only information from small local regions from the training data to synthesise each voxel, meaning intensity relationships learnt from other regions of the brain will not be applied.
Preprocessing. As voxel directly between scans it is important that all images of a respective modality are on the same intensity scale. We employ the following steps to ensure that the distributions of intensities within tissue classes are the same across all images of that modality.
Each T\(_1\) -weighted image is bias field corrected [11], skull stripped [12] and anatomically segmented [13]. GM and white matter (WM) masks are generated from these segmentations and a transformation from native to MNI space using free form deformations (FFD) [14] is computed.
Intensity normalisation is a key step that is particularly challenging in the presence of pathology, as it needs to be ensured that varying levels of pathology have no impact on intensity mappings. To do this we use the method employed in [15] using the previously computed WM and GM masks. This approach establishes a robust fixed point as the mean of the average intensities of the WM and GM which is then set to a common value. This method has the advantage of only using information from regions in which we are highly confident the tissue type is either healthy WM or GM and is therefore unaffected by WMH.
FLAIR images are also bias corrected and masked using the brain mask derived from the T\(_1\) -weighted image, rigidly transformed into the native space of the FLAIR image. The GM and WM masks are also transformed into FLAIR space and used for intensity normalisation.
Synthesis Training. The training set consists of pairs of T\(_1\) -weighted and FLAIR images, \(\mathbf T ^\mathrm {train}\) and \(\mathbf F ^\mathrm {train}\) respectively. All images are aligned to MNI space and re-sampled on a 1 mm\(^3\) voxel lattice using linear interpolation. The T\(_1\) -weighted image intensities are rescaled to the range [0; m], where m is the number of points the model will be evaluated at. The value of m will ultimately control the size and training time of the model, with a larger value leading to more accurate results. A kernel regression model with bandwidth h is generated at each voxel \(\mathbf x \) relating the T\(_1\) -weighted and FLAIR intensities in an s-by-s-by-s patch around x. The result of evaluating the model at each k in the range [1, m] is stored in vector \(\mathbf{M}_\mathbf{x}\) (1) using the regression model outlined below.
where \(N(\mathbf x ;\mathbf T ,s)\) and \(N(\mathbf x ;\mathbf F ,s)\) return a vector containing the voxels in a patch around voxel x of size s-by-s-by-s from each image in \(\mathbf T \) and \(\mathbf F \) respectively.
Synthesis Testing. To estimate the synthetic FLAIR image, the intensities of the T\(_1\) -weighted image, T, are rescaled to be between 0 and m and transformed into the native space of the FLAIR image along with mapping M. The synthetic image S at voxel \(\mathbf x \) is then calculated,
where \(L_{\mathrm {FT}}\) denotes the rigid transformation between FLAIR and T\(_1\) -weighted image spaces and \(L_{\mathrm {FM}}\) the FFD transformation between FLAIR and MNI spaces.
2.2 Lesion Segmentation
We identify lesions by detecting regions which are hyperintense in the FLAIR image relative to the synthetic image. A consequence of using kernel regression is a tendency for synthesised image intensities to be closer to the mean intensity in the respective regions, resulting in reduced image contrast. The method used for intensity normalisation determines two values corresponding to the mean intensities of healthy GM and WM. To correct tissue contrast we scale the synthetic image such that these two values match those of the acquired FLAIR images.
The confidence \(\varvec{\mathrm {\Sigma }}\) in the intensity-normalised synthesised images is computed by calculating the standard deviation of the errors achieved on the training images in MNI space. This yields a spatial variance map, which is used to assign a relative confidence to the synthesised intensities at each voxel. A z-score corresponding to the likelihood of the intensity of a voxel x falling outside of what is expected is then computed, \(\mathbf Z _\mathbf x ^\mathrm {S} = (\mathbf F _\mathbf x -\mathbf S _\mathbf x ) / \varvec{\mathrm {\Sigma }}_\mathbf{x '}\) where \(\mathbf x ' = L_{\mathrm {FM}}(\mathbf x ),\) which is turned into a p-value, \(\mathbf P ^\mathrm {S}_\mathbf x \). Another set of p-values \(\mathbf P ^\mathrm {F}\) are computed to reflect areas of hyperintensity in the FLAIR image. An individual image based z-score will be affected by the volume of hyperintense regions in the image. Therefore, the mean and standard deviation required to compute \(\mathbf P ^\mathrm {F}\) are estimated from intensity histograms of the normalised training images.
We combine the previously computed anatomical segmentations to create a binary mask \(\mathbf B \) to constrain the search for WMH to areas of the brain where they are expected to be present. This mask includes the WM and a number of cortical and deep GM structures which are close to areas where WMH is commonly found. The final WMH likelihood \(\mathbf L \) at voxel \(\mathbf x \) was thus computed by the multiplication of the three likelihood maps at \(\mathbf x \), \( \mathbf L _\mathbf{x } = \mathbf P _\mathbf x ^\mathrm {F} \mathbf P _\mathbf x ^\mathrm {S} \mathbf B _\mathbf x . \)
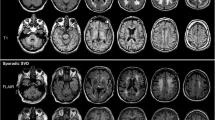
There are two main types of WMH. Small punctate lesions such as those visible in Fig. 2, and larger, lower intensity regions, such as those seen in Fig. 3. To account for both types, a low threshold \(t_\mathrm {l}\) is first used to binarize \(\mathbf L \) and only large (>200 mm\(^3\)) areas are kept. A higher threshold \(t_\mathrm {h}\) is then used and the initial segmentation taken to be the union of these two segmentations.
A refinement step is then carried out in which segmentations are repeatedly grown into neighbouring voxels with an intensity which lies above the lowest intensity in the original segmentation. A 5 mm limit is imposed to prevent the growth of incorrect “lesions”. Finally, small (<20 mm\(^3\)) segmentations are removed as these are often visually indistinguishable from noise.
3 Experiments and Results
Experiments were carried out to evaluate PHI-Syn against two widely used segmentation methods, and to investigate its applicability in a clinical setting.
3.1 Data
In the first experiment, we used a stroke dataset of 42 patients (mean age 64.9 years (SD 10)) from a study of mild stroke [16], obtained as described in [17]. Images were acquired with an in plane resolution of 0.94-by-0.94 mm and slice thickness 4 mm. Reference WMH segmentations were obtained semi-automatically. In a second experiment we used a dementia dataset of 468 subjects from VUMC, Amsterdam, which were provided for the PredictND studyFootnote 1. This clinical dataset contains MRI scans of varying resolutions and field strengths along with clinical scores for patients with a diagnosis of either subjective memory complaints (110), Alzheimer’s Disease (204), Frontotemporal Dementia (88), Lewy Body Dementia (47) and Vascular Dementia (19). Clinical Fazekas scores were visually assessed. Of the 468 subjects, 173 had a Fazekas score of 0, 205 (score 1), 61 (2) and 29 (3). Images were acquired at 3T (295), 1.5T (91) and 1T (82).
For both experiments, the synthesis model was trained on 31 subjects selected from the dementia dataset as the visually least pathological. However, a consequence of training on subjects from an elderly dataset is that most subjects have a small degree of periventricular WMH due to their age. These were, undesirably, reproduced in the synthetic images. An additional post-processing step on the synthetic images was added to address this: Voxels located up to 15 mm from the ventricular wall were capped at a maximum intensity value equal to the average between the mean FLAIR intensities of GM and WM. A special case must then be made for the region around posterior prolongations of the ventricles where non-pathological low level hyperintense streaks are often seen. A squaring of the probabilities in these regions was sufficient to ensure that true lesions would still be segmented, whilst the probabilities corresponding to low level hyperintensities would be suppressed. This additional step would not be required if a set of pathology free subjects were available for a particular application. Free parameters for synthesis were chosen empirically for all experiments as: \(m = 100\), \(s = 7\), \(h = 5\) as they balanced model size and computational speed with visually appealing synthesised images.
3.2 Evaluation Against Reference Segmentations
In this experiment we employ the stroke dataset to compare the proposed method against two standard methods from the Lesion Segmentation Toolbox v.2.0.12Footnote 2 - the Lesion Growth Algorithm (LST-LGA) [18] and the Lesion Prediction Algorithm (LST-LPA). The former requires a T\(_1\) -weighted image as well as a FLAIR image. White matter, grey matter and CSF segmentations are obtained from the T\(_1\) -weighted image and used to create a lesion belief map from the FLAIR image. This is first thresholded at a value \(\kappa \) and the resulting segmentations are grown along hyperintense voxels. LST-LPA is a supervised method for which a logistic regression model was trained on 53 Multiple Sclerosis patients with severe white matter lesion loads. Both methods output a lesion probability map, which the documentation suggests should be thresholded at 0.5. For LST-LGA, a \(\kappa \) of 0.3 is the default but it is strongly suggested that this is optimised. For each method, we provide results for both the suggested parameters and parameters selected through a grid search which maximised Dice Similarity Coefficient (DSC). These were found to be: LST-LGA*, \(\kappa = 0.07\), threshold \(=0.10\). LST-LPA*, threshold \(=0.10\). PHI-Syn*, \(t_\mathrm {l}=0.76\), \(t_\mathrm {h}=0.85\).
Segmentations were compared across a set of quantitative measures used previously in the ISLES 2015 segmentation challengeFootnote 3: Average Symmetric Surface Distance (ASSD, mm), DSC, Hausdorff Distance (HD, mm), Precision and Recall. A further metric, Load Correlation (LC) defined as the correlation between automatic and reference segmentation volumes over all subjects was also used with results shown in Table 1.
3.3 Relation to Clinical Scores
The Fazekas score is a commonly used four point clinical score derived from FLAIR images relating to the presence and degree of WMH [19]. It has particular use in the diagnosis of VD as it relates to the most significant pathological changes in the patient’s brain.
In this experiment we predicted synthetic Fazekas scores from the segmentations given by PHI-Syn and compared them to clinical Fazekas scores. The experiment was carried out using 1000 runs of 10-fold cross validation. Three features were extracted from the PHI-Syn segmentations: volume of lesions as a percentage of WM, volume of lesions greater than 15 mm from the ventricles as a percentage of WM, and volume of the largest lesion. At each fold, the training set was balanced by oversampling under-represented Fazekas scores classes. A set of support vector machine (SVM) classifiers using an error-correcting output code schema for multi-class classification (classifier A) were trained on the training set to predict a synthetic Fazekas score. A further binary SVM (classifier B) was trained on data balanced with respect to disease to predict a diagnosis of VD or not-VD from the clinical Fazekas scores. Synthetic Fazekas scores were then calculated for subjects in the test set using classifier A and diagnoses were predicted from both the true and synthetic Fazekas scores using classifier B.
The balanced accuracy for predicting a synthetic Fazekas score using classifier A was 61.5 %, with only 4 %/0.25 % being predicted a score of more than 1/2 points from their respective true clinical score. The balanced accuracy for predicting a diagnosis was 83.3 % from the true Fazekas scores and 83.9 % from the synthetic Fazekas scores with standard deviations of 1.2 % and 3.3 % respectively.
4 Discussion
The conducted experiments show that PHI-Syn achieves the highest or statistically joint highest scores in ASSD, DSC, HD, Recall and LC. Figure 3 shows three sample segmentations. Visual examination confirms superior ability of PHI-Syn, as compared to LST-LPA*, to locate smaller lesions distant from the ventricles (A and C). A lower HD score supports this observation. Instances in which PHI-Syn tends to be outperformed by LST-LPA* include cases of large areas of low intensity (B). Objective measurements and visual inspection both suggest PHI-Syn performs well in the majority of situations. A limitation of this experiment is that only WMH are included in the reference segmentation, and other hyperintense appearing pathologies such as stroke lesions, are not. All methods tested will identify all hyperintensities and as such the results of these experiments can only be used to compare methods relative to each other, and should not be used as an indicator of expected performance on another dataset.
A sample of two FLAIR images (bottom) and segmentations (top). Reference (blue), LST-LPA* (green) and PHI-Syn (red) segmentations are shown. Colours are additively mixed where segmentations overlap. e.g. purple indicates overlap between PHI-Syn and the reference, cyan: LST-LPA* and reference, yellow: LST-LPA* and PHI-Syn, white: all methods. Arrows draw attention to regions of particular interest. (Color figure online)
The balanced accuracy of predicted diagnoses from the synthetic Fazekas scores is comparable to those predicted when using the clinically assessed Fazekas scores, however the data is highly imbalanced and as such the balanced accuracy can be unstable and susceptible to noise. Future work involves using more VD cases to further investigate using synthetic over true Fazekas scores. However, these initial results suggest that a synthesised score is a valuable marker in cases where a clinical Fazekas score is not available.
We have shown that effective synthesis of pseudo-healthy images can be carried out using voxel-wise kernel regression, and that these images can be used to reliably identify WMH. We have also shown that the resulting segmentations can predict a Fazekas score which discriminates between vascular and non-vascular cases of dementia comparably to labour-intensive clinical scores.
References
Debette, S., Markus, H.S.: The clinical importance of white matter hyperintensities on brain magnetic resonance imaging: systematic review and meta-analysis. BMJ 341, c3666 (2010)
Cao, T., Zach, C., Modla, S., Powell, D., Czymmek, K., Niethammer, M.: Registration for correlative microscopy using image analogies. In: Dawant, B.M., Christensen, G.E., Fitzpatrick, J.M., Rueckert, D. (eds.) WBIR 2012. LNCS, vol. 7359, pp. 296–306. Springer, Heidelberg (2012)
Roy, S., Carass, A., Prince, J.: A compressed sensing approach for MR tissue contrast synthesis. In: Székely, G., Hahn, H.K. (eds.) IPMI 2011. LNCS, vol. 6801, pp. 371–383. Springer, Heidelberg (2011). doi:10.1007/978-3-642-22092-0_31
Tsunoda, Y., Moribe, M., Orii, H., Kawano, H., Maeda, H.: Pseudo-normal image synthesis from chest radiograph database for lung nodule detection. Adv. Intell. Syst. Comput. 268, 147–155 (2014)
Ye, D.H., Zikic, D., Glocker, B., Criminisi, A., Konukoglu, E.: Modality propagation: coherent synthesis of subject-specific scans with data-driven regularization. In: Mori, K., Sakuma, I., Sato, Y., Barillot, C., Navab, N. (eds.) MICCAI 2013, Part I. LNCS, vol. 8149, pp. 606–613. Springer, Heidelberg (2013)
Fischl, B., Salat, D.H., van der Kouwe, A.J.W., Makris, N., Ségonne, F., Quinn, B.T., Dale, A.M.: Sequence-independent segmentation of magnetic resonance images. Neuroimage 23, S69–S84 (2004)
Nguyen, H., Zhou, K., Vemulapalli, R.: Cross-domain synthesis of medical images using efficient location-sensitive deep network. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9349, pp. 677–684. Springer, Heidelberg (2015). doi:10.1007/978-3-319-24553-9_83
Kroon, D.-J., Slump, C.H.: MRI modalitiy transformation in demon registration. ISBI 2009, 963–966 (2009)
Roy, S., Carass, A., Prince, J.: Magnetic resonance image example based contrast synthesis. IEEE Trans. Med. Imaging 32(12), 2348–2363 (2013)
Roy, S., Carass, A., Shiee, N., Pham, D.L.: MR contrast synthesis for lesion segmentation. ISBI 2010, 932–935 (2010)
Tustison, N.J., Avants, B.B., Cook, P.A., Zheng, Y., Egan, A., Yushkevich, P.A., Gee, J.C.: N4ITK: improved N3 bias correction. IEEE Trans. Med. Imaging 29(6), 1310–1320 (2010)
Heckemann, R.A., Ledig, C., Gray, K.R., Aljabar, P., Rueckert, D., Hajnal, J.V., Hammers, A.: Brain extraction using label propagation, group agreement: pincram. PloS ONE 10(7), e0129211 (2015)
Ledig, C., Heckemann, R.A., Hammers, A., Lopez, J.C., Newcombe, V.F., Makropoulos, A., Lötjönen, J., Menon, D.K., Rueckert, D.: Robust whole-brain segmentation: application to traumatic brain injury. Med. Image Anal. 21(1), 40–58 (2015)
Rueckert, D., Sonoda, L.I., Hayes, C., Hill, D.L., Leach, M.O., Hawkes, D.J.: Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans. Med. Imaging 18(8), 712–721 (1999)
Huppertz, H.J., Wagner, J., Weber, B., House, P., Urbach, H.: Automated quantitative FLAIR analysis in hippocampal sclerosis. Epilepsy Res. 97(1), 146–156 (2011)
Heye, A.K., Thrippleton, M.J., Chappell, F.M., Hernández, M., Armitage, P.A., Makin, S.D., Maniega, S.M., Sakka, E., Flatman, P.W., Dennis, M.S., Wardlaw, J.M.: Blood pressure, sodium: association with MRI markers in cerebral small vessel disease. J. Cereb. Blood Flow Metab. 36(1), 264–274 (2016)
Hernández, M.D.C.V., Armitage, P.A., Thrippleton, M.J., Chappell, F., Sandeman, E., Maniega, S.M., Shuler, K., Wardlaw, J.M.: Rationale, design and methodology of the image analysis protocol for studies of patients with cerebral small vessel disease and mild stroke. Brain Behav. 5(12), e00415 (2015)
Schmidt, P., Gaser, C., Arsic, M., Buck, D., Förschler, A., Berthele, A., Hoshi, M., Ilg, R., Schmid, V.J., Zimmer, C., Hemmer, B.: An automated tool for detection of FLAIR-hyperintense white-matter lesions in multiple sclerosis. Neuroimage 59(4), 3774–3783 (2012)
Fazekas, F., Chawluk, J.B., Alavi, A., Hurtig, H.I., Zimmerman, R.A.: MR signal abnormalities at 1.5 T in Alzheimer’s dementia and normal aging. Am. J. Neuroradiol. 8(3), 421–426 (1987)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing AG
About this paper
Cite this paper
Bowles, C. et al. (2016). Pseudo-healthy Image Synthesis for White Matter Lesion Segmentation. In: Tsaftaris, S., Gooya, A., Frangi, A., Prince, J. (eds) Simulation and Synthesis in Medical Imaging. SASHIMI 2016. Lecture Notes in Computer Science(), vol 9968. Springer, Cham. https://doi.org/10.1007/978-3-319-46630-9_9
Download citation
DOI: https://doi.org/10.1007/978-3-319-46630-9_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-46629-3
Online ISBN: 978-3-319-46630-9
eBook Packages: Computer ScienceComputer Science (R0)