Abstract
The basic processes in molecules of biological interest induced by the absorption of VUV and soft X-rays are reviewed. The study of excitation, ionisation and dissociation in the gas phase on the one hand provides detailed information on the electronic structure and geometry that determine the functioning of these molecules in macroscopic systems and, on the other hand, sheds light on the microscopic effects of radiation damage in living cells.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
The electronic structure and geometrical arrangement (conformation, isomerisation, tautomerisation) of atoms and molecules, the basic constituents of matter, determine the functioning of systems at the macroscale. This is particularly true, in the case of biomatter, where for example the functionality of complex molecules, like enzymes and proteins built up by 20 different amino acids up to a size in the nanometer to micrometre range, is in close relation to the details of their conformation. Another example is the radiation damage where the macroscopic effects induced in living cells by the absorption of ionizing radiation are closely related to the structural and chemical properties of their molecular constituents, with processes initiated at the atomic and molecular level. It is well known, for example that substantial damage to DNA/RNA can be produced by slow electrons with energy of a few eV [1], produced either directly or via ionisation of the medium they are immersed in. In this field, atomic and molecular physics can provide a valuable contribution both experimentally and theoretically. Gas phase studies enable to disentangle the intrinsic properties of the molecules from those due to the interaction with the environment. Indeed the approach which begins with the characterisation of the building blocks of complex biological systems (bases, nucleosides and nucleotides for DNA, amino acids and peptides for proteins) and the understanding of the very basic physical chemical processes due to the interaction with ionising sources to systems of increasing complexity (clusters, hydrated clusters, nanoaggregates) provides important benchmark data for studies in liquid solutions or in the solid state. Moreover, studies of isolated biomolecules can benefit from the armoury of all theoretical techniques developed for polyatomic molecules, such as ab initio calculations and DFT methods.
The understanding of the physics and chemistry of isolated molecules of biological interest can also provide relevant contribution to biotechnological applications, such as sensors and molecular electronics, astrochemistry and astrobiology, where for example key information on the origin of life in the universe is provided by the understanding of the chemistry of relatively simple molecules in an environment subject to ionizing radiation.
This chapter is devoted to the description of photon induced processes in molecules of biological interest in gas phase. Several radiation sources have been used to investigate the interaction of “light” with biomolecules, from IR and UV lasers, to VUV and X-ray sources at fixed wavelength as well as synchrotron radiation. Synchrotron radiation with its tunability over a broad energy spectrum and synchrotron based spectroscopic techniques represent a unique combination to investigate the absorption of a defined amount of energy by the molecule, sometimes even at a specific bond or molecular site, and then to probe the effects of this excitation on the electronic structure and stability against fragmentation of the molecule. Thus most of this chapter will be devoted to the study of photoprocesses excited by synchrotron radiation. It can be easily predicted that the advent of Free Electron Lasers, FELs, and High Harmonic Generation, HHG, sources with intense pulses, whose duration is only a few fs or even hundreds of as, will produce a step forward in the understanding of the dynamics and energy flow in biomolecules. Indeed these sources allow a time-resolved study of the processes occurring between the energy absorption and the manifestation, for example of the damage or de-excitation and ‘repair’. This will lead to the control and proper handling of the process itself.
The chapter is organised as follows. Section 2 is devoted to the experimental methods used in the studies of molecules of biological interest. In Sect. 3 the applications of the different techniques to nitrogenous bases, amino acids and peptides are reported briefly. The subsections address the different wavelength regimes and one of them is devoted to time dependent studies based on pump-probe experiments, which are benefitting of the advent of FEL and HHG sources. Finally Sect. 4 is devoted to some perspectives and conclusions.
2 Methods
Most biomolecules, including even the smallest such as nucleobases and amino acids, are solid at room temperature and they have to be brought into the gas phase to be studied at the single molecule level. This poses new challenges compared with the investigation of atoms or smaller and more volatile molecules. Thermal evaporation has the advantage of producing a beam of neutral molecules. In the case of DNA/RNA bases a solid ring structure makes these molecules (except for guanine) quite resistant to thermal decomposition, so that they can be evaporated from ovens to produce effusive or supersonic beams of neutral molecules. In other cases, e.g. some nucleosides and amino acids, great care has to be taken to characterise the working conditions that guarantee the evaporation of intact molecules. For most of the more complex systems, especially if they contain reactive side groups such as carboxylic acid, hydroxyl, and sulfhydryl thermal evaporation cannot be employed, due to the fragility of the targets. In such cases, alternative and more elaborated approaches like Electrospray Ionisation (ESI) [1], Matrix-Assisted Laser Desorption Ionisation (MALDI) [2] and Laser-Induced Acoustic Desorption (LIAD) [3] have been successfully used instead of thermal evaporation. These techniques allow to bring large species intact in the gas phase. However, they also present some drawbacks, as the formation of protonated/de-protonated or multiply charged biomolecules in the ESI, possible contaminations from the matrix molecules and solvents in the MALDI, a pulsed source in the LIAD and, in all cases, the low density of the sample.
The approaches to produce the target beam are briefly described in Sect. 2.1, while the typical spectroscopic techniques regarding the detection of electrons and ions produced in the photo-induced processes, detected both separately and in time coincidence, are briefly reviewed in Sect. 2.2.
2.1 The Targets
The typical experimental set-up used for the evaporation of small biomolecules is composed of a resistively heated oven that heats a crucible containing powder of the target molecule. The heating is often provided by ceramic insulated wires or commercial Thermocoax heaters [4], which provide up to several tens of Watts of power. The winding of the oven should be non-inductive to avoid spurious magnetic fields in the set-up. Furthermore, the top and the bottom of the crucible are normally held at slightly different temperatures, with the top being hotter than the bottom in order to prevent condensation and blockage of the narrow orifice at the exit of the crucible. The oven is contained in a high or, even better, ultrahigh vacuum chamber, bakeable and equipped with a cold finger facing the oven, to trap the vapour. The cold trap limits the contamination of the set-up and helps maintaining a low background pressure, which is particularly important in mass spectrometric studies of biomolecules, where the relatively low density of the vapour beam requires low background pressure in order to produce a good signal-to-noise ratio. Efficient trapping is also necessary to prevent deposition of an insulating layer on the electrodes of the electron and ion analysers, which can seriously affect their efficiency and stability. Last but not the least, baking the entire vacuum system is an efficient way to clean the set-up of residual contaminations; this is a standard procedure for Ultra High Vacuum systems and it is best to provide this capability even for high vacuum systems. Design considerations for ovens have been published for evaporation of metal vapours [5] and similar ideas have been recently adapted and optimised for the evaporation of fragile biomolecules [6,7,8,9,10,11]. These more specific approaches are based on the use of non-metallic and more inert materials for the crucible, strict control of the working conditions and a careful characterisation of the thermal decomposition of the target molecules. The biomolecular sample can also be applied as saturated solution onto pre-cleaned fibreglass wool, which can be then packed tightly inside, for example an externally heated glass tube. This method serves to increase the surface area, thus enhancing the thermal desorption rate over the thermal decomposition rate [12].
2.2 Analytical Methods
Despite laboratory sources, like rare gases discharge lamps, have been used in the early photoionisation experiments on valence shell, synchrotron radiation has become the most ‘popular’ and effective radiation source for these studies due to the broad range of tunability of the photon energy, continuous from the UV to hard X-rays . This allows for photoionisation and photoexcitation experiments, able to map out both occupied and empty states respectively, of the valence as well as inner shell orbitals of the target. Photo Emission and X-ray Photoemission Spectroscopies (PES and XPS ), where the kinetic energy (KE) of the photoionised electron is measured at a fixed photon energy (hv), allows reconstruction of the electronic distribution of the molecular orbitals of binding energy BE = hv – KE. The typical PES/XPS set-up is based on electron optics principles and uses well defined geometries and electric fields to guide and select the electrons according to their kinetic energy and angle of emission. One of the most common, for example is the hemispherical deflector analyser, typically composed of an electrostatic lens, the energy selector and the detector. The electrostatic lens is responsible for the acceptance, transport and focusing of the electrons from the interaction region to the entrance of the hemispherical analyser, where the electron trajectories are ‘deflected’ according to their kinetic energy passing through a radial field. One or more detectors placed at the exit of the hemispheres detect the ‘selected’ electron, and their kinetic energy is reconstructed according to well-known equations.
In the valence shell, see Fig. 1, especially from comparison with theoretical predictions, important information about the electronic charge distribution of the orbitals in the molecule can be derived. In the inner shell the localised nature of the core electrons implies that each atom is affected by its surrounding chemical environment and site-selective information can be obtained. The use of the binding energy shifts to extract chemical information is also known as Electron Spectroscopy for Chemical Analysis (ESCA) [13]. Subtle differences among families of similar molecules (e.g. isomers or analogues), or the effect of functionalisation can be identified, assessed and discussed in terms of the measured and calculated inner shell chemical shifts.
Complementary information to that provided by electron emission, which probes the occupied states, can be obtained by probing the empty states. Near Edge X-ray Absorption Fine Structure (NEXAFS) spectroscopy provides electronic structure data, promoting core electrons to empty (excited) states [14]. The experiments, which necessarily require tunable photon energy, can be performed by scanning the photon energy across an inner shell threshold and measuring a signal that is proportional to the absorption cross section, for example the total ion yield, see Fig. 2. In biomolecules, C, N, O and S are typically the atoms that can be probed in the soft X-ray region, near their respective K-edges. A simple method of measuring the NEXAFS spectrum is to use an ion detector, placed at the magic angle, consisting of a channel electron multiplier or channel plate, and to measure the current, or number of ions (pulse counting).
Electron energy distributions measured over a broad range of kinetic energies from zero up to hv can be used to characterise the complete electron emission spectrum due to the photoemission as well as to the Auger decay of core ionic states or autoionisation of neutral excited states. If the electron spectrum is measured at a photon energy corresponding to a resonant core excited state, previously determined using NEXAFS, the technique is known as Resonant Auger spectroscopy. Similarly, autoionisation photoelectron spectra are due to excitation to a resonant valence excited state, which subsequently decays by emitting an electron. They differ from normal Auger spectroscopy because they involve the decay of a neutral excited state to a singly charged ionic state, whereas in the normal Auger process a singly charged ion decays to a doubly charged ion. Specially on biomolecules, the full electron emission spectra , including photoelectron, autoionisation, Auger and Resonant Auger electron characterised at different photon energies are useful benchmark data for Monte Carlo and Ion Tracking simulation codes, used to evaluate the direct and indirect radiation damage due to the interaction of ionising radiation with a biological medium. The valence and core electron spectra are also useful as benchmark data for calculations of molecular properties based on the electronic structure, particularly in the valence. If the calculation can reproduce accurately the measured valence and core spectra, and also the geometric structure (usually the calculated structure is compared with crystallographic data), then it is reasonable to expect that other calculated properties are accurately predicted, for example dipole and multipole momenta, appearance energies, electron and proton affinities, reaction rates, etc.
If the ion detection is performed using a technique that disperses the fragments according to their mass, then even more information is obtained, and the technique is known as photoionisation mass spectrometry (PIMS). The most common detectors are time of flight (TOF) [15] and quadrupole mass spectrometers (QMS). In the TOF spectrometer the ions are dispersed according to their mass over charge ratio (m/z), which determines their flight time to the detector. In the QMS, a radiofrequency field provides the m/z selection, where only a specific m/z, within the instrumental resolution, will travel to the detector while the other ones are lost through the QMS bars. With these detectors, the molecular fragmentation pattern is measured at fixed photon energy, and it is possible to relate the absorbed photon energy to the products of dissociation. The NEXAFS spectrum can be reconstructed by scanning the photon energy and reporting the total ion signal at each photon energy. Valence band ionisation can leave the molecule in its ground ionic state, which may or may not be stable, or in other excited ionic states up to a binding energy equal to the exciting photon energy, hv. Since excited electronic states contain more internal energy than the ground ionic state, they have a greater tendency to fragment. As the photon energy increases, more of these excited ionic states are produced, leading to stronger dissociation and to the opening of new fragmentation channels, with the possibility for molecular rearrangements and multiple bond breakings. These processes can be investigated and quantified in the study of the appearance energy (AE), which is the minimum energy required to produce a particular fragment ion in a specific molecule. Experimentally, the AE is determined by selecting a fragment and measuring its ion yield versus photon energy, see inset of Fig. 1. By fitting the spectrum with two straight lines, the first onset of the ion signal is determined. Interestingly, while the experimental determination of the AE is unique, the theoretical value depends not only on the geometry of the charged fragment, but also on the structure of the neutral fragments and on the fragmentation mechanism, providing essential information in the dynamics of molecular fragmentations.
In the soft X-ray region, excitation of core level resonances most likely gives rise to fast electron decay via Resonant Auger emission on the fs timescale, followed by molecular fragmentation on the ps or longer timescale. The situation is complex and the molecular fragmentation hard to predict, because even if the excitation was initially localised on a specific molecular site, after the electronic decay a delocalised valence excited state is produced in which the energy flows through the molecule before fragmentation. For ionisation, rather than excitation of a core level, the core hole decays via an Auger process , again on the timescale of a few fs, and most likely produces a doubly charged ionic state with both holes in the valence band (KVV decay). The fragmentation is then very rapid (if the molecule is not too large) as there is a strong Coulomb repulsion between the two holes. However, this situation is also rather complicated, as there are very many two-hole states, and it may be difficult to identify them.
The PEPICO set-up used at the Gas Phase photoemission beamline of Elettra is composed of a hemispherical electron analyser and a TOF mass spectrometers mounted opposite to each other at the magic angle for synchrotron radiation experiments. The superimposed schematics show the electron (blue) and ion (green) analysers as well as the photon beam path (violet arrow) [16, courtesy of Maclot]
The simultaneous detection of electrons and ions in time coincidence, i.e. from the same ionisation event, allows for a better control over the many variables in the physical process, adding further insights into molecular fragmentation. In photoelectron photoion [17] and photoelectron photoion photoion coincidence (PEPICO and PEPIPICO) [18] experiments, electrons and ions are detected in time coincidence in order to identify the ones which are generated in the same ionisation event. Figure 3 shows the example of a set-up used in electro-ion coincidence experiments. The basic idea of a coincidence experiment is that correlated particles are generated at the same time and they have a fixed time delay in their arrival time to the detectors, while uncorrelated particles are generated in different ionisation events and therefore will have a random distribution in their arrival times. Therefore the measurement of the time distribution of the difference in the arrival time of these particles will display correlated events, called ‘true coincidences’, as a peak on a flat background of uncorrelated events, called ‘random coincidences’. In practice, in these measurements the ionisation rate is reduced to a sufficiently low value that in a given time interval (the time resolution of the detection system), there is likely to be only one ionisation event. In these experiments the electron kinetic energy determines which valence or inner shell orbital has been ionised, providing state- and site-selectivity (for valence and core ionisation respectively), while the measurement of the mass spectrum in coincidence gives information about the molecular fragmentation of that specific state. In the PEPIPICO experiments, also the detailed description of the dissociation process of doubly/multiply charged ions and the determination of kinetic energy released in the process (available from TOF spectrometers) can be achieved providing a very comprehensive picture of the dynamics of fragmentation.
3 Applications to Nitrogenous Bases, Amino Acids and Peptides
In this section some applications of the above-mentioned techniques to a few classes of biomolecules like nucleobases, their parent compounds, derivatives and analogues, amino acids and their derivatives and oligomers, pharmaceuticals and other molecules with biochemical roles like antibiotics, sugar or lipids are collected. Among the very many excellent results that can be found in the literature concerning the study of biomolecules in the gas phase, the reported literature is far from being a comprehensive review, and we will display in some more details just a few cases in order to provide very brief examples of the implementation of the spectroscopic techniques previously illustrated.
3.1 Excitation and Ionisation in the VUV Range (laboratory Source and Synchrotron)
In the VUV range the electronic charge distribution of the empty orbitals below the first ionisation potential and of the first few molecular orbitals in the valence and inner-valence shells can be accessed. The comparison with theoretical predictions helps to understand the electronic properties and to model the chemical behaviour of the investigated molecules. PES and PEPICO experiments in the valence region can be performed by both rare gas lamps laboratory source (see, for example [17, 19,20,21,22]) as well as synchrotron radiation in DNA bases [23,24,25,26], amino acids [27,28,29,30,31], pharmaceuticals and other related biomolecules [32,33,34,35,36,37,38,39], while the tunability of the synchrotron radiation is compulsory for the study of empty orbitals [40, 41].
The valence photoelectron spectra allow the understanding of the nature of the outer orbitals and how the molecule is bound. Due to the complexity of the target these spectra generally exhibit broad overlapping bands, often resulting from several unresolved tautomers and conformers, with little evidence of resolved vibrational structure so that their interpretation much relies on ab initio and sophisticated theoretical calculations, like for example ADC(3) [25, 26] and TDDFT B-spline LCAO methods [24, 40], for both the outer and inner valence regions. Figure 4 shows an example of the joint experimental and theoretical study of the photoelectron spectra of halogenated pyrimidines, where the role of different halogen substitutions to the pyrimidine ring is investigated. These molecules are prototype radiosensitisers for selectively enhanced radioterapic effect in cancer treatment. Even though belonging to the same family and therefore very similar, the spectra clearly display some differences due to the presence of the halogen substitution. The substituent effects on the orbitals of the pyrimidine ring can be discussed as a function of the identity and position of the halogen atom. The shifts of the binding energies of these orbitals can be accounted for by a combination of the inductive and resonance effects of the halogen atoms of the ring orbitals [21].
An experimental approach to support the theoretical assignment and overcome the limited resolution in PES spectra is based on the measurement of the photoelectron angular distribution [25, 42] that, based on the different behaviour of the asymmetry parameter of \(\uppi \)- and \(\sigma \)-type orbitals, is able to distinguish between orbital types and is particularly useful in binding energy regions containing overlapping photoelectron bands. The photoelectron angular distribution displays also a chiroptical effect, observed as a strong forward/backward asymmetry, with respect to the photon propagation axis, when circularly polarised radiation is used to photoionise gas phase enantiomers. It has been shown that the new observable in the photoelectron angular distribution with circularly polarised radiation, the chiral parameter, is extremely sensitive to static molecular structures, chemical substitution, conformers, dimerisation and clustering [43]. The application to the amino acid alanine [44] showed that the technique provides a plausible conformer population in a genuine biological floppy system and it can be a precious tool for the study the electronic structure as well as molecular structures of biopolymer building blocks in a bottom/up approach of biomolecular complexity.
An alternative method to traditional PES measurements consists of high-resolution Threshold PhotoElectron (TPES) and Threshold PhotoElectron Photoion Coincidence (TPEPICO) spectroscopy. These techniques have been recently used to measure vibrationally resolved photoelectron spectra of DNA/RNA bases and amino acids [45,46,47,48,49], for both the ground and lowest electronic states, and allowed precise determination of vertical ionisation energies.
From the simple molecules of DNA/RNA and amino acids, the natural evolution in the study of elementary biomolecules leads to the investigation of their more complex structures, the nucleosides and peptides for DNA chain and proteins, respectively.
He I photoelectron spectra of pyrimidine and some halopyrimidines. The sticks indicate the ab initio ionisation energies calculated using the B3LYP [21] while the Hartree–Fock molecular orbitals refer to the case of pyrimidine
In the two cases, the glycosidic and the peptide bonds play a fundamental role in the formation of long and complex chains of biomolecules that, beginning from relatively few and simple building blocks provide the essential mean for the development of life, with storage and replication of the genetic information as well as for the most differentiated functions performed by proteins. Therefore, it is extremely interesting to investigate the electronic structure of these more complex compounds, also to understand how the properties of the single, isolated molecule is affected by the presence of neighbouring molecules present in their natural biological environment. Unfortunately, due to the fragility of these relatively complex molecules against thermal decomposition, only some selected targets have been investigated in their neutral form, mostly by mass spectrometry, which requires less density than photoemission experiments. However, several photoemission studies are reported in the case of dipeptides containing glycyl [29, 50] and cyclic dipeptides like Glycyl-Glycyl (cGG), Leucyl-Prolyl (cLP), Phenylalanyl-Prolyl (cPP) [30], Histidyl-Glycyl (cHisGly), Tyrosyl-Prolyl (cTyrPro) and Phenylalanyl-Phenylalanyl (cPhePhe) [31]. These last compounds have a solid ring structure, which makes them resistant in the evaporation. These joined experimental and theoretical studies via valence and inner shell photoemission spectroscopy compared with similar results obtained for their constituent isolated molecules, concluded that in most cases the side-chains interact weakly with the central moiety. Thus a building block approach, in which the chemical properties can be considered to be the sum of those of the functional groups making up the cyclic dipeptide, can be substantially justified.
3.2 Excitation and Ionisation in the Soft X-Ray
In the inner shell, the localised nature of the core electrons implies that each atom is affected by its surrounding chemical environment and site-selective information can be obtained. Subtle differences among families of similar molecules (e.g. isomers or analogues), or the effect of functionalisation can be identified, assessed and discussed in terms of the measured and calculated inner shell chemical shifts. Even though XPS measurements can be performed also with laboratory sources [51] as analytical tool, most of the recent XPS studies on biomolecules in the gas as well as the NEXAFS experiments have been performed with synchrotron radiation for DNA bases [52,53,54,55,56,57,58,59,60], amino acids [28, 30, 46, 61,62,63] and peptides [29,30,31, 64, 65] as well as other related biomolecules like pharmaceuticals or neurotransmitters [35,36,37,38, 59, 66,67,68,69]. Further than a conventional analytical tool, XPS spectroscopy has also been used to unravel intriguing structural and dynamical effects peculiar of biomolecules, like the interplay of different tautomeric forms in DNA bases, the population of a variety of conformers in amino acids or the interplay of resonant and inductive effects in the shielding of core holes in aromatic pyrimidinic rings.
The structure of DNA bases plays a fundamental role in the proper base paring mechanism in the DNA chain [70]. Indeed, in the gas phase and at temperatures of several hundreds Kelvin, a few tautomers of guanine and cytosine are significantly populated, whereas thymine, adenine and uracil exist in a single form [55, and references therein]. The group of Prince et al. at Elettra proved that core level photoemission spectroscopy can be used to study tautomers in thermally evaporated DNA bases, observing chemical shifts up to several eV [54, 55]. The advantage of this technique is that, having a precise control over the evaporation temperature of the experiment, the free energies of the sample can also be estimated and relative abundance of the different tautomers predicted and compared to the experimental results.
C(1s) XPS spectra of pyrimidine and halogenated pyrimidines: experimental results (open dots), fits with asymmetric Gaussian lineshapes (full line) and DTF theoretical calculations [66]
Amino acids are floppy molecules, presenting a rich variety and complexity in the possible geometric and electronic structural forms that they may adopt, depending upon the interplay of a variety of intramolecular hydrogen bonding and electron correlations. These different geometries, often achieved by rotation around a single bond (conformers) are sometimes so close in energy that even slightly different evaporation temperatures can populate several of them, so that they normally coexist during the experimental measurements. The different conformers can be difficult to be clearly identified in PES spectra, due to the delocalised nature of the valence orbitals. However, it has been proved that in XPS spectra, where the core electrons are very sensitive to the chemical environment, the binding energies of different conformers can be as distant as 1 eV [62] so that XPS can be used to identify and determine the relative populations of conformers of amino acids in the gas phase and reveal important information on their ‘shape’. Conformer effects, clearly observable in photoemission , appear to be more difficult to resolve in photoabsorption, probably due to partial cancellation of the energy shifts of opposite signs from the core hole states and unoccupied orbitals [52]. Nevertheless, the NEXAFS spectroscopy remain a useful tool in the study of the molecular electronic structure, providing insights about the nature of empty orbitals as well as some geometrical information on molecular bond lengths [67].
As a last example, Fig. 5 reports the results of the XPS study of a series of the halopyrimidine molecules [66] showing that, due to the higher electronegativity of the halogen atom compared to the substituted hydrogen, all the carbon atoms but in particular the one where the substitution has taken place are affected by halogenation and their binding energies shift towards higher energies. However, deeper insights regarding the resonant and inductive effects ruling the charge distribution in these aromatic molecules can be gained with the support of DFT calculations. By analysing the displacement of the electron charge density in molecules with a C(1s) core hole (see Fig. 6) it can be clearly observed that, regardless of the location of the carbon site, inductive effect drugs electron charge from neighbouring atoms while resonance contribution (\(\uppi \)) drugs electron charge from the atom in para-position. Therefore, rearrangements of the electronic charge occurring in the pyrimidine derivatives to screen a core hole estimated by ab initio methods are in very good agreement with the qualitative picture given by the inductive resonance model.
In biomolecules C, N, O and S are the typically constituent atoms. They can be probed efficiently in the soft X-ray region near their K-edges. Thus it is not surprising that there is no literature on the application of hard X-rays to isolated biomolecules, due to the low absorption cross section for low Z elements.
Difference density maps (top and side views) of “standard” versus “core hole” calculations. Panels (1), (2) and (3) represent the case of a core hole localised on the C5, C4 and C2 site, respectively, see Fig. 4 for the numbering of atoms [66]. Blue and red zones represent positive and negative isosurfaces, respectively
However, nanoparticles are increasingly used in the biological field for drug delivery due to their functional surface, which gives them the ability to bind, adsorb and carry other compounds [71]. Moreover, their enhanced permeability and retention and the easiness with which they are taken up by cells favours them with respect to other carrier systems. Among nanoparticles, noble metal nanoparticles are preferred due to their optical properties, non-toxicity and biocompatibility compared to the other metals. Wang et al. [72] have presented a comprehensive review of synchrotron radiation based techniques using radiation up to several keV for the study of nanomaterials at cellular and subcellular interfaces and their transformation it in vitro as well as of the molecular mechanism of the reaction of nanomaterials with biomolecules, but to our knowledge no studies of isolated nanoparticles have been yet reported.
3.3 Photofragmentation Studies
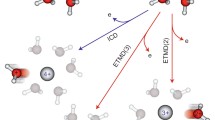
When the ionisation involves orbitals deeper than the ground state the charged molecule begins to fragment. Thus mass spectrometry, AE as well as the PEPICO measurements can characterise the molecular decomposition and link the production of certain fragments to specific photon energies and ionic states. In the most simple experiments, the fragmentation mass spectrum is measured at a fixed photon energy displaying all the fragmentation channels already opened. These experiments can be performed either at the fixed wavelength provided by discharge lamp laboratory sources [27, 34] or by tunable synchrotron radiation [73,74,75,76]. When a TOF spectrometer is used in conjunction with a continuous ionisation source, then the trigger for the measurement of the fragment flight time is typically given by the detection of an unresolved photoelectron in a PEPICO experiment. When the detected photoelectron is energy dispersed, then the PEPICO measurement also carry information of the state-selected fragmentation of the different molecular orbitals [17], which display marked selectivity, see Fig. 7.
A different approach to photofragmentation is represented by multi-photon ionisation, MPI, or Resonant Enhanced Multiphoton Ionisation, REMPI. In these cases a laser in the wavelength range 220–270 nm, a region typical of the \(\uppi \)* excitation in most of the molecules of biological interest, is used and the absorption of two or more photons leads to the ionisation and fragmentation of the sample. This method has been used by Barc et al. [77, 78] to investigate fragmentation of uracil, uracil–water and adenine–water clusters.
The mass spectra of DNA bases and amino acids measured at different photon energies have shown how the DNA bases, thanks to their cyclic structure, have a higher survival probability to radiation exposition with respect to amino acids [74], even though molecular fragmentation increases dramatically as the photon energy increases [73]. Furthermore, it has been observed that photons seem to be more harmful than electrons inducing molecular decomposition. Interestingly, peptides have been observed to be more radiation resistant that their constituent amino acids [79, 80] and synchrotron radiation VUV mass spectrometric studies of free polypeptides and proteins in the gas phase [81,82,83,84,85,86,87,88,89,90,91] were able to shine some light in this observation. The study of the protonated pentapeptide (leu–enk) measured in the VUV photon energy range 8–20 eV [84] has identified a possible mechanism to explain such increased resistance of peptides. Specially in the higher photon energy range, Bari et al. [84] observed that the fast intramolecular flow of the electron charge leads to the loss of a charged side chain (tyrosine in leu–enk case), which could be an efficient mechanism to ‘cool’ the remaining peptide, facilitating the survival of functional peptide substructures after absorption of very energetic photons. González-Magaña [81], in a photoionsation study of protonated synthetic peptides of increasing length, YGnF (n = 0, 1, 3, 5, 10), observed that up to n = 5 fragment ions related to the side-chains of the aromatic terminal amino acids Y and F dominate the fragmentation patterns, demonstrating an efficient hole migration towards the terminal amino acids upon photoionisation of the peptide backbone. However, beyond a certain peptide length (n = 10) they also observed significant reduction in fragmentation, with large dications and large singly charged ions that was attributed to a quenching of the charge migration mechanism and the subsequent increased stability typical of the large peptide regime [82]. In the case of the cytochrome c protein Milosavljevic et al. [82] observed a strong connection between ionisation potential and protein conformation, ruled by the charged state, on one hand suggesting that the conformation plays a crucial role in the protein photostability and, the other hand, proposing a novel experimental approach to investigate protein structure in the gas phase [65].
A key feature of synchrotron radiation is the tunability of the photon beam over a large energy range, which opens up the possibility of continuously monitoring the consequences of photon irradiation on the mass spectra as a function of the wavelength, that is often called ‘action spectroscopy’ [86, 91]. In the VUV range, this allows for the measurement of the AE of biomolecules, which are very important in order to explore their degradation pathways. In the small molecules, this information permits assessment, on thermochemical grounds, of the possible ionic and neutral products, as well as to propose the likely dissociation processes. Joint experimental and theoretical investigations of DNA/RNA bases [92, 93] and their functionalised analogues as halosubstituted DNA bases [32] and amino acids [49, 94, 95] for example, have allowed to discuss the role that these biomolecules could have played in the origin and development of life on earth as well as in applications like radiosensitisers in radiotherapy for cancer treatment, providing deep insights in the very basic mechanism of their decomposition. In the inner shell regions, where basically all fragmentation channels are opened, the resonant excitation of core electrons can be used to selectively deposit the energy on specific atoms in the molecules, performing a site-selective study of the molecular fragmentation. The mass spectra measured at several photon energies across inner shell thresholds display significant differences depending on the localisation of the core hole [58, 96, 97]. The more selective PEPICO and Resonant Auger-Ion Coincidence experiments allow for both site- and state-selective mass spectroscopy . The experiments performed in pyrimidine [96] and 2-Bromopyrimidine [97] have clearly demonstrated that fragmentation following inner shell excitation and valence photoionisation is exactly the same as long as the final ion state reached by the decay of the core excited state or electron ionisation are the same, see Fig. 8. This can be explained [97] by the fact that the fragmentation occurs on a timescale longer than the non-radiative relaxation of the inner shell vacancy. Thus, it is the charge distribution of the final singly charged ion that controls the fragmentation, and the enhancements observed in the intensity of some fragments of the mass spectra measurement across inner shell thresholds of pyrimidine and 2-Bromopyrimidine is due to a combination of the state-selective fragmentation of the molecular orbitals and an altered branching ration in the population of the valance orbitals due to Resonant Auger decay. For the large biomolecules [63, 64, 98], the mass resolved NEXAFS spectra of protonated peptides and proteins in the gas phase qualitatively resemble the ones in the condensed phase, carrying information on the inner shell excitations localised at different sites within the peptide.
The energy selected mass spectra of 2-Bromopyrimidine at a few BEs for the case of the C\(_{2}\) (1s \(\rightarrow \uppi \)*) excitation (b) and valence photoionisation at 100 eV photon energy (c). In panels (a) and (d) the C\(_{2}\) (1s \(\rightarrow \uppi \)*) RAE and PES spectra, respectively, are shown. The bars labelled 1 to 5 in panels (a and d) represent the selected energies, with corresponding energy resolution, for the PEPICO measurements [97]
At photon energies above the core ionisation, the most likely de-excitation process is the Auger decay that efficiently populates fast dissociating doubly and multiply charged ion state. In this case, in order to ‘localise’ the energy deposition into a specific molecular site, and have a complete control over the decay and fragmentation, a multiple coincidence experiment, where the photoelectron, the Auger electron ad the two charged fragments are detected in coincidence, should be performed. However, due to the extremely differential information to be measured, such experiments are at the limit of feasibility even for the most efficient set-ups. In alternative, several experiments have been performed by detecting only one of the two ejected electrons, either the photoelectron in nucleobases [57, 99], nucleosides [59, 100], ribose [69] and amino acids [101], or the Auger electron [102] in electron energy resolved photoion photoion coincidence (PEPIPICO) measurements. These experiments provide detailed information of the dynamics of fragmentation and bond cleavage patterns in doubly ionised molecules. These PEPIPICO experiments on nucleobases have shown that the final fragments are produced directly by simple ring bond fractures with some possibility of hydrogen migration without involving complex geometrical rearrangements of the parent molecule, while in ribose a very strong damage is observed following core ionisation, with the residual of very small fragments. In nucleosides, due to redistribution of the final valence shell holes, the fragmentation does not exhibit any significant site-specific fragmentation and indeed cannot be described by the fragmentation of separated nucleobase and ribose dications.
3.4 Time-Dependent Studies
The absorption of a photon from the IR to X-ray can initiate processes and chemical reactions in molecules of biological interest. How these processes depend on the time scales, energetics, and molecular distances has been the subject of considerable research effort. The advent of radiation sources with tunable energy and duration, which is comparable with the time scale of the nuclei, and, more recently, electronic dynamics has disclosed the possibility to perform time-resolved studies with the aim to understand as well as to control these processes. The technique adopted in these studies, named pump-probe spectroscopy, involves the excitation of the sample by one pulse train (pump) and the monitoring of the changes induced in the sample by the second pulse train (probe), which is suitably delayed with respect to the pump.
The two examples reported below have been chosen considering the new challenges and opportunities disclosed by the advent of new excitation sources like the soft X-ray Free Electron Lasers (FEL) and the ultra fast lasers.
UV(\(\lambda<\) 400 nm)-induced damage to DNA has profound biological consequences, including photocarcinogenesis [103,104,105], thus it is not surprising that the excited states of the nucleic acid are highly stable to photochemical decay, perhaps as a result of natural selection during a long period of molecular evolution. This photostability due to the rapid decay for electronic energy, has attracted and is attracting a lot of interest. These studies benefit a lot by the development of fs lasers, VUV and soft X-ray FEL’s and molecular beam techniques that allow the study of isolated DNA bases despite their low vapour pressure and easy thermal decomposition, as well as advances in quantum chemistry that have made possible the modelling of excited states. Crespo-Hernandez et al. [106] reviewed in 2005 the impressive armoury of experimental and theoretical techniques that have been used to study excited states in nucleic acids and their constituents. All that work has shown convincingly that the fluorescence lifetimes of single DNA bases in solution at room temperature are in the subpicosecond range. For base monomers, the lack of solvent effects in the condensed phase and the ultrashort lifetimes in supersonic jet experiments suggest [103] that internal conversion is not the result of strong solute–solvent interactions, but the inevitable outcome of nonadiabatic dynamics on the complex potential energy landscape of the bases, leading to the redistribution of both charge and energy within the molecule via the coupling of vibrational with electronic degrees of freedom. As photoelectron spectroscopy is sensitive to both molecular orbital configurations and vibrational dynamics, Ullrich et al. [12] used Time-Resolved PhotoElectron Spectra (TRPES) to study the electronic relaxation processes in DNA and RNA bases adenine, cytosine, thymine and uracil in a molecular beam. As opposed to other experimental approaches, like for example time-resolved ion yield measurements [107], TRPES allows for direct identification of the states involved in the electronic relaxation process through projection of the excited state dynamics onto cationic states. Hence photoelectron spectroscopy can provide information on the character of the excited state and, since ionisation is always allowed, states appearing as ‘dark’ in absorption can be probed. In the experiment pump wavelengths between 250 and 277 nm lead to initial excitation of the bright \(S_2{(\uppi \uppi ^*)}\) state and ionisation was then produced by a pulse at 200 nm. The instrument response function of 160 fs Full Width at Half Max (FWHM) allowed the determination of time constants down to about 20 % of the cross-correlation FWHM, i.e. 40 fs. The main attention has been placed on adenine and the results show that the initially prepared bright S\(_{2}\) (\(\uppi \uppi \)*) state decays rapidly (<50 fs) to the S\(_{1}\) (n\(\uppi \)*) state, which has a lifetime of 750 fs. There are also indications for an additional decay pathway consistent with the theoretically predicted S\(_{3}(\uppi \sigma \)*). The results for the pyrimidine bases, Fig. 9, displayed a dynamics with a multi-exponential decay: an ultrashort decay <50 fs followed by a slower one in the range of several hundreds of fs (820 fs for cytosine, 490 fs for thymine, 530 fs for uracil) and a ps channel (3.2 ps for cytosine, 6.4 ps for thymine, 2.4 ps for uracil).
Comparison of decay traces extracted from two dimensional global fits to TRPES measurements of cytosine, thymine and uracil [12]. Black circles represent the energy-integrated photoelectron signal for each channel and the red line is the best fit. Four channels are assumed: a Gaussian component (blue), a short (green) and a long-lived (purple) exponential decay, and a probe–pump signal (grey). See text for details
These time constants are consistent with calculations of the potential energy surface [108,109,110,111,112], but the assignment of the transients to electronic or nuclear relaxation varies among theoretical models. Approaches based on linear interpolation [110] or minimal energy paths [111] predict a barrierless fast decay to the electronic ground, thus an electronic relaxation. Dynamic simulations [108] predict an indirect path [112] where the initial ultrafast nuclear relaxation into the \(\uppi \uppi \)* state involves the C–O stretch, with the population trapped for picoseconds behind a reaction barrier. This suggests an interpretation of the 100 fs constant as a nuclear relaxation. Ultrafast pump-probe optical spectroscopies due to the reduced Franck–Condon overlap upon vibrational relaxation are unable to disentangle among these models. An alternative is provided by XPS or Auger electron spectroscopies, which can core ionise the molecule at any nuclear geometry. Time-resolved studies in the core shell region needed the advent of X-ray FELs where the evolution of the wavepacket in the valence excited states is followed by monitoring its effects on the inner shell binding energies, in a direct way by XPS and indirectly by Auger electron spectroscopy. Due to the state- and site-selectivity of the core spectroscopy the dynamics of the excited molecule can be observed by a ‘local’ point of view. A first experiment [113] exploited the short duration (50–80 fs) of the LCLS at the SLAC National laboratory to ionise the O1s in thymine following the excitation of a 70 fs UV pulse of 266 nm. The O KVV Auger spectrum, measured as a function of the delay between the two pulses, showed a dominant 200 fs electronic relaxation of the photoexcited \(\uppi \uppi \)* state and lead to the conclusion that, under the conditions of that experiment, the majority of the excited-state population is not efficiently trapped by an excited-state reaction barrier. On the other hand some preliminary results on the time-resolved XPS of C1s on uracil [114] also measured at LCLS appear to support the existence of a reaction barrier as previously predicted [108]. Thus, still there are many remaining puzzles to completely disentangle the nature of the photostability of nucleobases to UV radiation damage.
Transfer of electronic charge within a single molecule plays a key role in catalysis, DNA damage by ionizing radiation, photosynthesis, respiration. The ability of molecules such as peptides and DNA to act as charge conduits is an intrinsic part of many biological processes. Photodissociation studies with ns UV laser radiation has provided information on the charge transfer, for example in protonated peptides isolated in vacuo, and how, depending on the initial site of photoexcitation, this can lead to a slow and non-hazardous statistical dissociation or a prompt cleavage of peptide bonds [115]. However, only pump-probe experiments can fully elucidate the charge location during the process. In their pioneer works with nanosecond laser in the 1990s Weinkauf and coworkers showed that if an electron is selectively ionised from a chromophore at a terminal end of a peptide, then the location of the charge could be probed using the shift in absorption of the chromophore, so that the passage of the charge through up to 12 sigma bonds in a quadrapeptide was observed [116, 117]. Later experiments were able to extract a 80 fs lifetime for charge transfer from an ionised chromophore to the amine group in 2phenylethyl-N,Ndimethylamine molecule and more recent calculations [118] predicted that, depending on the cationic states contributing to the wavepacket and the conformation of the neutral molecule, the charge migration across the full extent of the molecule could take 5 fs or less. This represents therefore one of the most suited processes to exploit the performances of attosecond pulse trains in the extreme ultraviolet [119]. Calegari et al. [120] investigated ultrafast charge dynamics in the amino acid phenylalanine after prompt ionisation induced by isolated attosecond pulses. A probe pulse then produced a doubly charged molecular fragment by ejection of a second electron, and charge migration manifested itself as a sub-4.5 fs oscillation in the yield of this fragment as a function of pump-probe delay, Fig. 10. This temporal scale is definitely shorter than the vibrational response of the molecule. Numerical simulations of the temporal evolution of the electronic wave packet created by the attosecond pulse supported the interpretation of the experimental data in terms of charge migration, resulting from a periodic variation of the charge density around the amine group.
Pump-probe measurements in phenylalanine amino acid [120]. (a) Yield of doubly charged immonium ion (m/z 60) as a function of pump-probe delay, measured with 3 fs temporal steps. The red line is a fitting curve with an exponential rise time of 10 fs and an exponential relaxation time of 25 fs. (b) Yield of doubly charged immonium ion versus pump-probe delay measured with 0.5-fs temporal steps, within the temporal window shown as dotted box in (a). The red line is the fitting curve given by the sum of the fitting curve shown in (a) and a sinusoidal function of frequency 0.234 PHz (4.3 fs period). (c) Difference between the experimental data and the exponential fitting curve displayed in (a)
4 Conclusions
In this chapter it has been shown how the electromagnetic radiation sources, mainly synchrotron radiation, combined with electron spectroscopies and mass spectrometry can be used for the characterisation of the electronic structure, the conformations and the dynamic processes that lead to the redistribution of the energy absorbed by molecules of biological interest, such as amino acids, peptides and the building blocks of nucleic acids. The advent of more intense and short pulse sources will enlarge the number of molecules to be investigated, overcoming the limitation of the low density of the beams produced by novel methods.
While most of the work performed up to now has been restricted to thermal evaporation, and therefore to small molecules, a promising research area for the future is towards larger molecules, brought into the gas phase by sophisticated and softer methods. Among others, the state of the art technique to bring unfragmented nucleotides, proteins or peptides from solution into the gaseous phase is represented by the ESI technique [121, 122]. The ESI source overcomes the difficulties of introducing large molecules prepared in solution into a mass spectrometer, which works in a high vacuum environment. Since the commercial development of ESI sources, mass spectrometry has become the most popular tool for the study of very large organic molecules as analytical tool as well as for innovative research applications. The coupling of this versatile ion source with spectroscopic techniques implies that the mass selected ions generated in the gas phase by an ESI source are collected in an ion trap and then excited/ionised by the radiation. The combination of an ESI with powerful fs laser has provided a much better sequence analysis of the protein structure and despite the low density of the target beam, which hampers the achievement of a good signal-to-noise ratio, with the available photon flux in third generation synchrotron sources, some pioneering works using mass spectrometric techniques have been reported [64, 82, 88, 90, 98, 123,124,125]. Using similar methods, Giuliani et al. [90] produced free protonated and charge state-selected cytochrome ions (consisting of 104 amino acids) and performed VUV photoionisation mass spectrometry. An improvement of the throughput of the sources is needed in order to make them suitable for electron spectroscopies. The high intensity of the new FEL sources will certainly favour the use of ESI sources.
Another subject in fast development is related to the investigation of the effects of the environment in the ionisation/fragmentation of biomolecules. The natural environment is an aqueous solution. Thus sources of dry or hydrated clusters of biomolecules are being developed [126,127,128]. In this way a solvated environment can be mimicked in the gas phase and its effects on the structure and properties of the biomolecular system investigated.
Furthermore, the new radiation sources will made possible, as described in Sect. 3.4, to follow the evolution of the energy flow in the molecule as well as processes like H migration, molecular dynamics (rotation or bending of bonds, etc.) which can strongly affect the chemical behaviour of these molecules or their fragments.
References
Bodaïffa B, Cloutier P, Hunting D, Huels MA, Sanche L (2000) Science 287:1658
Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM (2003) Science 246 (1989) 64. Fenn Angew JB Chem Int Ed 42:3871
Karas M, Bachmann D, Hillenkamp F (1985) Anal Chem 57:2935
Golovlev VV, Allman SL, Garrett WR, Taranenko NI, Chen CH (1997) Int J Mass Spectrom 169–170:69
Ross KJ, Sonntag B (1995) Rev Sci Instrum 66:4409
Denifl S, Matejcik S, Gstir B, Hanel G, Probst M, Scheier P, Märk TD (2003) J Phys Chem 118:4107
Kopyra J, König-Lehmann C, Szamreja I, Illenberger E (2009) Int J Mass Spectrom 285:131
Bergen T, Biquard X, Brenac A, Chandezon F, Huber BA, Jalabert D, Lebius H, Maurel M, Monnand E, Opitz J, Pesnelle A, Pras B, Ristori C, Rocco JC (1999) Rev Sci Instrum 70:3244
Lago AF, Countinho LH, Marinho RRT, Naves de Brito A, de Souza GGB (2004) Chem Phys 307:9
Tabet J, Eden S, Feil S, Abdoul-Carime H, Farizon B, Farizon M, Ouaskit S, Märk TD (2010) Nucl Instrum Methods Phys Res Sect B 268:2458
Levola H, Kooser K, Rachlew E, Nömmiste E, Kukk E (2013) Int J Mass Spectrom 353:7
Ullrich S, Schultz T, Zgierski MZ, Stolow A (2004) Phys Chem Chem Phys 6:2796
Siegbahn K (ed) (1969) ESCA applied to free molecules. North-Holland, ISBN 0720401607
Stöhr J (1992) NEXAFS spectroscopy. Springer Science and Business Media. ISBN 978-3-662-02853-7
Wiley WC, McLaren IH (1955) Rev Sci Instrum 25:1150
Maclot S (2014) Ph.D. thesis
Plekan O, Coreno M, Feyer V, Moise A, Richter R, de Simone M, Sankari R, Prince KC (2008) Phys Scr 78:58105
Kukk E, Sankari R, Huttula M, Sankari A, Pilling S, Aksela S (2007) J Electron Spectrosc Relat Phenom 155:141
Kimura K, Katsumata S, Achiba Y, Yamazaki T, Iwata S (1981) Handbook of He I photoelectron spectra of fundamental organic molecules. Halsted Press, New York
Debies TP, Rabalais JW (1974) J Electron Spectrosc Relat Phenom 3:315
O’Keeffe P, Bolognesi P, Casavola AR, Catone D, Zema N, Turchini S, Avaldi L (2009) Mol Phys 107:2025
Gerson SH, Worley SD, Bodor N, Kaminski JJ, Flechtner TW (1978) J Electron Spectrosc Relat Phenom 13:421
Schwell M, Hochlaf M (2014) Topics of current chemistry. Springer, Berlin, Heidelberg
Holland DMP, Potts AW, Karlsson L, Stener M, Decleva P (2011) Chem Phys 390:25
Trofimov AB, Schirmer J, Kobychev VB, Potts AW, Holland DP, Karlsson L (2006) J Phys B: At Mol Opt Phys 39:305
Zaytseva IL, Trofimov AB, Schirmer J, Plekan O, Feyer V, Richter R, Coreno M, Prince KC (2009) J Phys Chem A 113:15142
Plekan O, Feyer V, Richter R, Coreno M, Prince KC (2008) Mol Phys 106:1143
Plekan O, Feyer V, Richter R, Coreno M, de Simone M, Prince KC, Carravetta V (2007) J Phys Chem A 111:10998
Feyer V, Plekan O, Richter R, Coreno M, Prince KC, Carravetta V (2009) J Phys Chem A 113:10726
Wickrama Arachchilage AP, Wang F, Feyer V, Plekan O, Prince KC (2010) J Chem Phys 133:174319
Wickrama Arachchilage AP, Wang F, Feyer V, Plekan O, Prince KC (2012) J Chem Phys 136:124301
Dampc M, Mielewska B, Siggel-King MRF, King GC, Zubek M (2009) Chem Phys 359:77
Castrovilli MC, Bolognesi P, Cartoni A, Catone D, O’Keeffe P, Casavola AR, Turchini S, Zema N, Avaldi L (2014) J Am Soc Mass Spectrom 25:351
Feyer V, Plekan O, Richter R, Coreno M, Prince KC (2009) Chem Phys 358:33
Maris A, Melandri S, Evangelisti L, Caminati W, Giuliano BM, Plekan O, Feyer V, Richter R, Coreno M, Prince KC (2012) J Electron Spectros Relat Phenom 185:244
Ahmed M, Ganesan A, Wang F, Feyer V, Plekan O, Prince KC (2012) J Phys Chem A 116:8653
Ahmed M, Wang F, Acres RG, Prince KC (2014) J Phys Chem A 118:3645
Feketeova L, Plekan O, Goonewardane M, Ahmed M, Albright AL, White J, O’Hair RAJ, Horsman MR, Wang F, Prince KC (2015) J Phys Chem A 119:9986
Giuliani A, Limão-Vieira P, Duflot D, Milosavljevic AR, Marinkovic BP, Hoffmann SV, Mason N, Delwiche J, Hubin-Franskin M-J (2009) Eur Phys J D 51:97
Stener M, Decleva P, Holland DMP, Shaw DA (2011) J Phys B: At Mol Opt Phys 44:075203
Ferreira da Silva F, Almeida D, Martins G, Milosavljevic AR, Marinkovic BP, Hoffmann SV, Mason NJ, Nunes Y, Garcia G, Limao-Vieira P (2010) Phys Chem Chem Phys 12:6717
Potts AW, Holland DMP, Trofimov AB, Schirmer J, Karlsson L, Siegbahn K (2003) J Phys B: At Mol Opt Phys 36:3129
Nahon L, Garcia GA, Powis I (2015) J Electron Spectros Relat Phenom 204:322 (and references there in)
Tia M, Cunha De Miranda B, Daly S, Gaie-Levrel F, Garcia GA, Powis I, Nahon L (2013) J Phys Chem Lett 4:2698
Touboul D, Gaie-Levrel F, Garcia GA, Nahon L, Poisson L, Schwell M, Hochlaf M (2013) J Chem Phys 138:094203
Powis I, Rennie EE, Hergenhahn U, Kugeler O, Bussy-Socrate R (2003) J Phys Chem A 107:25
Hochlaf M, Pan Y, Lau KC, Majdi Y, Poisson L, Garcia GA, Nahon L, Al-Mogren MM, Schwell M (2015) J Phys Chem A 119:1146
Majdi Y, Hochlaf M, Pan Y, Lau K-C, Poisson L, Garcia GA, Nahon L, Al-Mogren MM, Schwell M (2015) J Phys Chem A 119:5951
Jochims H-W, Schwell M, Chotin JL, Clemino M, Dulieu F, Baumgärtel H, Leach S (2004) Chem Phys 298:279
Richer G, Sandorfy C, Chaer Nascimento MA (1984). J Electron Spectrosc Relat Phenom 34:327
Slaughter AR, Banna MS (1988) J Phys Chem 92:2165
Feyer V, Plekan O, Richter R, Coreno M, Prince KC, Carravetta V (2008) J Phys Chem A 112:7806
Feyer V, Plekan O, Kivimaki A, Prince KC, Moskovskaya TE, Zaytseva IL, Soshnikov DY, Trofimov AB (2011) J Phys Chem A 115:7722
Plekan O, Feyer V, Richter R, Coreno M, Vall-llosera G, Prince KC, Trofimov AB, Zaytseva IL, Moskovskaya TE, Gromov EV, Schirmer J (2009) J Phys Chem A 113:9376
Feyer V, Plekan O, Richter R, Coreno M, Vall-llosera G, Prince KC, Trofimov AB, Zaytseva IL, Moskovskaya TE, Gromov EV, Schirmer J (2009) J Phys Chem A 113:5736
Plekan O, Feyer V, Richter R, Coreno M, de Simone M, Prince KC, Trofimov AB, Gromov EV, Zaytseva IL, Schirmer J (2008) Chem Phys 347:360
Itälä E, Ha DT, Kooser K, Huels MA, Rachlew E, Nõmmiste E, Joost U, Kukk E (2011) J Electron Spectrosc Relat Phenom 184:119
Lin Yi-Shiue, Lin Huei-Ru, Liu Wei-Lun, Lee Yuan T, Tseng Chien-Ming, Ni Chi-Kung, Liu Chen-Lin, Tsai Cheng-Cheng, Chen Jien-Lian, Wei-Ping Hu (2015) Chem Phys Lett 636:146
Itälä E, Kooser K, Rachlew E, Levola H, Ha DT, Kukk E (2015) J Chem Phys 142:194303
Milosavljevic AR, Cerovski VZ, Canon F, Rankovic ML, N. Škoro, Nahon L, Giuliani A (2014) J Phys Chem Lett 5:1994
Zhang W, Carravetta V, Plekan O, Feyer V, Richter R, Coreno M, Prince KC (2009) J Chem Phys 131:035103
Plekan O, Feyer V, Richter R, Coreno M, de Simone M, Prince KC, Carravetta V (2007) Chem Phys Lett 442:429
Marinho RRT, Lago AF, Homem MGP, Coutinho LH, de Souza GGB, Naves de Brito A (2006) Chem Phys 324:420
Gonzalez-Magaña O, Reitsma G, Tiemens M, Boschman L, Hoekstra R, Schlathölter T (2012) J Phys Chem A 116:10745
Milosavljević AR, Nicolas C, Ranković ML, Canon F, Miron C, Giuliani A (2015) J Phys Chem Lett 6:3132
Bolognesi P, Mattioli G, O’Keeffe P, Feyer V, Plekan O, Ovcharenko Y, Prince KC, Coreno M, Amore Bonapasta A, Avaldi L (2009) J Phys Chem A 113:48
Bolognesi P, O’Keeffe P, Ovcharenko Y, Coreno M, Avaldi L, Feyer V, Plekan O, Prince KC, Zhang W, Carravetta V (2010) J Chem Phys 133:034302
Plekan O, Feyer V, Richter R, Moise A, Coreno M, Prince KC, Zaytseva IL, Moskovskaya TE, Soshnikov DY, Trofimov AB (2012) J Phys Chem A 116:5653
Ha DT, Huels MA, Huttula M, Urpelainen S, Kukk E (2011) Phys Rev A 84:033419
Watson JD, Crick F (1953) Nature 171:964
Nivethaa EAK, Dhanavel S, Narayanan V, Vasuc CA, Stephen A (2015) RSC Adv 5:1024 (and reference therein)
Wang B, Feng W, Chai Z, Zhao Y (2015) Sci China 58:768
Pilling S, Lago AF, Coutinho LH, De Castilho RB, De Souza GGB, De Brito AN (2007) Rapid Commun Mass Sp 21:3646
Pilling S, Andrade DPP, de Castilho RB, Cavasso-Filho RL, Lago AF, Coutinho LH, de Souza GGB, Boechat-Roberty HM, Naves de Brito A (2008) Org Matter Space Proc IAU Symp No 251:371
de Souza GGB, Coutinho LH, Nunez C, Bernini R, Castilho RB, Lago AF (2007) J Phys: Conf Ser 88:012005
Levola H, Kooser K, Itälä E, Kukk E (2014) Int J Mass Spectrom 370:96
Barc B, Ryszka M, Poully J-C (2014) Al Maalouf EJ, el Otell Z, Tabet J, Parajuli R, van der Burgt PJM, Limão-Vieira P, Cahillane P, Dampc M, Mason NJ, Eden S. Int J Mass Spectr 194:365–366
Barc B, Ryszka M, Spurrell J, Dampc M, Limão-Vieira P, Parajuli R, Mason NJ, Eden S (2013) J Chem Phys 139:244311
Barbier B, Chabin A, Chaput D, Brack A (1998) Planet Space Sci 46:391
Boillot F, Chabin A, Bure C, Venet M, Belsky A, Bertrand-Urbaniak M, Delmas A, Brack A, Barbier B (2002) Orig Life Evol Biosph 32:359
González-Magaña O, Reitsma G, Bari S, Hoekstra R, Schlathölter T (2012) Phys Chem Chem Phys 14:4351
Milosavljevic AR, Nicolas C, Lemaire J, Dehon C, Thissen R, Bizau J-M, Refregiers M, Nahon L, Giuliani A (2011) Phys Chem Chem Phys 13:15432
Brunet C, Antoine R, Allouche A-R, Dugourd P (2011) J Phys Chem A 115:8933
Bari S, Gonzalez-Magaña O, Reitsma G, Werner J, Schippers S, Hoekstraand R, Schlathölter T (2011) J Chem Phys 134:024314
Milosavljević AR, Nicolas C, Gil JF, Canon F, Réfrégiers M, Nahon L, Giuliani A (2012) J Synch Rad 19:174
Giuliani A, Milosavljević AR, Canon F, Nahon L (2014) Mass Spectrom Rev 33:424
Canon F, Milosavljević AR, van der Rest G, Réfrégiers M, Nahon L, Sarni-Manchado P, Cheynier V, Giuliani A (2013) Angew Chem 125:8535
Canon F, Milosavljević AR, Van Der Rest G, Réfrégiers M, Nahon L, Sarni-Manchado P, Cheynier V, Giuliani A (2013) Angew Chem Int Ed 52:8377
Giuliani A, Milosavljević AR, Hinsen K, Canon F, Nicolas C, Réfrégiers M, Nahon L (2012) Angew Chem 124:9690
Giuliani A, Milosavljević AR, Hinsen K, Canon F, Nicolas C, Réfrégiers M, Nahon L (2012) Angew Chem Int Ed 51:9552
Canon F, Milosavljević AR, Nahon L, Giuliani A (2015) Phys Chem Chem Phys 17:25725
Schwell M, Jochims H-W, Baumgärtel H, Leach S (2008) Chem Phys 353:145
Jochims H-W, Schwell M, Baumgärtel H, Leach S (2005) Chem Phys 314:263
Gaie-Levrel F, Garcia GA, Schwell M, Nahon L (2011) Phys Chem Chem Phys 13:7024
Schwell M, Jochims H-W, Baumgärtel H, Dulieuc F, Leach S (2006) Planet Space Sci 54:1073
Bolognesi P, O’Keeffe P, Avaldi L (2012) Soft X-ray interaction with organic molecules of biological interest. In: Garcia Gomez-Tejedor G, Fuss MC (eds) Radiation damage in biomolecular systems. Springer Science + Business Media B.V, pp 165
Bolognesi P, Kettunen JA, Cartoni A, Richter R, Tosic S, Maclot S, Rousseau P, Delaunay R, Avaldi L (2015) Phys Chem Chem Phys 17:24063
Milosavljević AR, Canon F, Nicolas C, Miron C, Nahon L, Giuliani A (2012) J Phys Chem Lett 3:1191
Itälä E, Ha DT, Kooser K, Nõmmiste E, Joost U, Kukk E (2011) Int J Mass Sp 306:82
Itälä E, Kooser K, Rachlew E, Huels MA, Kukk E (2014) J Chem Phys 140:234305
Itälä E, Huels MA, Rachlew E, Kooser K, Hägerth T, Kukk E (2013) J Phys B: At Mol Opt Phys 46:215102
Ha DT, Wang Y, Alcamí M, Itälä E, Kooser K, Urpelainen S, Huels MA, Kukk E, Martín F (2014) J Phys Chem A 118:1374
Miller DL, Weinstock MA (1994) J Am Acad Dermatol 30:774
Kraemer KH (1997) 94:11
Young AR (1997) Br J Clin Pract 89:10
Crespo-Hernandez CE, Cohen B, Hare PM, Kohler B (2004) Chem Rev 104:1977
Canuel C, Mons M, Piuzzi F, Tardivel B, Dimicoli I, Elhanine M (2005) J Chem Phys 122:074316
Hudock HR, Levine BG, Thompson AL, Satzger H, Townsend D, Gador N, Ullrich S, Stolow A, Martínez TJ (2007) J Phys Chem A 111:8500
Szymczak JJ, Barbatti M, Soo Hoo JT, Adkins JA, Windus TL, Nachtigallová D, Lischka H (2009) J Phys Chem A 113:12686
Perun S, Sobolewski AL, Domcke WJ (2006) J Phys Chem A 110:13238
Merchan M, González-Luque R, Climen T, Serrano-Andrés L, Rodríguez E, Reguero M, Peláez D (2006) J Chem Phys B 110:26471
Asturiol D, Lasorne B, Robb MA, Blancafort LJ (2009) Phys Chem A 113:10211
McFarland BK, Farrell JP, Miyabe S, Tarantelli F, Aguilar A, Berrah N, Bostedt C, Bozek JD, Bucksbaum PH, Castagna JC, Coffee RN, Cryan JP, Fang L, Feifel R, Gaffney KJ, Glownia JM, Martinez TJ, Mucke M, Murphy B, Natan A, Osipov T, Petrović VS, Schorb S, Schultz Th, Spector LS, Swiggers M, Tenney I, Wang S, White JL, White W, Gühr M (2014) Nature. Communication 5:4235
Bolognesi P et al (2015) J Phys Conf Ser 635:112062
Skinnerup Byskov C, Jensen F, Jørgensenc TJD, Brøndsted Nielsen S (2014) Phys Chem Chem Phys 16:15831
Weinkauf R, Aicher P, Wesley G, Grotemeyer J, Schlag EW (1994) J Phys Chem 98:8381
Weinkauf R, Schanen P, Metsala A, Schlag EW, Bürgle M, Kessler H (1996) J Phys Chem 100:18567
Lünnemann S, Kuleff AI, Cederbaum LS (2013) Chem Phys 414:100
Krausz F, Ivanov M (2009) Rev Mod Phys 81:163
Calegari F, Ayuso D, Trabattoni A, Belshaw L, De Camillis S, Anumula S, Frassetto F, Poletto L, Palacios A, Decleva P, Greenwood JB, Martín F, Nisoli M (2014) Science 346:336
Yamashita M, Fenn JB (1984) J Phys Chem 88:4451
Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM (1989) Science 246:64
Reitsma G, Boschman L, Deuzeman MJ, González-Magaña O, Hoekstra S, Cazaux S, Hoekstra R, Schlathölter T (2014) Phys Rev Lett 113:053002
Milosavljević AR, Giuliani A, Nicolas C, Gil J-F, Lemaire J, Réfrégiers M, Nahon L (2010) J Phys: Conf Ser 257:012006
Milosavljević AR, Cerovski VZ, Canon F, Nahon L, Giuliani A (2013) Angew Chem Int Ed 52:7286
Maclot S, Capron M, Maisonny R, Ławicki A, Méry A, Rangama J, Chesnel J-Y, Bari S, Hoekstra R, Schlatholter T, Manil B, Adoui L, Rousseau P, Huber BA (2011) Chem Phys Chem 12:930
Zimmermann U, Malinowski N, Näher U, Franck S, Martin TP (1994) Z Physik D 31:85
Khistyaev K, Golan A, Bravaya KB, Orms N, Krylov AI, Ahmed M (2013) J Phys Chem A 117:6789
Acknowledgements
Work partially supported by the COST Action Nano-IBCT (CM 1002) and XLIC (CM1204), MAE-CI Italia-Serbia ‘A nanoview of radiation-biomatter interaction’.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Bolognesi, P., Avaldi, L. (2017). Photoprocesses with Biomolecules in the Gas Phase. In: Solov’yov, A. (eds) Nanoscale Insights into Ion-Beam Cancer Therapy. Springer, Cham. https://doi.org/10.1007/978-3-319-43030-0_6
Download citation
DOI: https://doi.org/10.1007/978-3-319-43030-0_6
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-43028-7
Online ISBN: 978-3-319-43030-0
eBook Packages: Physics and AstronomyPhysics and Astronomy (R0)