Abstract
Microsatellites, also referred to as short tandem repeats (STRs) or simple sequence repeats (SSRs) are short sequences of tandem repeats of 1–6 bp in length in clusters of less than 150 bp flanked by sections of non-repetitive unique sequences that are scattered throughout the nuclear genome. These markers are co-dominant and hypervariable, revealing many alleles per locus; they are inherited in Mendelian fashion making them useful for detecting genetic variability within species. Once isolated and characterized, microsatellites can also be used in closely related taxa. Microsatellites can be amplified, even from highly degraded DNA, and are very simple to score. More importantly, these markers are highly polymorphic due to the plethora of variations in the repeat motifs. Several studies have endorsed microsatellite markers as an effective genetic tool to determine the historical distinctiveness of populations, and hence, the designation of species. Being highly polymorphic and selectively neutral, microsatellite markers offer a powerful genetic tool for investigating population structure, colonization processes, temporal and spatial population dynamics and evolutionary trends of insect pests. Furthermore, these markers have been successfully applied to different invasive fruit fly species to infer the evolutionary aspects underlying their invasive processes. Microsatellite markers have offered an analytical tool for the study of fruit fly invasion genetics as exemplified for the Mediterranean fruit fly, Ceratitis capitata. Herein, a detailed utility of microsatellite markers in inferring invasion histories of key fruit flies of economic importance is given, with a special focus on invasion into Africa of Bactrocera dorsalis.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction to the Use of Molecular Markers
In Africa, and sub-Saharan Africa in particular, the agricultural sector is the backbone of most economies in pursuit of sustainable development. Specifically, the horticultural sub-sector is a precious tool that is contributing to poverty alleviation by increasing food security and income generation for continued economic growth. However, this sub-sector is constrained by a number of biotic and abiotic factors. Amongst the former is infestation by tephritid fruit flies that are well recognised as a group of pests of economic significance (Ekesi and Billah 2007). Sub-Saharan Africa is the native home to about 915 fruit fly species out of which 299 species belong to the genera Ceratitis MacLeay, Trirhithrum Bezzi and Dacus Fabricius (White and Elson-Harris 1992). Due to the globalisation of trade, the emergent tourism industry, porous borders and poor phytosanitary expertise, the likelihood of inadvertent introduction and spread of exotic fruit fly species across the continent is escalating. Therefore, measures to strengthen the phytosanitary/quarantine infrastructure are of paramount importance to avert the establishment of alien invasive pests.
Knowledge of the genetic structure and geographical variability of invasive fruit fly species is a vital pre-requisite to implementing quarantine, control and eradication measures (Roderick and Navajas 2003; Malacrida et al. 2007). In the past, morphological characters were considered sufficient to describe species. However, morphological characters for species delineation have several limitations. The existence of homoplasy amongst characters and cryptic speciation in some insect families, as in the tephritid fruit flies, make species-level descriptions based on adult and larval morphology extremely difficult (Armstrong et al. 1997; De Meyer 1998; McPheron 2000). This has led taxonomists and quarantine officials to seek alternative ways to identify tephritid fruit flies, including the use of molecular markers (Sonvinco et al. 1996; Armstrong et al. 1997; Morrow et al. 2000). Indeed, biochemical (allozymes) and DNA molecular markers have been used to elucidate the variability in population structure of several tephritid species (Baruffi et al. 1995; Malacrida et al. 1996). In addition to this, molecular markers have advantages over biochemical tools and hence have become the current tool of choice. Molecular markers are site specific DNA sequences that are easily detected in the genome. These markers are neutral and can be utilised in a number of ways including, but not limited to: the analysis of genetic variability; to make inferences on population genetic structure; for DNA fingerprinting; for chromosome mapping; and for the identification and description of species. There are a number of molecular markers that have been developed and are in use: RAPDs, RFLPs, PCR-RFLPs, microsatellites, minisatellites and SNPs amongst others (Baruffi et al. 1995; Barr et al. 2006; Malacrida et al. 1996; Khamis et al. 2008). The best molecular markers have a combination of the following properties: they should occur frequently in the genome; be co-dominant; be highly polymorphic; be vastly reproducible and transferable to many taxa; be cheap to develop and apply; and not be affected by environmental conditions.
In some frugivorous tephritid fruit fly species, diagnostic morphological characters for the identification of adult flies have been made available (Adsavakulchai et al. 1999; De Meyer 2005; Drew et al. 2006, 2008). However, inconsistencies in the limits of fruit fly species identification based on conventional adult morphological features together with overlapping host and geographical ranges, have profound effects on quarantine, management and biological studies of these species (Clarke et al. 2005). These limitations have led to the development and improvement of molecular tools for identification and classification of the fruit flies pest species, and for understanding their population structure (Armstrong et al. 1997; Malacrida et al. 1998; Han and McPheron 1997; Manni et al. 2015). Furthermore, because molecular markers are phenotypically neutral and resistant to environmental cues (unlike morphological characters) they can be used as a single reliable taxonomic tool. Several molecular markers are available to discriminate amongst tephritid fruit fly species. These markers have successfully been used to validate species (Khamis et al. 2012; Schutze et al. 2014), infer phylogenetic relationships (Boykin et al. 2014), verify intra-specific variation between populations (Bonizzoni et al. 2000; Baliraine et al. 2004) and trace the routes of invasion of pest fruit fly species (Aketarawong et al. 2007, 2014; Khamis et al. 2009).
2 Application of Microsatellites: Advantages and Limitations
Several studies have endorsed microsatellite markers as an effective genetic tool to determine the historical distinctiveness of populations, and hence, the designation of species (Hedrick et al. 2001; Wang et al. 2001). Microsatellites are short sequences of tandem repeats of 1–6 bp in length in clusters of less than 150 bp in length flanked by sections of non-repetitive unique sequences that are scattered throughout the nuclear genome and mostly associated with conserved loci containing coding regions (Loxdale and Lushai 1998). Microsatellites have also been referred to as short tandem repeats (STRs) (Edwards et al. 1991) or simple sequence repeats (SSRs) (Jacob et al. 1991). These markers are co-dominant and hypervariable, revealing many alleles per locus; they are inherited in Mendelian fashion making them useful for detecting genetic variability within species. Once these markers have been isolated and characterized, they may also be used in closely related taxa. Microsatellites can be amplified even from highly degraded DNA and are very simple to score (Bruford and Wayne 1993). More importantly, these markers are highly polymorphic due to the plethora of variations in the repeat motifs. In 1989, Weber and May developed a universal method for isolating microsatellites. The polymorphism of these markers was confirmed by Litt and Luty (1989) who detected allelic variants amongst individuals using amplification of the (TG)n microsatellites in the human actin gene. Although the origin of microsatellite polymorphism is still debated, it is likely to be due to slipped-strand mispairing (Levinson and Gutman 1987), slippage events during DNA replication/repair/recombination (Schlötterer and Tautz 1992) or asymmetrical cross-over between sister chromatids (Innan et al. 1997).
Despite the uncertainty surrounding microsatellite evolution, they have been adopted widely and applied in many fields of study since their initial description by Hamada et al. (1982). Microsatellites markers are an invaluable method for genome mapping in many organisms (Schuler et al. 1996), and are also applicable in various fields ranging from ancient and forensic DNA studies, to population genetics and conservation/management of biological resources (Jarne and Lagoda 1996). Furthermore, microsatellites can be used for species identification, genetic tagging, breeding studies, reproductive biology, taxonomy, phylogenetic studies, disease diagnostics and genetic diversity studies (Abdul-Muneer 2014). Moreover, these markers offer a diagnostic tool that can differentiate between species not easily separated by morphological traits (Kinyanjui et al. 2016). The usefulness of microsatellite markers is evidenced by the rising numbers of mapped genes based on microsatellites (Schuler et al. 1996).
The wide applicability of microsatellite markers is associated with their many advantages. Microsatellite markers are robust and very informative compared with other markers such as RFLPs, RAPDs and AFLPs (He et al. 2003; Lee et al. 2004). Importantly, microsatellite markers are PCR-based and therefore require low quantities of DNA. The primer lengths and high annealing temperatures of microsatellite markers in genotyping guarantee their reproducibility. Once isolated, microsatellite markers can be cross amplified amongst closely related species (Baliraine et al. 2002). Furthermore, these markers can be multiplexed hence reducing the time and cost of analysis. Although microsatellites are a successful tool in genetics, they do have some drawbacks. Firstly, microsatellite markers need to be isolated de novo for most species being analysed for the first time; this isolation is expensive, laborious and time consuming (Zane et al. 2002). Secondly, the likelihood of null alleles occuring when using ancient or degraded DNA is very high, leading to difficulties in estimating allelic frequencies and heterozygosity (Kumar et al. 2009). Last but not least, homoplasy in some organisms is a common problem for the application of microsatellite markers in phylogenetics, leading to false identification of species descents (Estoup et al. 2002).
3 Microsatellite Markers: Isolation and Characterization
There are several methods that have been described for the isolation of microsatellite loci; the first was the de novo isolation described by Rassmann et al. (1991). This protocol has become the traditional method for microsatellite isolation and it involved identification of microsatellite-containing clones by colony hybridization with their respective specific probes. Following subsequent advances in methodologies and DNA sequencing techniques, isolation of microsatellites can now be achieved either by: (i) Constructing and screening either enriched or non-enriched genomic libraries or by utilizing the products generated by other molecular markers or by the application of next-generation sequencing systems; (ii) Making use of the EST sequences already deposited in the public domain databases or sequencing PCR products generated by consensus/universal primers; and (iii) Testing the amplification potential of other microsatellite markers developed in other related species (i.e. cross-species amplification).
3.1 Isolation of Microsatellites from Enriched Genomic DNA Libraries
This can be achieved through selective hybridization methods where the genomic DNA undergoes fragmentation either by use of restriction enzymes, sonication or, less frequently, nebulisation (Senan et al. 2014). Fragments of DNA in the range of 300–700 bp in length are selected and ligated into a common vector. Ligation can be done directly or after ligation to specific adapters (Zane et al. 2002). The DNA is then denatured and subjected to enrichment by hybridization with either: (i) biotinylated oligos followed by capture of biotinylated hybrids (oligo-bound DNA fragments) in a vectrex-aridin matrix (Kandpal et al. 1994); (ii) oligonucleotides (oligos) bound to nylon membranes (Karagyozov et al. 1993; Chen et al. 1995; Edwards et al. 1996) (iii) 5’ biotinylated repeat oligos and subsequent capture by biotinylated hybrids by streptavidin coated magnetic beads (Aketarawong et al. 2006; Khamis et al. 2008); (iv) biotinylated microsatellite-probe-streptavidin coated magnetic bead complex (White and Powell 1997). Screening for positive clones is achieved by means of southern hybridization using the probes mentioned above and after blotting the bacterial colonies onto nylon membranes. Colony transfer is done either by classical replica plating or by picking single colonies and ordering them in new arrayed plates. After successful identification of positive microsatellite-containing clones, specific primers are designed and PCR conditions are optimized to allow the amplification of each locus from different individuals of a population. The fragments are then amplified, cloned and sequenced directly and probed for the presence of microsatellites. The efficiency of this approach entirely depends on the specific binding of streptavidin coated beads to biotin.
Ostrander et al. (1992) and Paetkau (1999) described protocols that allow the selective amplification of microsatellites containing genomic DNA using very specific primers. This is known as the primer extension method and it relies on the construction of a primary genomic DNA library in a phagemid vector to recover the library as single stranded DNA which is then subjected to primer extension using repeat specific non-biotinylated oligos or 5’biotinylated oligos. Ostrander et al. (1992) further demonstrated primer extension steps that selectively generated only double-stranded products from vectors containing the tandem repeats of interest, and then transformed them in to E. coli cells. Streptavidin coated magnetic beads were used to selectively pick out the 5’biotinylated hybrids and convert them in to double-stranded DNA via a second round of primer extension for transformation. Pandolfo (1992) described the ligation of a vectorette (i.e. a linker containing a non-complementary region) to restricted yeast artificial chromosome (YAC) DNA. Using microsatellite-specific primers and universal vector primers, the vectorette-ligated DNA could be amplified and the products cloned and sequenced to probe for the desired repeat loci.
There are plenty of enrichment protocols available but the selective hybridization stands out as it allows for enrichment and selection prior to cloning, thereby providing a faster and easier method to handle multiple samples (Senan et al. 2014; Glenn and Schable 2005). This method is very simple, reproducible and cost effective for isolating microsatellites.
3.2 Isolation of Microsatellites from Non-Enriched Genomic DNA Libraries
Golein et al. (2006) demonstrated that genomic DNA could be restricted, ligated in to vectors and transformed to generate a non-enriched genomic DNA library. The clones were then spotted on to gridded nylon filters and screened with radio -labelled microsatellite probes or subjected to enrichment with biotin labelled-probes-streptavidin and sequenced.
3.3 Other Methods for Isolation of Microsatellites
A number of other techniques have been used to isolate microsatellite loci. These include isolation from RAPDs which involves the blotting of RAPD products on to nitrocellulose membranes which are then screened, using digoxygenin-labelled probes, for positive clones that could be detected using autoradiography. Another technique was described by Zane et al. (2002) and is known as FIASCO (Fast isolation by AFLP of sequences containing repeats). In this protocol the AFLP bands were hybridized with biotinylated probes which were then selectively probed using streptavidin-coated beads followed by cloning and sequencing of the enriched DNA fragments, to generate new microsatellite markers. However, due to the labour intensive process required for the de novo isolation of microsatellites, and in view of recent advances in DNA sequencing technologies (e.g. next generation sequencing [NGS] and better bioinformatics), these methods represent powerful alternatives to conventional methods for isolation of microsatellite markers. Large amounts of data can be produced via NGS and screening can be done using bioinformatics tools; this avoids the need for construction of microsatellite-enriched DNA libraries and provides a rapid approach for the large-scale generation of microsatellite loci. Reductions in sequencing costs will make the rapid identification of microsatellite markers even easier and cheaper. There are plenty of EST sequences that have been deposited in public domain databases (Rudd 2003). Several tools are available to mine for microsatellite loci and these include TROLL (Castelo et al. 2002), MISA (Thiel et al. 2003), SciRoKo (Kofler et al. 2007), Msatcommander (Faircloth 2008) amongst others. However, generation of these markers is limited to the availability of EST sequences, particularly if EST sequences are not deposited in publicly accessible domains/databases. Closely related individuals tend to have greater DNA conservation in their coding regions hence EST microsatellite markers cannot be used in differentiating such individuals since they show less polymorphism and are therefore less efficient (Gupta et al. 2003). Recently, the high-throughput genomic-sequencing technique has produced millions of base pairs and short fragment reads which can be screened using bioinformatics tools to identify primers that amplify a large number of polymorphic microsatellite loci (Abdelkrim et al. 2009).
4 Prospects for Tracing the Routes of Fruit Fly Invasions out of Africa and the Population Structure of Fruit Flies Using Microsatellite Markers: The Case of Ceratitis Species and Bactrocera oleae (Rossi)
Being highly polymorphic and selectively neutral, microsatellite markers offer a powerful genetic tool for investigating population structure, colonization processes, temporal and spatial population dynamics and evolutionary trends of insect pests (Wu et al. 2009). Furthermore, these markers have been successfully applied to different invasive fruit fly species to infer the evolutionary aspects underlying their invasive processes (Bonizzoni et al. 2001, 2004; Baliraine et al. 2004; Khamis et al. 2009). Due to their polymorphic nature these markers have also been utilized in the analyses of fruit fly population structure across different geographical areas, and in tracing the origins of adventive populations (Bonizzoni et al. 2000, 2001; Meixner et al. 2002). Moreover, these markers have proven to be useful for cross-species amplification to study the population structure of tephritid species when no previous genetic information was available (Baliraine et al. 2003, 2004; Shearman et al. 2006).
Globally, phytophagous members of the family Tephritidae are amongst the most important pests of fruits and vegetables. With more than 4000 species described, this family is the most diverse and contains 500 genera of which the four most economically important genera are: Ceratitis, Bactrocera, Anastrepha and Rhagoletis (White and Elson-Harris 1992). All the genera have native distribution ranges. For example, Ceratitis is an Afro-tropical genus, Bactrocera is mainly confined to the Oriental and Australasian regions, Anastrepha to South and Central America and the West Indies, while Rhagoletis has representatives in the Americas, Europe and temperate Asia (White and Elson-Harris 1992). In addition to the polyphagous nature of some species belonging to this family, several are considered highly invasive; aided by globalization of trade and poor quarantine infrastructure in the invaded countries. In recent years a member of these genera have been reported outside their native ranges. The pattern and routes of invasions of these species is of paramount importance for their management hence governments and NRI’s have mobilized extensive ecological and evolutionary genetic research on these invasive pest species (Aluja and Norrbom 2000).
Microsatellite markers have offered an analytical tool for the study of fruit fly invasion genetics as exemplified for the Mediterranean fruit fly (medfly), Ceratitis capitata (Weidemann) (Diptera: Tephritidae) (Bonizzoni et al. 2000). Ceratitis capitata is a tephritid fruit fly of global economic significance (Malacrida et al. 2007). In the past century, this pest has spread from its native Afro-tropical range to several countries including the Mediterranean basin, parts of South and Central America, and Australia (Fletcher 1989). Bonizzoni et al. (2000) isolated 43 microsatellite markers of which they used ten to unravel polymorphisms amongst C. capitata samples from six geographical populations (Kenya, La Réunion, Madeira, South Italy, Greece and Peru) from its native and invaded ranges. These markers detected a decrease in the number of polymorphisms from tropical Africa to the Mediterranean basin and to South America. Comparison of the Kenyan population with other populations in the study showed that the Kenyan population had the highest average number of alleles per locus, of which many were at low frequency, and most were private confirming an African origin for C. capitata. These results were consistent with the colonization history of C. capitata and indicative of a hierarchical migration structure through Spain and subsequently along the Mediterranean basin to the East (Malacrida et al. 1998; Gomulski et al. 1998).
Furthermore, a number of studies have demonstrated the utility of cross amplification of microsatellite markers to closely related species for population genetic studies (Baliraine et al. 2002, 2004; Shearman et al. 2006). For instance, Baliraine et al. (2002) screened 24 microsatallite markers from C. capitata for cross species amplification in the Natal fruit fly, Ceratitis rosa Karsch, Ceratitis fasciventris (Bezzi) and the mango fruit fly, Ceratitis cosyra (Walker). The sequence analysis indicated that most C. capitata-based microsatellite markers were useful for population genetic studies in the various species tested, a fact that will facilitate the tracing of the geographical origin of adventive pest populations, and assessment of their invasive potential and risk (Baliraine et al. 2002). In a similar study, Baliraine et al. (2004) compared genetic variability data from C. rosa and C. fasciventris with those derived from C. capitata to determine the geographical origin of the Ceratitis species. The results from this study confirmed the hypothesis of an East African origin for Ceratitis species (De Meyer et al. 2002).
A study by Nardi et al. (2005) used microsatellite markers and mitochondrial sequences to examine the population structure and colonization history of the olive fruit fly, Bactrocera oleae (Rossi). Their study revealed that Africa, and not the Mediterranean, is the origin of B. oleae infesting cultivated olives, which is supported by the significantly greater genetic diversity of microsatellite loci from samples collected in Africa compared with samples collected from the Mediterranean region. The results also indicated that the recent invasion of B. oleae in to the Americas most likely originated from the Mediterranean area.
5 Potential for Use of Microsatellite Markers to Infer Fruit Fly Invasion Histories: The Case of Bactrocera dorsalis (Hendel) in Africa
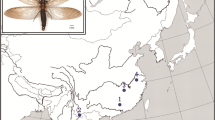
Dacine fruit flies of the genus Bactrocera Macquart (Diptera: Tephrtitidae) are also economically important fruit fly pests (White and Elson-Harris 1992). With life-history traits that include high mobility and dispersive powers, high reproductive rates and extreme polyphagy, Bactrocera species are well documented invaders and rank high on quarantine lists worldwide (European and Mediterranean Plant Protection Organization 2009). Several Bactrocera species have been introduced accidentally in to different parts of the world. This is due to the globalisation of horticultural trade and has had major economic consequences (Clarke et al. 2005; De Meyer et al. 2009). In 2003, a new fruit fly pest was detected in Kenya (Lux et al. 2003) soon after the completion of a programme of monthly fruit collections carried out between 1999 and 2003 (Copeland et al. 2004). The insect was described as Bactrocera invadens (Drew et al. 2005) and was rated as “a devastating quarantine pest” by the Inter-African Phytosanitary Council in 2005 (French 2005). Within two years of its detection in the coastal region of Kenya, the species was recorded in several other countries on the African mainland (Mwatawala et al. 2004; Drew et al. 2005; Ekesi and Billah 2007). It is now known to be present in tropical Africa from Senegal to Mozambique, as well as in the Comoro Islands in the Indian Ocean. Through integrative multidisciplinary research efforts this species has now been synonymised with the oriental fruit fly, Bactrocera dorsalis (Hendel) (Schutze et al. 2014).
Since this species was a new invader in the African continent, the timing and the pathway of its invasion were unknown. The fact that the first historical records of this pest in Africa were from the East coast may indicate that this area was the port of entry of B. dorsalis in Africa, but this hypothesis had not been tested. Moreover the native range was not well defined: it has been suggested that it ranges from Sri Lanka to the Southern Indian sub-continent from where the species may have invaded Africa (Mwatawala et al. 2004; Drew et al. 2005; Khamis et al. 2009). The detrimental effects of this invasive species stimulated several studies to define its ecological niche and invasion potential (Ekesi et al. 2006; De Meyer et al. 2009; Mwatawala et al. 2006). However, due to the ‘novelty status’ of this fruit fly as a dispersive invader, no data were available on its genetic diversity or on the degree of co-ancestry amongst African populations and the supposed native populations from Southern India and Sri Lanka. Consequently, no inferences based on genetic data were possible concerning the invasion route of B. dorsalis. To unravel the mystery surrounding this new invader, a set of 11 polymorphic microsatellite markers were isolated, characterised and utilised to evaluate the level of genetic diversity and the extent of common ancestry amongst several African populations collected across the actual invaded area in tropical Africa from East to West (Khamis et al. 2008; 2009).
Using these markers it was possible to successfully infer the dynamic aspects of the invasion of Africa by B. dorsalis, confirm its Asian origin, assess the diversity of African populations and its invasion routes in to Africa. The genetic data generated by microsatellite markers left no doubt that Sri Lanka was within the native range of B. dorsalis as the sample from there was characterized by all the genetic features expected in a large population from a native area; these included a large number of alleles coupled with a large number of private alleles occurring at high frequency. The Sri Lankan sample was also clearly genetically separate from the African samples and only a small percentage of genomes from Sri Lankan flies could be found in African flies (Fig. 4.1). From throughout the invaded range of East, Central and West Africa, the genetic data also suggested the presence of populations with relatively high levels of genetic diversity associated with limited geographic structure. Furthermore, although the invasion was a relatively recent event in Africa, there was no genetic footprint of bottlenecks although populations appeared large enough to maintain, especially in the West, a relatively large number of alleles with a low frequency. All these genetic features suggest a process of rapid population growth and expansion. The markers also identified genotypes that, when analysed using Structure 2.2 (a program that infers genetic clustering of populations using the Bayesian clustering algorithm; Pritchard et al. 2000) were present throughout Africa, i.e. some genotypes of East African flies could not be distinguished from West African flies (Fig. 4.1). Instead, the populations clustered into four genetic groups, three African clusters and the Sri Lankan cluster. Likewise, the genetic data demonstrated that the invasion and dispersal pattern of B. dorsalis in Africa was rapid and apparently chaotic, with the potential for multiple introductions as suggested from hypothetical outbreaks. Also analysis of the genotypes identified by the programs Structure 2.2 and GenClass 2.0 (which assigns or excludes reference populations as possible origins of individuals, on the basis of multi-locus genotypes) (Piry et al. 2004), allowed the main pathway of dispersal of B. dorsalis in to Africa to be inferred. Two results were found: (a) a higher or equal rate of co-ancestry of eastern and western flies in two African clusters (Fig. 4.1); (b) the major average assignment probability of eastern flies to the west than vice-versa (Table 4.1) (Khamis et al. 2009). These two results support the fact that the invasion of this pest began in East Africa. Based on their high values of co-ancestry, coastal regions of East Africa, where B. dorsalis was first found (i.e. Kenya and / or Tanzania), were consistently identified as the places from where the African invasion probably started. Another result was the major, although low, average assignment probability of East African flies to Sri Lanka compared with West Africa to Sri Lanka. These genetic data were consistent with the supposition that East Africa was the port of entry for B. dorsalis. Another very important aspect of the study concerned the Nigerian population of B. dorsalis which occupied its own cluster, suggesting that this outbreak in the West arose at the same time as the other two outbreaks in the East.
6 Conclusion
This review, although focused on only a few species of tephritid fruit flies, provides evidence that microsatellite variation can play an important role in the study of fruit fly population dynamics. These markers enable researchers to scrutinize variability within populations and the rate of divergence amongst populations. The information gained provides insights into the population structure of tephritid species present in Africa, allowing inferences to be made concerning their source areas and invasion histories. Moreover, these population data are of paramount importance for implementation of eco-friendly sustainable management strategies against fruit flies. Establishing the origin of pest species has facilitated the identification and introduction of biological control agents (i.e. parasitoids) from the pest’s native region, for use in Africa.
References
Abdelkrim J, Robertson BC, Stanton J-A L, Gemmell NJ (2009) Fast, cost-effective development of species-specific microsatellite markers by genomic sequencing. BioTechniques 46:185–192
Abdul-Muneer PM (2014) Application of Microsatellite Markers in Conservation Genetics and Fisheries Management: Recent Advances in Population Structure Analysis and Conservation strategies. Hindawi Publishing Corporation, Gen Res Intern Volume 2014, Article ID 691759, 11. http://dx.doi.org/10.1155/2014/691759
Adsavakulchai A, Baimai V, Prachyabrued W, Grote PJ, Lertlum S (1999) Morphometric study for identification of the Bactrocera dorsalis complex (Diptera: Tephritidae) using wing image analysis. Biotropica 13:37–48
Aketarawong N, Bonizzoni M, Malacrida AR, Gasperi G, Thanaphum S (2006) Seventeen novel microsatellite markers from an enriched library of the pest species Bactrocera dorsalis sensu stricto. Mol Ecol Notes 6:1138–1140
Aketarawong N, Bonizzoni M, Thanaphum S, Gomulski LM, Gasperi G, Malacrida AR, Gugliemino CR (2007) Inferences on the population structure and colonization process of the invasive oriental fruit fly, Bactrocera dorsalis (Hendel). Mol Ecol 16:3522–3532
Aketarawong N, Guglielmino CR, Karam N, Falchetto M, Manni M, Scolari F, Gomulski LM, Gasperi G, Malacrida AR (2014) The oriental fruit fly Bactrocera dorsalis s.s. in East Asia: disentangling the different forces promoting the invasion and shaping the genetic make-up of populations. Genetica 142:201–213
Aluja M, Norrbom AL (2000) Fruit flies (Tephritidae): phylogeny and evolution of behavior. CRC Press, Boca Raton
Armstrong KF, Cameron CM, Frampton ER (1997) Fruit fly (Diptera: Tephritidae) species identification: a rapid molecular diagnostic technique for quarantine application. Bull Entomol Res 87:111–118
Baliraine FN, Bonizzoni M, Osir EO, Lux SA, Mulaa FJ, Zheng L, Gomulski LM, Gasperi G, Malacrida AR (2002) Comparative analysis of microsatellite loci in four fruit fly species of the genus Ceratitis (Diptera: Tephritidae). In: Proceedings of 6th international fruit fly symposium, 6–10 May 2002, Stellenbosch, South Africa
Baliraine FN, Bonizzoni M, Osir EO, Lux SA, Mulaa FJ, Zheng L, Gomulski LM, Gasperi G, Malacrida AR (2003) Comparative analysis of microsatellite loci in four fruit fly species of the genus Ceratitis (Diptera: Tephritidae). Bull Entomol Res 93:1–10
Baliraine FN, Bonizzoni M, Guglielmino CR, Osir EO, Lux SA, Mulaa FJ, Gomulski LM, Quilici S, Gasperi G, Malacrida AR (2004) Population genetics of the potentially invasive African fruit fly species, Ceratitis rosa and Ceratitis fasciventris (Diptera: Tephritidae). Mol Ecol 13:683–695
Barr NB, Copeland RS, De Meyer M, Masiga D, Kibogo HG, Billah MK, Osir E, Wharton RA, McPheron BA (2006) Molecular diagnostics of economically important Ceratitis fruit fly species (Diptera: Tephritidae) in Africa using PCR and RFLP analyses. Bull Entomol Res 96:505–521
Baruffi L, Damiani G, Guglielmino CR, Malacrida AR, Bandi C, Gasperi G (1995) Polymorphism within and between populations of Ceratitis capitata: comparison between RAPD and multilocus enzyme electrophoresis. Heredity 74:425–437
Bonizzoni M, Malacrida AR, Guglielmino CR, Gomulski LM, Gasperi G, Zheng L (2000) Microsatellite polymorphism in the Mediterranean fruit fly Ceratitis capitata. Insect Mol Biol 9:251–261
Bonizzoni M, Zheng L, Guglielmino CR, Haymer DS, Gasperi G, Gomulski LM, Malacrida AR (2001) Microsatellite analysis of medfly bioinfestations in California. Mol Ecol 10:2515–2524
Bonizzoni M, Guglielmino CR, Smallridge CJ, Gomulski LM, Malacrida AR, Gasperi G (2004) On the origins of medfly invasion and expansion in Australia. Mol Ecol 13:3845–3855
Boykin LM, Schutze MK, Krosch MN, Chomic A, Chapman TA, Englezou A, Armstrong KF, Clarke AR, Hailstones D, Cameron SL (2014) Multi-gene phylogenetic analysis of south-east Asian pest members of the Bactrocera dorsalis species complex (Diptera: Tephritidae) does not support current taxonomy. J Appl Entomol 138:235–253
Bruford MW, Wayne RK (1993) Microsatellite and their application to population genetic studies. Curr Opin Genet De 3:939–943
Castelo AT, Martins W, Gao GR (2002) TROLL-tandem repeat occurrence locator. Bioinformatics 18:634–636
Chen H, Pulido JC, Duyk GM (1995) MATS: A rapid and efficient method for the development of microsatellite markers from YACs. Genomics 25:1–8
Clarke AR, Armstrong KF, Carmichael AE, Milne JR, Raghu S, Roderick GK, Yeates DK (2005) Invasive phytophagous pests arising through a recent evolutionary radiation: the Bactrocera dorsalis complex of fruit flies. Ann Rev Entomol 50:293–319
Copeland RS, White IM, Okumu M, Machera P, Wharton RA (2004) Insects associated with fruits of the Oleaceae (Asteridae, Lamiales) in Kenya, with special reference to the Tephritidae (Diptera). Bishop Mus Bull Entomol 12:135–164
De Meyer M (1998) Revision of the subgenus Ceratitis (Ceratalaspis) Hancock, 1918 (Diptera: Tephritidae, Ceratitini). Bull Entomol Res 88:257–290
De Meyer M (2005) Phylogenetic relationships within the fruit fly genus Ceratitis MacLeay (Diptera: Tephritidae), derived from morphological and host plant evidence. Insect Syst Evol 36:459–480
De Meyer M, Copeland RS, Wharton RA, McPheron BA (2002) On the geographical origin of the medfly, Ceratitis capitata (Wiedemann). Abstract presented at the 6th international symposium on fruit flies of economic importance, Stellenbosch, South Africa, 6–10 May 2002.
De Meyer M, Robertson MP, Mansell MW, Ekesi S, Tsuruta K, Mwaiko W, Vayssières JF, Peterson AT (2009) Ecological niche and potential geographic distribution of the invasive fruit fly Bactrocera invadens (Diptera, Tephritidae). Bull Entomol Res 100:35–48
Drew RAI, Tsuruta K, White IM (2005) A new species of pest fruit fly (Diptera: Tephritidae: Dacinae) from Sri Lanka and Africa. Afri Entomol 13:149–154
Drew RAI, Dorji C, Romig MC, Loday P (2006) Attractiveness of various combinations of colors and shapes to females and males of Bactrocera minax (Diptera: Tephritidae) in a commercial mandarin grove in Bhutan. J Econ Entomol 99:1651–1656
Drew RAI, Raghu S, Halcoop P (2008) Bridging the morphological and biological species concepts: studies on the Bactrocera dorsalis (Hendel) complex (Diptera: Tephritidae: Dacinae) in South-east Asia. Biol J Linn Soc 93:217–226
Edwards A, Civitello A, Hammond HA, Caskey CT (1991) DNA typing and genetic mapping with trimeric and tetrameric tandem repeats. Am J Hum Genet 49:746–756
Edwards KJ, Barker JHA, Daly A, Jones C, Karp A (1996) Microsatellite libraries enriched for several microsatellite sequences in plants. Biotechniques 20:758–760
Ekesi S, Billah MK (2007) A field guide to the management of economically important Tephritid fruit flies in Africa. 2nd ed, icipe Science press B1–I4
Ekesi S, Nderitu P, Rwomushana I (2006) Field infestation, life history and demographic parameters of the fruit fly Bactrocera invadens (Diptera: Tephritidae) in Africa. Bull Entomol Res 96:379–386
Estoup A, Jarne P, Cornuet JM (2002) Homoplasy and mutation model at microsatellite loci and their consequences for population genetics analysis. Mol Ecol 11:1591–1604
European and Mediterranean Plant Protection Organization, EPPO (2009) Bactrocera invadens (Diptera: Tephritidae) http://www.eppo.org/QUARANTINE/Alert_List/insects/BCTRIN.htm
Faircloth BC (2008) MSATCOMMANDER: detection of microsatellite repeat arrays and automated, locus-specific primer design. Mol Ecol Resour 8:92–94
Fletcher BS (1989) Movements of tephritid fruit flies. In: Robinson AS, Hooper GH (eds) Fruit flies: their biology, natural enemies and control, vol 3B. Elsevier Science Publications, Amsterdam, pp 209–219
French C (2005) The new invasive Bactrocera species. pp. 19–20 In: Insect pest control newsletter, No. 65. International Atomic Energy Agency, Vienna, Austria
Glenn TC, Schable NA (2005) Isolating microsatellite DNA loci. Method Enzymol 395:202–222
Golein B, Talaie A, Zamani Z, Moradi B (2006) Development and characterization of new microsatellite loci from lemon (Citrus limon). Int J Agr Biol 8:172–174
Gomulski LM, Bourtzis K, Brogna S, Morandi PA, Bonvicini C, Sebastiani F, Torti C, Guglielmino CR, Savakis C, Gasperi G, Malacrida AR (1998) Intron size polymorphism of the Adh1 gene parallels the worldwide colonization history of the Mediterranean fruit fly, Ceratitis capitata. Mol Ecol 7:1729–1742
Gupta PK, Rustgi S, Sharma S, Singh R, Kumar N, Balyan HS (2003) Transferable EST-SSR markers for the study of polymorphism and genetic diversity in bread wheat. Mol Genet Genomics 270:315–323
Hamada HM, Petrino MG, Kakunaga T (1982) A novel repeated element with Z-DNA forming potential is widely found in evolutionarily diverse eukaryotic genomes. PNAS 79:6465–6469
Han HY, McPheron BA (1997) Molecular phylogenetic study of Tephritidae (Insecta: Diptera) using partial sequences of the mitochondrial 16S ribosomal DNA. Mol Phyl Evol 7:17–32
He GH, Meng RH, Newman M, Gao G, Pittman RN, Prakash CS (2003) Microsatellites as DNA markers in cultivated peanut (Arachis hypogaea L.). BMC Plant Biol 3:3
Hedrick PW, Parker KM, Lee RN (2001) Using microsatellite and MHC variation to identify species, ESUs, and Mus in endangered Sonoran topminnow. Mol Ecol 10:1399–1412
Innan H, Terauchi R, Miyashita NT (1997) Microsatellite polymorphism in natural populations of the wild plant Arabidopsis thaliana. Genetics 146:1441–1452
Jacob HJ, Lindpaintner K, Lincoln SE, Kusumi K, Bunker RK, Mao YP, Ganten D, Dzau VJ, Lander ES (1991) Genetic mapping of a gene causing hypertensive rat. Cell 67:213–224
Jarne P, Lagoda PJL (1996) Microsatellites, from molecules to populations and back. TREE 11:424–429
Kandpal RP, Kandpal G, Weissman SM (1994) Construction of libraries enriched for sequence repeats and jumping clones, and hybridization selection for region-specific markers. PNAS 91:88–92
Karagyozov L, Kalcheva ID, Chapman VM (1993) Construction of random small-insert genomic libraries highly enriched for simple sequence repeats. Nucleic Acids Res 21:3911–3912
Khamis F, Karam N, Guglielmino CR, Ekesi S, Masiga D, De Meyer M, Kenya EU, Malacrida AR (2008) Isolation and characterization of microsatellite markers in the newly discovered invasive fruit fly pest in Africa, Bactrocera invadens (Diptera: Tephritidae). Mol Ecol Res 8:1509–1511
Khamis FM, Karam N, Ekesi S, De Meyer M, Bonomi A, Gomulski LM, Scolari F, Gabrieli P, Siciliano P, Masiga D, Kenya EU, Gasperi G, Malacrida AR, Guglielmino CR (2009) Uncovering the tracks of a recent and rapid invasion: the case of the fruit fly pest Bactrocera invadens (Diptera: Tephritidae) in Africa. Mol Ecol 18:4798–4810
Khamis FM, Masiga DK, Mohamed SA, Salifu D, De Meyer M, Ekesi S (2012) Taxonomic identity of the invasive fruit fly pest, Bactrocera invadens: concordance in morphometry and DNA barcoding. PLoS One 7:e44862
Kinyanjui G, Khamis FM, Mohamed S, Ombura LO, Warigia M, Ekesi S (2016) Identification of aphid (Hemiptera: Aphididae) species of economic importance in Kenya using DNA barcodes. Bull Entomol Res 106:63–72. doi:10.1017/S0007485315000796
Kofler R, Schlotterer C, Lelley T (2007) SciRoKo: a new tool for whole genome microsatellite search and investigation. Biogeosciences 23:1683–1685
Kumar P, Gupta VK, Misra AK, Modi DR, Pandey BK (2009) Potential of molecular markers in plant biotechnology. Plant Omics J 2:141–162
Lee JM, Nahm SH, Kim YM, Kim BD (2004) Characterization and molecular genetic mapping of microsatellite loci in pepper. Theor Appl Genet 108:619–627
Levinson G, Gutman GA (1987) Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol 4:203–221
Litt M, Luty JA (1989) A hypervariable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle actin gene. Am J of Hum Gen 44:397–401
Loxdale HD, Lushai G (1998) Molecular markers in entomology. Bull Entomol Res 88:557–600
Lux SA, Copeland RS, White IM, Manrakhan A, Billah MK (2003) A new invasive fruit fly species from the Bactrocera dorsalis (Hendel) group detected in East Africa. Insect Sci Appl 23:355–361
Malacrida AR, Guglielmino CR, D’Adamo P, Torti C, Marinoni F, Gasperi G (1996) Allozyme divergence and phylogenetic relationships among species of tephritid flies. Heredity 76:592–602
Malacrida AR, Marinoni F, Torti C, Gomulski LM, Sebastiani F, Bonvicini C, Gasperi G, Guglielmino CR (1998) Genetic aspects of the worldwide colonization process of Ceratitis capitata. J Hered 89:501–507
Malacrida AR, Gomulski LM, Bonizzoni M, Bertin S, Gasperi G, Guglielmino CR (2007) Globalization and fruit fly invasion and expansion: the medfly paradigm. Genetica. doi:10.1007/s10709-006-9117-2
Manni M, Lima KM, Guglielmino CR, Lanzavecchia SB, Juri M, Vera T, Cladera J, Scolari F, Gomulski L, Bonizzoni M, Gasperi G, Silva JG, Malacrida AR (2015) Relevant genetic differentiation among Brazilian populations of Anastrepha fraterculus (Diptera, Tephritidae). ZooKeys 540:157–173
McPheron BA (2000) Population genetics and cryptic species. In: Tan KH (ed) Area-wide control of fruit flies and other insect pests. Penerbit Universiti Sains Malaysia, Penang, pp 483–490
Meixner MD, McPheron BA, Silva JG, Gasparich GE, Sheppard WS (2002) The Mediterranean fruit fly in California: evidence for multiple introductions and persistent populations based on microsatellite and mitochondrial DNA variability. Mol Ecol 11:891–899
Morrow J, Scott L, Congdon B, Yeates D, Frommer M, Sved J (2000) Close genetic similarity between two sympatric species of tephritid fruit fly reproductively isolated by mating time. Evolution 54:899–910
Mwatawala MW, White IM, Maerere AP, Senkondo FJ, De Meyer M (2004) A new invasive Bactrocera species (Diptera: Tephritidae) in Tanzania. Afri Entomol 12:154–156
Mwatawala MW, De Meyer M, Makundi RH, Maerere AP (2006) Biodiversity of fruit flies (Diptera, Tephritidae) at orchards in different agro-ecological zones of the Morogoro region, Tanzania. Fruits 61:321–332
Nardi F, Carapelli A, Dallai R, Roderick GK, Frati F (2005) Population structure and colonization history of the olive fly, Bactrocera oleae (Diptera, Tephritidae). Mol Ecol 14:2729–2738
Ostrander EA, Jong PM, Rine J, Duyk G (1992) Construction of small-insert genomic DNA libraries highly enriched for microsatellite repeat sequences. PNAS 89:3419–3423
Paetkau D (1999) Microsatellites obtained using strand extension: an enrichment protocol. Biotechniques 26:690–697
Pandolfo M (1992) A rapid method to isolate (GT)n repeats from yeast artificial chromosomes. Nucleic Acids Res 20:1154
Piry S, Alapetite A, Cornuet JM, Paetkau D, Baudouin L, Estoup A (2004) Geneclass 2: a software for genetic assignment and first s – generation migrant detection. J Hered 95:536–539
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotypic data. Genetics 155:945–959
Rassmann K, Schlotterer C, Tautz D (1991) Isolation of simple sequence loci for use in polymerase chain reaction-based DNA fingerprinting. Electrophoresis 12:113–118
Roderick G, Navajas M (2003) Genotypes in novel environments: genetics and evolution in biological control. Nat Rev Gen 4:889–899
Rudd S (2003) Expressed sequence tags: alternative or complement to whole genome sequences? Trends Plant Sci 8:321–329
Schlötterer C, Tautz D (1992) Slippage synthesis of simple sequence DNA. Nuc Acids Res 20:211–215
Schuler GD, Boguski MS, Stewart EA, Stein LD, Gyapay G, Rice K, White RE, Rodriguez-Tomé P, Aggarwal A, Bajorek E, Bentolila S, Birren BB, Butler A, Castle AB, Chiannilkulchai N, Chu A, Clee C, Cowles S, Day PJ, Dibling T, Drouot N, Dunham I, Duprat S, East C, Edwards C, Fan JB, Fang N, Fizames C, Garrett C, Green L, Hadley D, Harris M, Harrison P, Brady S, Hicks A, Holloway E, Hui L, Hussain S, Louis-Dit-Sully C, Ma J, MacGilvery A, Mader C, Maratukulam A, Matise TC, McKusick KB, Morissette J, Mungall A, Muselet D, Nusbaum HC, Page DC, Peck A, Perkins S, Piercy M, Qin F, Quackenbush J, Ranby S, Reif T, Rozen S, Sanders C, She X, Silva J, Slonim DK, Soderlund C, Sun WL, Tabar P, Thangarajah T, Vega-Czarny N, Vollrath D, Voyticky S, Wilmer T, Wu X, Adams MD, Auffray C, Walter NA, Brandon R, Dehejia A, Goodfellow PN, Houlgatte R, Hudson JR Jr, Ide SE, Iorio KR, Lee WY, Seki N, Nagase T, Ishikawa K, Nomura N, Phillips C, Polymeropoulos MH, Sandusky M, Schmitt K, Berry R, Swanson K, Torres R, Venter JC, Sikela JM, Beckmann JS, Weissenbach J, Myers RM, Cox DR, James MR, Bentley D, Deloukas P, Lander ES, Hudson TJ (1996) A gene map of the human genome. Science 274:540–546
Schutze MK, Aketarawong N, Amornsak W, Armstrong KF, Augustinos AA, Barr N, Bo W, Bourtzis K, Boykin LM, Cáceres C, Cameron SL, Chapman TA, Chinvinijku S, Chomiˇc A, De Meyer M, Drosopoulou E, Englezou A, Ekesi S, Gariou- Papalexiou A, Geib SM, Hailstones D, Hasanuzzaman M, Haymer D, Hee AKW, Hendrichs J, Jessup A, Ji Q, Khamis FM, Krosch MN, Leblanc L, Mahmood K, Malacrida AR, Mavragani-Tsipidou P, Mwatawala M, Nishida R, Ono H, Reyes J, Rubinoff D, San Jose M, Shelly TE, Srikachar S, Tan KH, Thanaphum S, Haq I, Vijaysegaran S, Wee SL, Yesmin F, Zacharopoulou A, Clarke AR (2014) Snonymization of key pest species within the Bactrocera dorsalis species complex (Diptera: Tephritidae): Taxonomic changes based on a review of 20 years of integrative morphological, molecular, cytogenetic, behavioural and chemoecological data. System Entomol. doi:10.1111/syen.12113
Senan S, Kizhakayil D, Sasikumar B, Sheeja TE (2014) Methods for development of microsatellite markers: an overview. Not Sci Biol 6:1–13
Shearman DCA, Gilchrist AS, Crisafulli D, Graham G, Lange C, Frommer M (2006) Microsatellite markers for the pest fruit fly, Bactrocera papayae (Diptera: Tephritidae) and other Bactrocera species. Mol Ecol Notes 6:4–7
Sonvinco A, Manso F, Quesada-Allue LA (1996) Discrimination between the immature stages of Ceratitis capitata and Anastrepha fraterculus (Diptera: Tephritidae) populations by random amplified polymorphic DNA polymerase chain reaction. J Econ Entomol 89:1208–1212
Thiel T, Michalek W, Varshney RK, Graner A (2003) Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet 106:411–422
Wang R, Zheng L, Touré YT, Dandekar T, Kafatos F (2001) When genetic distance matters: measuring genetic differentiation at microsatellite loci in whole-genome scans of recent and incipient mosquito species. PNAS 98:10769–10774
Weber JL, May PE (1989) Abundant class of human DNA polymorphism which can be typed using polymerase chain reaction. Am J Hum Gen 44:388–396
White IM, Elson-Harris MM (1992) Fruit flies of economic significance: their identification and bionomics. C.A.B International and Australian Centre for International Agricultural Research, Canberra, p 601
White G, Powell W (1997) Isolation and characterization of microsatellite loci in Swietenia humilis (Meliaceae): an endangered tropical hardwood species. Mol Ecol 6:851–860
Wu Y, Li Z, Wu J (2009) Polymorphic microsatellite markers in the melon fruit fly, Bactrocera cucurbitae (Coquillett) (Diptera: Tephritidae). Mol Ecol Resour 9:1404–1406
Zane L, Bargelloni L, Patarnello T (2002) Strategies for microsatellite isolation: a review. Mol Ecol 11:1–16
Acknowledgements
Research activities were funded by grants from GIZ/BMZ, Biovision, the EU, IAEA, DFID and DAAD to the African Fruit Fly Programme (AFFP) of icipe.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Khamis, F.M., Malacrida, A.R. (2016). Role of Microsatellite Markers in Molecular Population Genetics of Fruit Flies with Emphasis on the Bactrocera dorsalis Invasion of Africa. In: Ekesi, S., Mohamed, S., De Meyer, M. (eds) Fruit Fly Research and Development in Africa - Towards a Sustainable Management Strategy to Improve Horticulture. Springer, Cham. https://doi.org/10.1007/978-3-319-43226-7_4
Download citation
DOI: https://doi.org/10.1007/978-3-319-43226-7_4
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-43224-3
Online ISBN: 978-3-319-43226-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)