Abstract
We present a procedure that detects myocardial infarction by analyzing left ventricular shapes recorded at end-diastole and end-systole, involving both shape and statistical analyses. In the framework of Geometric Morphometrics, we use Generalized Procrustes Analysis, and optionally an Euclidean Parallel Transport, followed by Principal Components Analysis to analyze the shapes. We then test the performances of different classification methods on the dataset.
Among the different datasets and classification methods used, we found that the best classification performance is given by the following workflow: full shape (epicardium+endocardium) analyzed in the Shape Space (i.e. by scaling shapes at unit size); successive Parallel Transport centered toward the Grand Mean, in order to detect pure deformations; final statistical analysis via Support Vector Machine with radial basis Gaussian function. Healthy individuals show both a stronger contraction and a shape difference in systole with respect to pathological subjects. Moreover, endocardium clearly presents a larger deformation when contrasted with epicardium. Eventually, the solution for the blind test dataset is given. When using Support Vector Machine for learning from the whole training dataset and for successively classifying the 200 blind test dataset, we obtained 96 subjects classified as normal and 104 classified as pathological. After the disclosure of the blind dataset this resulted in 95 % of total accurracy with sensitivity at 97 % and specificity at 93 %.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
We present a procedure able to detect myocardial infarction by analyzing Left Ventricular (LV) shapes, under the assumption that statistical shape analysis can predict a patient disease status. Such a procedure involves two main issues: shape analysis and statistical analysis
As regards shape analysis, Geometric Morphometrics offers the most used tool, very effective when shape data are based on homologous landmarks, i.e. Generalized Procrustes Analysis (GPA) [1, 2]. GPA may be performed in both Size-and-Shape Space (SSS) or Shape Space (SS); it centers and optimally rotates shapes, optionally scaling to unit size, in order to remove non-shape informed attributes. Usually, GPA is followed by a Principal Components Analysis (PCA) performed on aligned coordinates, which gives a ranking of the main shape-change modes; PCA can be linear or non-linear, and allows visualizing main shape-change modes.
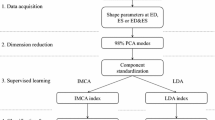
Data handling: Given the raw data, we consider as representative of the LV configurations three different shape-data: the full shape, the endocardial shape, and the epicardial shape. On each of the three datasets, containing 800 shapes, we perform different types of shape analyses whose outcome is split in end-diastolic (ED) or end-systolic (ES) data, numbered from 1 to 6 in the Figure, and then submitted to classification procedures.
A major issue in motion sampling is the generation of homologous landmarks for each time frame, and the selection of homologous time instants along the cardiac cycle; as example, in [3] end-diastolic (ED) and end-systolic (ES) data were analyzed, in [4, 5] the entire cardiac revolution was analyzed, and evaluated at homologous electro-mechanical times.
Another key issue is the discrimination among shape differences and motion differences: two left ventricles having quite different ED and ES shapes may beat in the same way, that is the deformation from ED to ES is the same. A fundamental distinction should be made in the context of shape analysis: is the between-groups shape difference the objective of the analysis or rather the deformation differences occurring between them? This question applies only if, for the same subject, at least two different shapes corresponding to two different times such as ED and ES, appear in the dataset [3]. Optionally, a complete sequence of shapes, representing the entire cardiac revolution, might be included in the analysis as in [4, 5]. Shape differences can be gauged with standard GPA+PCA; its drawback is the mixing of inter- and intra- individual variations, thus preventing the detection of deformation patterns.
When a dataset contains many individuals, each represented by several shapes varying in time, the filtering of inter-individual differences is important. This point underlies many analytical consequences impacting the strategies aimed at exploring the shape data. In fact, while shape differences can be evaluated using standard GPA+PCA, the motion differences among groups should imply the eradication of inter-individual differences. This is necessary because, if a dataset contains different individuals, each represented by several shapes varying in time, standard GPA+PCA ineluctably mix inter- and intra- individual variations, thus preventing the appreciation of pure motion patterns. This problem can be solved by estimating deformations occurring within each individual, and by applying them to a mannequin, which can be the Grand Mean (GM) of the entire dataset, or another appropriately chosen configuration. Of course, the mannequin represents the same shape for all individuals, and in correspondence of it shape differences literally disappear, as it does not vary among individuals. In [4, 5] the GM was used, thus making both ED and ES recognizable as deformed states, but another option could be that of centering in ED, depending upon the disease under study, while ES should never be erased since it contains information about inotropic state.
The geometrical tool needed for such operation is the Levi Civita Parallel Transport (PT) and the workflow of shape analysis becomes GPA+PT+PCA. It is possible to prove that once all the shapes have been optimally aligned (via GPA), an Euclidean translation can well approximate PT on the Riemannian manifold [6]. This kind of PT has the desirable property to maintain the size increment when transporting deformations in SSS (size is here defined as the square root of the summed squared distances of each landmark to the centroid).
As regards statistical analysis, classification problems are a central topic in clinical practice for a wide range of medical fields [7, 8]. However, both the nature of the data and the statistical procedures used for classifying training versus test datasets make this step highly situation-specific. Recently, besides classical methods such as Linear Discriminant Analysis (LDA) and Logistic Regression (LR), several other methods entered classification practice e.g. Quadratic Discriminant Analysis (QDA), Support Vector Machine (SVM), Neural Networks (NN) and Random Forest (RF) among others, whose comparison could be challenging [9–11].
As a consequence, in any specific classification problem a comparison among different classification methods should be performed. In particular, both sensitivity and specificity should be evaluated as they have an unbalanced weight in clinics. For example, in [9] it was suggested that, while SVM shows high accuracy, its sensitivity is low with respect to other methods. In addition, a goodness of fit test, such as Hosmer Lomeshow test should be always performed in order to evaluate the distribution of predictions in deciles.
2 Methods
Raw data includes 400 left ventricles (200 for training, plus 200 blind), sampled with epi- and endo-cardial landmarks, at both ES and ED, for a total of 800 shapes, [12]. From the point of view of shape analysis, ES and ED must be regarded as different shapes; thus, we organize our dataset as a list of 800 shapes, and we consider three sub shape-data: (1) full shapes, consisting of both epicardial and endocardial data; (2) endocardial shapes; (3) epicardial shapes, see Fig. 1. We apply both the GPA+PCA and GPA+PT+PCA strategies to the three aforementioned shape-data, in both SSS and SS, and using PT with two different data centering (i.e. in the Grand Mean or in Diastole), thus totalling 30 different sub-analyses which provide the PC scores to be used for classification procedures, see Table 1; it is worth noting that all 800 shapes have to undergo a common GPA+PCA or GPA+PT+PCA; once shape analysis has been done, the 800 PC scores are split in ED or ES data, each with 400 cases: these will be the datasets to be used for classification procedures.
Once shape analysis has been done on the whole shape-dataset, only the training subset is considered for classification method selection; from the training subset we assemble a learning dataset, composed of a sub-train and a sub-test dataset (not blind), by randomly selecting 50 healthy and 50 pathological cases to fill each of the two sub-datasets. The assembly is repeated and analyzed 1000 times. ES=end-systolic data, ED=end-diastolic data, P=Pathology, H=Healthy.
Top left: Proportion of non significant H-L test over the 1000 classification simulations performed for the 5 methods on any type of shape analysis. Ordinal positions on x-axis correspond to shape analysis’ types in Table 1. Top right: Corresponding mean AUC over 1000 simulations. Best performances, in all panels, according to H-L test and ROC AUC is for type 22 (vertical, dashed line). Bottom left: Mean total misclassification over 1000 simulations. Bottom right: Mean sensitivity over 1000 simulations.
We use five classification procedures: LDA, LR, QDA, RF and SVM with Gaussian Radial Basis Kernel Function. To assess the performances of the five different procedures, we use only the labelled training dataset, that is, half of the 400 cases (100 healthy + 100 pathological) as follows:
-
1.
At first, using the whole training dataset, and for any of the 30 types of shape analyses, we perform an univariate association filtering via ANOVA on the first 100 PC scores. The design is a classical one-way ANOVA using the training affiliation healthy/pathological as a two-levels factor. P-value was set (conservatively) to 0.05. All significant PC scores are retained for classifications.
-
2.
We assemble a warm-up dataset by randomly extracting 50 healthy individuals, and 50 patients affected by Myocardial Infarction (MI) from the training dataset; this procedure yields a learning training subset and a pseudo-blind subset, each containing 100 cases (i.e. 50 healthy and 50 pathological), see Fig. 2.
-
3.
We use this training subset in the 5 classification methods and we employ their learning functions in order to predict the pseudo-blind dataset; given that this is not blind, we can evaluate the performance of each classification method.
Steps (2) and (3) are repeated 1000 times. Basing on the corresponding results we counted total misclassified cases and their two components, e.g. Sensitivity and Specificity. Hosmer Lomeshow test (H-L test) was used in order to assess the Goodness of Fit of any classification problem. Receiving Operating Curves (ROC) and the Area under the Curve (AUC) were also computed. The type of shape analysis and the classification method with the best global performance in classifying the 1000 random sub-test datasets were successively used for classifying the blind test dataset. As primary criterion, we choose the percentage of non significant H-L test (the higher the better) in order to select the best classification method.
The use of H-L test as a primary criterion deserves particular attention. In fact, two classifications could have identical AUCs, sensitivities and specificities but the distributions of probabilities corresponding to misclassified cases can be very different. H-L test divides subjects into deciles based on predicted probabilities, then computes a chi-square from observed and expected frequencies. For example, in two different classifications, the probabilities of misclassified cases can be around 0.5. Or they can have values (leading to wrong classification) close to 0 or 1. In the latter case the severity of misclassification is worse.
Only for the best type analysis selected with this method, we also re-run a non linear PCA based on the Relative Warps Analysis (RWA) [2]. RWA uses the Thin Plate Spline (TPS) interpolation function in order to compute and visualize the deformation occurring between a reference and a target shape. RWA, with the associated scores, yields a sequence of ordered subspaces onto which each single case is projected. Warping is parametrized by the \(\alpha \) parameter: for \(\alpha =1\), large-scale variations (variations among specimens in the relative positions of widely separated landmarks) are given more weight with respect to the small-scale ones; for \(\alpha =-1\), the opposite is true, and more weight is given to variation in the relative positions of landmarks that are close together. A value of \(\alpha =0\) yields to virtually identical results of a linear PCA on Procrustes coordinates. More details can be found in [13, 14]. To detect the importance of large- or small-scale variations, the exponent \(\alpha \) of the bending energy matrix was set equal to 1, 0 (corresponding to standard PCA), and \(-1\). On the resulting RW scores of these three types of RWA we run the best classification method found when using standard PCA. The results were compared with the standard PCA results and the absolute best result is used for classifying the blind dataset.
3 Results
In Fig. 3 results of performances of the five methods are shown. It appears evident that SVM with Gaussian Radial Basis kernel function is characterized, under the 1000 simulations, by a higher probability to present a non significant H-L test. Table 2 reports the results relative to the 30 types of analyses (only for SVM) that shows the higher percentage of non significant H-L test in comparison to other methods. Analysis type 22 is the best for H-L evaluation; it uses the full shape for shape analysis, subjected to a GPA-SS, plus PT-cGM (PT centered in Grand Mean), plus PCA; then, only systolic data are used for statistical analysis.
A: probability-density distributions emerging from our classification of the blind dataset. Only the results obtained with the optimal analysis method (type 22) and the first two sub-optimal ones (types 16 and 28) are presented. B: the corresponding histograms. C: healthy-subject distribution fitted by the sum of two normal distributions. D: pathological-subject distribution fitted by the sum of two normal distributions.
Among the firsts 100 PC scores for analysis type 22, the univariate association filtering found 1, 2, 3, 5, 6, 8, 12, 13, 21, 24, 31, 38, 57, significant (significance level = 0.05) by using the known healthy/pathological affiliation as binary factor. These PC scores were used to select the best classification method.
We found that, in comparison to results of Table 2, RWA performed on the type of analysis 22 with \(\alpha =-1\) yields a lowest percentage of misclassification (\(8\,\%\)), and of significant H-L test (\(1.9\,\%\)) after re-running the resampling procedure we described above. RW scores significant in the univariate association filtering were: 1\(\sim \)5, 7\(\sim \)10, 15, 16, 20, 38, 74. Moreover, mean AUC was slightly improved (99.3), the sensitivity reduced (\(3\,\%\)), as well as total misclassification (8 %). RWA on type of analysis 22 with \(\alpha =-1\) will then be used in order to predict the blind test dataset.
Figure 4 (left) shows the 800 shapes in PCA scatterplot corresponding to the type of analysis 22. Healthy individuals (green) clearly set apart from pathological ones (black), while blind (red) subjects are dispersed across the two distributions; see also Supplementary Figure S1 with dynamic 3D pdf (requires Adobe Reader).
Deformations associated to PC1 and PC2 extremes are also shown in Fig. 4: we plot the values of \(\Vert x_M-x\Vert \), with \(x_M\) the position of a point in the GM, and x its position at ED or ES; the colormap ranges from blue (min) to red (max). It is evident that PC1 represents contraction. This contraction is more evident on the endocardium than in epicardium and healthy individuals occupy a more extreme position, along PC1, than pathological individuals. This can be better appreciated in Fig. 5 where mean deformations (relative to the Grand Mean) of healthy and pathological individuals are illustrated conjointly. Clearly healthy subjects undergo a larger, thus more efficient, contraction than MI patients even in the epicardium. The larger shape differences occur in the middle of endocardial geometry. It is important to note here that we illustrated endocardium and epicardium separately for sake of clarity while actually in type of analysis 22 they were analyzed together as a whole geometry (thus one inside the other). See also Supplementary Figure S2.
Using SVM for learning from the whole training dataset and for successively classifying the blind test dataset, and RWA with \(\alpha =-1\), we obtained 96 subjects classified as normal and 104 classified as pathological. After the disclosure of the blind dataset this resulted in 95 % of total accurracy with sensitivity at 97 % and specificity at 93 %.
We are thus able to report, case by case, the resulting classification and the probability of being found pathological according to the specified learning function. Figure 6 shows the per-class density distributions of this probability. We illustrated the results coming from type of analysis 22 (our optimal result) together with the first two sub-optimal types, i.e. types 16 and 28. The two curves are pretty similar and the 0/1 classifications are much similar among the three types. It is evident that the two groups are well separated with very few cases possessing probabilities around 0.5.
The distributions of the two probabilities suggest that, within each class, more than one normal distribution is represented. This could be evidence of a few pre-clinical healthy individuals and a few only moderately pathological subjects.
The fact that RWA with \(\alpha =-1\) performs better in discriminating healthy from pathological subjects could be related to physiological evidences: myocardial infarction is a particularly localized pathology. It can be transmural or subendocardial, but in both cases a relatively small region of LV is interested. This region can be found at several LV locations, and \(\alpha =-1\) gives more importance to small scale variations; this is coherent with this particular type of pathology. On the opposite, for example, a pathology that moulds the entire LV shape, such as Aortic Regurgitation could be better discriminated using \(\alpha =1\). Testing this is beyond the scope of the present paper.
However, this result suggests that a tuned evaluation of deformation could correlate with the deep nature of pathology. Another result that should be commented is the evidence that the full shape (i.e. epicardium+endocardium) better discriminates than epicardium or endocardium alone. Given that the location of infarction was not known for the available sample of pathological individuals, we can only speculate that the lesser contraction of pathological condition evidenced in Fig. 5 is inevitably related to a lesser extent of myocardial thickness variation and that this feature can only be recognized by analyzing together epicardium and endocardium.
Finally, it has also to be pointed out that the relative age of infarction, whose information was missing in the blind dataset, might be a further factor contributing to diagnostic accuracy. It is in fact well known that LV undergoes a time-dependent overall shape change post-infarction which was variably interpreted and measured but is in general termed “remodeling”, just to underscore that not only the infarcted area but also the remaining still healthy LVs undergo modifications to adapt, globally, to the loss of viable contracting muscle. As remodeling might be minimal in some cases due to a little recent infarction or to a relatively old one and stabilized, the double normal distributions seen in Fig. 6 (pathological side) might represent these conditions. On the other hand, subclinical and localized ischemia might, on the “healthy” side, explain the double distribution there. Clearly, these are speculations and only the full disclosure of the blind database will enable adequate considerations.
4 Conclusions
Deformation analysis performs better than shape analysis alone in detecting pathology. This can be done by adding the PT to standard GPA+PCA. This allows recovering the attributes linked to the contraction process per-se that, ultimately, follows the mechanics of heart functioning. Filtering inter-individual shape differences becomes, thus, very important when exploring systo-diastolic shape changes occurring in a blind sample of healthy subjects and patients affected by Myocardial Infarction.
References
Adams, D.C., Rohlf, F.J., Slice, D.E.: Geometric morphometrics: ten years of progress following the revolution. Ital. J. Zool. 71, 5–16 (2004)
Dryden, I.L., Mardia, K.V.: Statistical Shape Analysis. Wiley, Chichester (1998)
Zhang, X., Cowan, B.R., Bluemke, D.A., Finn, J.P., Fonseca, C.G., Kadish, A.H., Lee, D.C., Lima, J.A.C., Suinesiaputra, A., Young, A.A., Medrano-Gracia, P.: Atlas-based quantification of cardiac remodeling due to myocardial infarction. PLoS ONE 9(10), e110243 (2014)
Piras, P., Evangelista, A., Gabriele, S., Nardinocchi, P., Teresi, L., Torromeo, C., Schiariti, M., Varano, V., Puddu, P.E.: 4D-analysis of left ventricular heart cycle using Procrustes motion analysis. Plos One 9, e86896 (2014)
Madeo, A., Piras, P., Re, F., Gabriele, S., Nardinocchi, P., Teresi, L., Torromeo, C., Chialastri, C., Schiariti, M., Giura, G., Evangelista, A., Dominici, T., Varano, V., Zachara, E., Puddu, P.E.: A new 4D trajectory-based approach unveils abnormal LV revolution dynamics in hypertrophic cardiomyopathy. PloS One 10(4), e0122376 (2015)
Varano, V., Gabriele, S., Teresi, L., Dryden, I., Puddu, P.E., Torromeo, C., Piras, P.: Comparing shape trajectories of biological soft tissues in the size-and-shape. BIOMAT 2014 Congress Book (in press, 2015)
Efron, B.: The efficiency of logistic regression compared to normal discriminant analysis. J. Am. Stat. Assoc. 70, 892–898 (1975)
Peter, C.A.: A comparison of regression trees, logistic regression, generalized additive models, and multivariate adaptive regression splines for predicting AMI mortality. Stat. Med. 26, 2937–2957 (2007)
Maroco, J., Silva, D., Rodrigues, A., Guerreiro, M., Santana, I., de Mendonça, A.: Data mining methods in the prediction of Dementia: a real-data comparison of the accuracy, sensitivity and specificity of linear discriminant analysis, logistic regression, neural networks, support vector machines, classification trees and random forests. BMC Res. Notes 4, 299 (2011)
Puddu, P.E., Menotti, A.: Artificial neural networks versus proportional hazards Cox models to predict 45-year all-cause mortality in the Italian Rural Areas of the Seven Countries Study. BMC Res. Methodol. 12, 100 (2011)
Goss, E.P., Ramchandani, H.: Comparing classification accuracy of neural networks, binary logit regression and discriminant analysis for insolvency prediction of life insurers. J. Econ. Finan. 19, 1–18 (1995)
Fonseca, C.G., Backhaus, M., Bluemke, D.A., Britten, R.D., Do Chung, J., Cowan, B.R., Dinov, I.D., Finn, J.P., Hunter, P.J., Kadish, A.H., Lee, D.C., Lima, J.A.C., Medrano-Gracia, P., Shivkumar, K., Suinesiaputra, A., Tao, W., Young, A.A.: The cardiac atlas project: an imaging database for computational modeling and statistical atlases of the heart. Bioinformatics 27(16), 2288–2295 (2011)
Bookstein, F.L.: Morphometric Tools for Landmark Data. Cambridge University Press, Cambridge (1991)
Rohlf, F.J.: Relative warp analysis and an example of its application to mosquito wings. In: Marcus L.F., Bello E., García-Valdecasa A., (eds.) Contributions to morphometrics, Museu Nacionale de Ciencias Naturales, pp. 131–159 (1993)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
1 Electronic supplementary material
Below is the link to the electronic supplementary material.

Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this paper
Cite this paper
Piras, P. et al. (2016). Systo-Diastolic LV Shape Analysis by Geometric Morphometrics and Parallel Transport Highly Discriminates Myocardial Infarction. In: Camara, O., Mansi, T., Pop, M., Rhode, K., Sermesant, M., Young, A. (eds) Statistical Atlases and Computational Models of the Heart. Imaging and Modelling Challenges. STACOM 2015. Lecture Notes in Computer Science(), vol 9534. Springer, Cham. https://doi.org/10.1007/978-3-319-28712-6_13
Download citation
DOI: https://doi.org/10.1007/978-3-319-28712-6_13
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-28711-9
Online ISBN: 978-3-319-28712-6
eBook Packages: Computer ScienceComputer Science (R0)