Abstract
Automated segmentation of intervertebral discs (IVDs) from magnetic resonance imaging has the potential to enhance the efficiencies of radiological investigations of large clinical and research imaging datasets. This work presents an automated method for localization and 3D segmentation of IVDs that is applied to magnetic resonance imaging of the thoraco-lumbar spine as part of the segmentation challenge at the 3rd MICCAI Workshop & Challenge on Computational Methods and Clinical Applications for Spine Imaging - MICCAI–CSI2015. Our initialization method involves multi-atlas registration and a hierarchical conditional shape regression for localization of all imaged lumbar and thoracic discs, and active shape model based 3D segmentation. Comparisons between manual (ground truth) and automated segmentation of 105 disc volumes (T11/T12 - L5/S1) revealed a mean Dice score of \(0.896\,{\pm }\,0.024\) and mean absolute square distance of \(0.642\,{\pm }\,0.169\) mm. Our automated segmentation approach provided accurate segmentation of IVDs from turbo spine echo images which are highly competitive with leading state-of-the-art 3D segmentation techniques.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Spine-related disorders account for the largest proportion of musculoskeletal complaints in industrialized countries [1, 2]. Magnetic resonance imaging (MRI) allows highly detailed, multiplanar investigations of spine pathologies, such as intervertebral disc (IVD) prolapse, herniation and degeneration [3]. Informatic tools offer significant opportunities for improving the efficiency of radiological assessment of the spine by reducing the time- and expertise-intensive encumbrances of tedious tasks as required in three-dimensional (3D) segmentation and measurement of anatomical structures. Precise segmentation of IVDs is a pre-requisite for many clinical applications (diagnosis, treatment planning and evaluation), and automated segmentation has the potential to enhance the efficiencies of radiological investigations of large clinical and research imaging datasets.
This work presents a fully automated algorithm for 3D segmentation of lumbar and thoracic IVDs from sagittal T2-weighted MRI scans and evaluates it on a publicly available dataset as part of the challenge on “Automatic IVD localization and segmentation from 3D T2 MRI data” at the 3rd MICCAI Workshop & Challenge on Computational Methods and Clinical Applications for Spine Imaging - MICCAI–CSI2015. The current method extends and fully automates our previous work based on segmentation of lumbar spine IVDs via active shape models (ASMs) [4]. This ASM approach has been applied successfully in a series of studies but requires further development of the pipeline for more generalized application in a clinical framework. Initially, our fully automated approach was developed using 3D sampling perfection with application optimized contrast using different flip angle evolution (SPACE) scans [4], while clinical examinations are routinely performed using two-dimensional (2D) turbo spin echo (TSE) images. In clinical TSE scans, this ASM segmentation scheme was successfully applied to the segmentation of IVDs in the lumbar region although a simple and quick manual initialization step was required in the form of point identification of individual vertebræ [5, 6]. The automated initialization on the 3D SPACE scans [4] made use of the detailed high-resolution imaging information in the axial plane that is not available in routine sagittal TSE images. Moreover, the validation of the 3D segmentation algorithm on clinical TSE datasets was performed through a simplified evaluation framework based on 2D manual segmentations in the mid-sagittal slice only. The purpose of the present work is to perform a volumetric validation of our 3D segmentation scheme against the manual segmentation of IVDs acquired from an entire MRI set of TSE sagittal slices, as offered through the MICCAI–CSI2015 segmentation challenge. Notably, two important advances are addressed in this current work: (i) an improved automated IVD localization to provide a fully automated pipeline well-suited to the processing of sagittal MRI scans acquired in routine clinical examinations, and (ii) a larger, external validation of our automated 3D IVD segmentation system on a publicly available MRI dataset having “ground-truth” manual segmentations of the IVDs performed in all sagittal slices in T2-weighted TSE examinations.
2 Methods
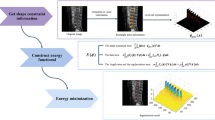
The central methodological innovations of this work lie in designing a novel approach for fully automated IVD localization from sagittal TSE images (Fig. 1), which provide a robust basis for subsequent 3D segmentation. The segmentation algorithm is based on a previously presented method of IVD segmentation using a 3D ASM-based approach [4]. The initialization pipeline for lumbar spine localization is summarized in Fig. 1 and explained in detail in the next sections.
Summary of the automated sacrum localization method. First, training 2D atlases (A) are rigidly registered to the inferior mid-sagittal, mid-coronal portion of the processed case (B). Second, the best (based on normalized mutual information) result is used to initialize subsequent registration of an average atlas (C). Finally, a 3D deformable template is used to pre-segment the L5 and S1 vertebral bodies (D).
2.1 Prior Knowledge
The initialization method employs prior knowledge from a training dataset of 2D spine atlases and their segmentations in the following three forms:
2D Atlases. Training atlases were extracted from 35 T2-weighted sagittal TSE examinations (in-plane resolution \(0.71\,{\times }\,0.71\) mm, image matrix \(448\,{\times }\,448\)) acquired from a heterogeneous sample of patients (different from the training data provided as part of the MICCAI–CSI2015 challenge). The selection of training atlases focused on including a spectrum of patients with varying anatomy and pathology. This procedure aimed to increase robustness for application to unseen datasets, which would be limited if atlases from the same datasets were used in the leave-one-out fashion. The mid-sagittal sections were individually cropped (to identical dimensions) to include the inferior mid-coronal section of the mid-sagittal training slice. The field-of-view (FoV) differed considerably among the training scans and therefore varying portions of the sacrum and coccyx were visualized in the atlases (see Fig. 1A for some examples). The heterogeneity of the FoV among atlases is important for enabling robustness for varying FoV which may be encountered across the spectrum of cases for segmentation.
Average 2D Atlas. The 2D atlases were also used to create an average atlas of the L5-S1-S2 region (Fig. 1C). The registration during atlas generation was performed using a robust inverse-consistent rigid registration algorithm [7], followed by non-rigid registration based on the method of diffeomorphic deamons [8].
3D Statistical Shape Model of the L5-S1 Vertebral Bodies. Automated segmentations of the 35 original (uncropped) training scans were used to create an ASM of the combined L5 and S1 vertebral bodies (Fig. 1D) [4].
2.2 Spine Localization
The atlases and deformable templates were used to automatically initialize the ASM-based IVD segmentation in the following fashion:
Approximate Sacrum Localization Using Multi-Atlas Registration. For each case to be segmented, all atlases were registered to the mid-sagittal slice using the robust inverse-consistent rigid registration algorithm [7]. The registration was initialized by automatically positioning each atlas to the inferior mid-coronal area of the mid-sagittal slice (Fig. 1A). The registration results were compared using normalized mutual information metric and the best atlas was selected for the subsequent localization step (Fig. 1B).
Refinement of the Sacrum Location by Average Atlas Registration. The position of the selected registered atlas was used to automatically place an average atlas template of the sacral portion of the spinal region (Fig. 1C), which was subsequently registered to the case to be segmented [7]. This step was found to positively complement the multi-atlas selection and increased the accuracy of the localization pipeline. This is likely due to the reduced dimensions of the average atlas that gives space to precisely fit the L5-S1-S2 region, compared to the larger individual atlases (Fig. 1).
Pre-Segmentation of the L5 and S1 Vertebral Bodies Using Deformable Template Registration. The registered 2D average atlas was used to automatically place a 3D deformable template combining the L5 and S1 vertebral bodies (Fig. 1D). The deformable template was 3D, unlike the image atlases that were registered in the mid-sagittal plane to increase the computational efficiency. The deformable template was laterally centered around the mid-sagittal slice and used for approximate segmentation of the S1 and L5 vertebral bodies in 3D. The deformable segmentation was based on an ASM strategy, similar to the one later applied to the IVDs [4].
Localization of the Neighboring Lumbar IVDs Using Conditional Shape Models. Automatic initialization of the ASM segmentation of the IVDs was performed hierarchically using the segmentation of the S1, L5 vertebral bodies and conditional shape models of de Bruijne et al. [9]. The conditional shape models describe relations between neighboring shapes \(S_1\) (shape to be estimated) and \(S_2\) (known shape) using a probability distribution based on Gaussian conditional density \(P(S_1|S_2)\). The most likely estimate \(\mu \) of the shape \(S_1\) can be obtained as:
where \(\mu _1\) and \(\mu _2\) are the mean shapes of the training data of \(S_1\) and \(S_2\), \(\varSigma \) is the combined covariance matrix, and \(\gamma \) is a ridge regression coefficient to improve numerical stability [9].
Using the conditional probabilities, the shapes of the L5/S1 and L4/L5 IVDs were estimated from the pre-segmented L5 and S1 vertebral bodies. These estimates served as an initialization for the ASM segmentation of these two lumbar IVDs. In the next step, the initial shape of the L3/L4 IVD was estimated from the segmentation of L4/L5 and L5/S1 IVDs. This iterative process continued superiorly until the end of the image FoV was reached.
2.3 Imaging Dataset
Our 3D ASM-based approach for segmentation of lumbar IVDs was validated against the publicly available manually segmented MRI dataset released via the SpineWeb initiativeFootnote 1. The datasets consisted of sagittal T2-weighted TSE scans acquired from 25 subjects from a 1.5T MRI scanner (Magnetom Sonata, Siemens Healthcare, Erlangen, Germany). The images (39 slices per case) were acquired with in-plane resolution \(1.25\,{\times }\,1.25\) mm (image matrix \(305\,{\times }\,305\)) and slice spacing 3.3 mm [10]. The data was split into three sets: a training dataset (15 cases), a testing dataset (five cases) and a dataset for live segmentation challenge (five cases). Manual “ground-truth” segmentations of seven IVDs (T10/T11 - L5/S1) were provided for 15 cases in the training dataset for quantitative evaluation and parameter tuning.
2.4 Implementation Details
The ASM segmentation in our study was run in two steps. The parameter specifications, enabling progressive refinement of the deformable models, are provided in Table 1 (see Neubert et al. [4] for fuller parameter descriptions).
3 Results
The segmentation results were evaluated using the Dice score coefficient (DSC) and mean absolute square distance (MASD) similarity metrics (Fig. 2).
The testing dataset of 15 cases was segmented with mean DSC value of \(0.896\,{\pm }\,0.024\) between our automated and the manual segmentation measures of IVD volumes. The overall mean MASD was \(0.642\,{\pm }\,0.169\) mm. The large majority (\(95\,\%\)) of the 105 IVDs were segmented with high accuracy based on having DSC\(\,{>}\,0.85\), MASD\(\,{<}\,1\) mm). Two outliers (\(1.9\,\%\)) with lower segmentation accuracy were identified - one L5/S1 IVD (subject 10) and one L4/L5 IVD (subject 4, Fig. 3). The fully automated initialization step worked successfully on 14 out of 15 cases (\(93\,\%\)). Only one case failed and required a simple manual initialization involving one-click identification of a “central” point in the S1 vertebral body in the mid-sagittal slice.
Evaluation on the testing dataset of five cases resulted into mean DSC of \(0.828\,{\pm }\,0.037\) and mean MASD of \(1.39\,{\pm }\,0.13\) mm. The live segmentation challenge on the remaining five cases was achieved with mean DSC of \(0.889\,{\pm }\,0.033\) and mean MASD of \(1.22\,{\pm }\,0.10\) mm.
Qualitative results on an example case (subject 4). The manual segmentation is shown in green, the automated segmentation in red, and the overlap in yellow. The L4/L5 IVD shows lower accuracy (DSC\(\,{=}\,0.809\), MASD\(\,{=}\,1.647\) mm) than the remaining IVDs. The automated segmentation extends beyond the IVD region anteriorly and laterally, as indicated by the arrows. (Color figure online)
4 Discussion
The automated initialization performed very well for 14 / 15 of the training cases providing an excellent basis for our fully automated pipeline. Overall, there was very good agreement between the IVD segmentations from the manual (ground truth) and our automated approaches. In this challenge for segmentation of IVDs from TSE images of the lumbar spine region, our fully automated segmentation approach delivered results highly competitive with leading state-of-the-art 3D segmentation techniques [4, 10, 11]. Only 2 / 105 (1 L5/S1, 1 L4/L5 IVD) outliers were identified (Fig. 2) with slightly lower segmentation accuracy (DSC\(\,{\sim }0.80\)). This was likely due to a combination of factors, such as specific anatomical features (e.g. unclear boundaries between an IVD and the hypo-intense closely apposed psoas muscle) as seen in Fig. 3.
To further evaluate our automated scheme, it would be beneficial to validate our approach on larger datasets of heterogeneous clinical populations. Additional work involving an increase in the number of atlases and varying the initial atlas positioning before the rigid registration may further improve our automated approach by increasing the (already high) percentage of cases initialized fully automatically. Importantly, our system offers a quick and simple “fall-back” option for manual initialization in the form of one mouse click, offering a very fast, robust clinical solution. This approach can also be used if fast segmentation of one isolated IVD is required as it overcomes the need for sequential segmentation starting from the S1, L5 vertebral bodies.
A limitation of the presented framework is the reliance on certain features related to the imaging protocol, e.g. sufficient coverage of the sacrum and a good left/right centering of the spine in the FoV. This plainly influences the applicability of our current models to other spinal regions (e.g. the cervical spine), which would involve the future addition of new imaging atlases. However, clinical MRI examinations of the lumbar spine have extremely similar imaging protocols for patients presenting with low back pain. The vast majority of clinical studies that we have encountered would satisfy the FoV requirements for the applicability of our fully automated method.
5 Conclusion
An automated approach for localization and segmentation of IVDs from T2-weighted sagittal TSE scans of the thoraco-lumbar spine was successfully validated on a publicly available dataset with manual “ground-truth” segmentations. The presented method was used in the segmentation challenge at MICCAI-CSI2015.
References
Woolf, A., Pfleger, B.: Burden of major musculoskeletal conditions. Bull. World Health Organ. 81(9), 646–656 (2003)
Hoy, D., Brooks, P., Blyth, F., Buchbinder, R.: The Epidemiology of low back pain. Best Pract. Res. Clin. Rheumatol. 24(6), 769–781 (2010)
Haughton, V.: Medical imaging of intervertebral disc degeneration: current status of imaging. Spine 29(23), 2751–2756 (2004)
Neubert, A., Fripp, J., Engstrom, C., Schwarz, R., Lauer, L., Salvado, O., Crozier, S.: Automated detection, 3D segmentation and analysis of high resolution spine MR images using statistical shape models. Phys. Med. Biol. 57(24), 8357–8376 (2012)
Neubert, A., Fripp, J., Engstrom, C., Walker, D., Weber, M., Schwarz, R., Crozier, S.: Three-dimensional morphological and signal intensity features for detection of intervertebral disc degeneration from magnetic resonance images. J. Am. Med. Inform. Assoc. 20(6), 1082–1090 (2013)
Neubert, A., Fripp, J., Engstrom, C., Gal, Y., Crozier, S., Kingsley, M.: Validity and reliability of computerized measurement of intervertebral disc height and volume from magnetic resonance images. Spine J. 14(11), 2773–2781 (2014)
Rivest-Hénault, D., Dowson, N., Greer, P., Fripp, J., Dowling, J.: Robust inverse-consistent affine CT-MR registration in MRI-assisted and MRI-alone prostate radiation therapy. Med. Image Anal. 23(1), 56–69 (2015)
Vercauteren, T., Pennec, X., Perchant, A., Ayache, N.: Diffeomorphic demons: efficient non-parametric image registration. NeuroImage 45(1 Suppl.), 61–72 (2009)
de Bruijne, M., Lund, M., Tankó, L., Pettersen, P., Nielsen, M.: Quantitative vertebral morphometry using neighbour-conditional shape models. Med. Image Anal. 11(5), 503–512 (2007)
Chen, C., Belavy, D., Yu, W., Chu, C., Armbrecht, G., Bansmann, M., Felsenberg, D., Zheng, G.: Localization and segmentation of 3D intervertebral discs in MR images by data driven estimation. IEEE Trans. Med. Imaging 34(8), 1719–1729 (2015)
Law, M., Tay, K., Leung, A., Garvin, G., Li, S.: Intervertebral disc segmentation in MR images using anisotropic oriented flux. Med. Image Anal. 17(1), 43–61 (2013)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this paper
Cite this paper
Neubert, A., Fripp, J., Chandra, S.S., Engstrom, C., Crozier, S. (2016). Automated Intervertebral Disc Segmentation Using Probabilistic Shape Estimation and Active Shape Models. In: Vrtovec, T., et al. Computational Methods and Clinical Applications for Spine Imaging. CSI 2015. Lecture Notes in Computer Science(), vol 9402. Springer, Cham. https://doi.org/10.1007/978-3-319-41827-8_15
Download citation
DOI: https://doi.org/10.1007/978-3-319-41827-8_15
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-41826-1
Online ISBN: 978-3-319-41827-8
eBook Packages: Computer ScienceComputer Science (R0)