Abstract
Physiological Computing augments the information bandwidth between a computer and its user by continuous, real-time monitoring of the user’s physiological traits and responses. This is especially interesting in a context of emotional assessment during human-computer interaction. The electroencephalogram (EEG) signal, acquired on the scalp, has been extensively used to understand cognitive function, and in particular emotion. However, this type of signal has several drawbacks, being susceptible to noise and requiring the use of impractical head-mounted apparatuses. For these reasons, the electrocardiogram (ECG) has been proposed as an alternative source to assess emotion, which is continuously available, and related with the psychophysiological state of the subject. In this paper we analyze morphological features of the ECG signal acquired from subjects performing an attention-demanding task. The analysis is based on various unsupervised learning techniques, which are validated against evidence found in a previous study by our team, where EEG signals collected for the same task exhibit distinct patterns as the subjects progress in the task.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Physiological Computing, as a research area, integrates psychophysiological information into computer systems by continuous, real-time monitoring of the user [1]. These systems augment the information bandwidth between the user and the computer, enabling a better interpretations of the user’s psychophysiological state. Indeed, in natural human communication, the speaker’s attitude, posture, tone, and facial expressions, among others, strongly influence the semantic interpretation done by the receiver [2].
Straightforward approaches to physiological computing, requiring no extra hardware, are, for example, keystroke dynamics [3], speech analysis [4], and automatic facial expression recognition [5]. However, all these examples exhibit serious problems to their usefulness. Keystroke dynamics and speech analysis both require continuous voluntary activity, while the usefulness of facial expression analysis for behavioral science has been recently questioned in [6]. One possible alternative to these modalities, although requiring extra hardware, is the use of the subject’s biosignals (e.g. electrodermal activity, peripheral temperature, blood volume pulse, electrocardiogram, electroencephalogram signals), acquiring them during normal human-computer interaction tasks [7, 8]. These signals have the twofold advantage of being always available, and measuring the natural physiological responses of the body to a given affective state, which cannot be voluntarily masked.
The electroencephalogram (EEG) signal, acquired on the scalp, has been extensively used to understand cognitive function, and in particular emotion [9, 10], being a noninvasive, cost-effective technique, with good temporal resolution [11]. However, it has various drawbacks, such as susceptibility to noise (in particular motion artifacts and eye blinks) and, most importantly, requires the use of some kind of head-mounted equipment to support the (typically wet) electrodes, which becomes impractical for continued use. In this context, the electrocardiogram (ECG) signal has been suggested as a possible option [12, 13]. Nevertheless, the usefulness of the EEG as source of ground-truth information has not been discarded [14].
In this paper, we make a morphological analysis, using unsupervised learning techniques, of the ECG acquired from subjects performing a task that demands high levels of attention over a long period of time. This experiment simulates what may happen, for instance, during an interactive educational game, extended work hours, repetitive daily tasks, or sleep deprivation, where attention levels fluctuate throughout the execution of the task. This is particularly important in various professions, such as doctors, pilots, drivers or industrial equipment operators, for which momentary or prolonged lapses of attention may be catastrophic [14]. In addition, we compare the results obtained with the ECG signal to our previous work using the EEG, which provided evidence that the subjects indeed exhibit distinct affective states throughout the completion of the task [15].
The remainder of the paper is organized as follows: Sect. 2 describes the experimental setup. Section 3 details the proposed methodology, including the description of the clustering methods used, as well as several clustering validation metrics. Section 4 presents the obtained results, which are discussed in Sect. 5. Finally, Sect. 6 concludes the paper.
2 Affective Elicitation and Data Acquisition
The ECG signal presents several attributes that make it especially interesting in a physiological computing framework. Specifically, it is continuously available, providing a rich wellbeing indicator, is related with the psychophysiological state of the subject, and is easy to acquire unobtrusively with wearable devices. This is further enhanced by following an off-the-person approach, where the sensors are seamlessly integrated into objects with which subjects regularly interact, such as a keyboard, a video game controller, or a mobile device, without the need to change normal interaction patterns [16].
Schematic representation of the heart compartments and its electric system, showing the contribution of each component to the prototypical heartbeat signal recorded at the body surface (used with permission from [18])
It is widely known that the basic function of the heart is to pump blood throughout the body, demanding a highly synchronized sequence of muscular contractions. These contractions are initiated by small electrical currents that propagate through the heart’s muscle cells, generating an electrical signal that can be recorded at the body surface (the ECG). In healthy individuals, the electrical activity of the heart is guided by the self-excitatory nature of the sinus node on the left atrium (see Fig. 1), which naturally produces electrical depolarizations at a rate of about 100 beats per minute. However, the sinus node is under systemic control by the endocrine system and the Autonomic Nervous System (ANS). The ANS is composed by two complementing, self-balancing subsystems, the Sympathetic and Parasympathetic Nervous Systems (SNS and PSNS, respectively). While the SNS is typically responsible for the promotion of fight-or-flight responses in the organism (e.g. by increasing the heart rate), the PSNS is responsible for the promotion of rest-and-digest responses, which induce relaxation and a return to normal function. As a whole, the ANS provides an access route to the affective state of a person [17], by analyzing the patterns of physiological activity initiated by both the SNS and PSNS. In particular for the ECG, the amplitude and latency of the P-QRS-T complexes is influenced by multiple psychophysiological factors, and some changes in the user’s behavior result in slight variations in the heart rate and waveform morphology.
The ECG and EEG signals analyzed here were acquired in the context of the HiMotion project [19], an experiment to acquire information related to human-computer interaction and physiological signals on different cognitive activities. During the experimental session, the subjects were asked to execute various interactive cognitive tasks. Particularly, a concentration task was performed, adapted from a similar test from the MENSA set [20]. In this test, the subject is presented with a matrix of 800 integers (20 lines by 40 columns), as shown in Fig. 2. The goal of the game is to identify, line by line, all the pairs of consecutive numbers that add to 10. This task requires high levels of attention, as the pairs may overlap (i.e. the same number may belong to two pairs), measuring the capacity of the subject to maintain an attentive state over a long period of time.
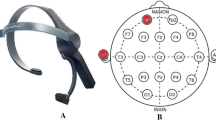
Biosignal data was obtained from 24 subjects (17 males and 7 females) with ages in the range \(23.3 \pm 2.4\) years, using a Thought Technology ProComp2 acquisition system, with a sampling rate of 256 Hz. The ECG was acquired with Ag/AgCl electrodes placed on the chest (4th intercostal space in the mid clavicular line), while the EEG was acquired at four scalp locations according to the 10–20 system (\(F_{p1}\), \(F_z\), \(F_{p2}\), and \(O_z\)), as shown in Fig. 3.
3 Proposed Methodology
It should be noted that each person has different characteristics in terms of attention span and, as such, aside from the temporal information regarding the start and the end of each line of the attention game, no more information is available for this data set. Particularly, there is no ground-truth information regarding the time instants in which the affective state of each test subject has in fact changed. For this reason, we propose the use of unsupervised learning techniques to analyze the ECG data.
The proposed methodology is presented in Fig. 4 and it is divided in three main stages: feature extraction, clustering, and validation of the clustering results. We start by filtering and segmenting the raw ECG, and then we apply clustering techniques to analyze the data. Subsequently, the results of those clustering algorithms are validated using several metrics, exploiting our previous analysis of the same data set with the EEG signal [15]. This somewhat follows the methodology proposed in [14], where the EEG signal is used as a benchmark against which the performance of attention recognition via the ECG is compared. All these stages are explained in the following subsections.
3.1 ECG Feature Extraction
Raw ECG signals are typically affected by various noise sources such as motion artifacts, power line interference, and electromyographic noise. To enhance the signal-to-noise ratio (SNR), and to reduce the influence of the cited noise sources, we used a band-pass Finite Impulse Response (FIR) filter with a Hamming window of 300 ms, and cutoff frequencies of 5–20 Hz. The filtered signal was then fed to a segmentation algorithm, with the purpose of identifying the locations of the R peaks. For that we used the algorithm by Engelse and Zeelenberg [21], with the modifications proposed in [22]. Individual heartbeat segments of 600 ms were extracted from the filtered signal, between 200 ms before and 400 ms after the R peak. Finally, in order to further improve the SNR, heartbeat templates were formed using sequences of 5 consecutive heartbeats, computing their element-wise mean (an example of these templates can be seen in Fig. 5). These templates form the feature space used by the clustering algorithms, described in Sect. 3.3.
3.2 EEG Feature Extraction
Our previous work, focusing on the EEG signal, is based on two distinct feature extraction techniques. The first follows the traditional approach of analyzing the various EEG frequency bands, the Band Power Features (BPF). Specifically, we used the theta (from 4 to 8 Hz), lower alpha (from 8 to 10 Hz), upper alpha (from 10 to 13 Hz), beta (from 13 to 25 Hz), and gamma (from 25 to 40 Hz) bands. The second approach uses a method of synchronization quantification, the Phase-Locking Factor (PLF), which leverages the fact that EEG signals exhibit an oscillatory behavior whose phase dynamics are modulated by the neurological tasks [23]. The PLF between two signals is defined as [24]:
where \(\phi _i[n]\) and \(\phi _k[n]\), \(n = 1,\,...,\,T\) are the phases of the signals, T is the number of discrete time samples, and \(j = \sqrt{-1}\) is the imaginary unit. This measure ranges from 0 to 1, with a value of \(\varrho _{ik} = 1\) corresponding to perfect synchronization between the two signals (constant phase lag), while the value \(\varrho _{ik} = 0\) corresponds to no synchronization. These two feature extraction methods form distinct feature spaces, upon which clustering methods were applied.
3.3 Unsupervised Learning
Clustering is one of the central problems in Pattern Recognition and Machine Learning. Hundreds of clustering algorithms exist, differently handling issues such as cluster shape, density, and noise, among other aspects. These techniques require the definition of a similarity measure between patterns, be it geometrical or probabilistic, which is not easy to specify in the absence of any prior knowledge about cluster shapes and structure.
One of the classical approaches for clustering is the use of hierarchical agglomerative algorithms [25], which produce a tree of nested objects (the dendrogram) that establishes the hierarchy between the clusters. These methods only require a measure of (dis)similarity and a linkage criterion between instances, while partitional methods (e.g. k-means or k-medoids) also require a priori the number of clusters, and an initial assignment of data to clusters. The linkage criterion specifies how intergroup similarity is defined. In particular, we apply the Average Link (AL) and Ward’s Linkage (WL) criteria [26]. Furthermore, to obtain a partition of the data from a dendrogram, we use the largest lifetime criterion [27].
Moreover, we use a new high order dissimilarity measure, called dissimilarity increments, proposed in [28]. This measure is computed over triplets of nearest neighbor patterns and is defined as:
where \(x_j\) is the nearest neighbor of \(x_i\), and \(x_k\) is the nearest neighbor of \(x_j\), different from \(x_i\). In equation (2), \(D_*(\cdot ,\cdot )\) can be any dissimilarity measure, such as the Euclidean distance. The dissimilarity increments measure can give more information about patterns belonging to the same cluster, since it changes smoothly if the patterns are in the same cluster. In [29], an agglomerative hierarchical algorithm, called SLDID, was proposed. This algorithm is a variant of the Single Link (SL) criterion using the dissimilarity increments distribution (DID), which was derived under mild approximations in [30], to modify the way that clusters are merged. In this paper we used a family of DID algorithms: ALDID and WLDID. They are variants of the traditional hierarchical clustering algorithms AL and WL, respectively. The main difference between AL and ALDID is that in AL, in each iteration the pair of clusters with the highest cohesion is always merged; in ALDID some tests are made using the minimum description length (MDL) criterion between two possibilities. These two possibilities consist in the DID of the two clusters combined, and the DID of the two clusters separated. One advantage in using an algorithm from this family is that it stops merging clusters before all the data is merged into one cluster, revealing intrinsic cluster structure in the data when the true number of clusters is unknown.
Consensus Clustering Consensus clustering, also known as Clustering combination, is a powerful technique that combines the information of multiple clustering partitions, forming a clustering ensemble (\(\mathbb {P}\)), and creating a consensus partition that leverages the results of individual clustering algorithms. Recent surveys present an overview on this research topic [31, 32]. One of the significant approaches is the Evidence Accumulation Clustering (EAC) [33]. This framework is based on the aggregation of object co-occurences, and the consensus partition is obtained through a voting process among the objects. Specifically, the consensus clustering problem is addressed by summarizing the information of the ensemble into a pairwise co-association matrix, where each entry holds the fraction of clusterings in the ensemble in which a given pair of objects is placed in the same cluster:
For the construction of the ensemble, we use the k-means algorithm [25] with different parameters and initializations. We created a set of \(N=100\) partitionsFootnote 1 by randomly choosing the number of clusters, following the work in [34] where the minimum and maximum number of clusters per partition depends on the number of objects n, and is bound to the interval \([\frac{\sqrt{n}}{2},\sqrt{n}]\).
The extraction of the consensus partition can be performed using several approaches based on the induced co-association matrix: (i) as a new (dis)similarity-based representation of objects, where the intrinsic structure of the data is enhanced through the evidence accumulation process, enabling the determination of the consensus partition using algorithms that explicitly use similarities as input, such as hierarchical linkage methods (as classically performed in [33]); (ii) as a new vector-based object description, considering each line of the matrix a new feature vector representation, and using it as input to a clustering algorithm such as the k-means [35]; (iii) as a new probabilistic distribution characterized by the probability of pairs of objects being in the same cluster [36].
Application to EEG and ECG The focus of this work was the unsupervised analysis of the ECG signals, and for that we applied all the described techniques: (i) hierarchical agglomerative algorithms; (ii) hierarchical agglomerative algorithms with dissimilarity increments; (iii) consensus clustering based on evidence accumulation clustering, using as extraction criterion the average linkage method with the number of clusters automatically determined by the life-time criterion.
The clustering of the ECG heartbeats was performed over the means of 5 consecutive heartbeats. Since we are willing to compare these partitions with the ones obtained on the context of EEG, where for each line of the test there is only one cluster, it was necessary to post-process the obtained partitions, choosing as representative cluster for each line the one with highest cardinality (largest time span).
In the context of EEG clustering, we applied the hierarchical agglomerative methods with and without dissimilarity increments.
3.4 Cluster Validation
Cluster validation techniques have been developed to guide the design of clustering experiments and to assess the quality of the outcome. There are three types of cluster validity measures [37–41]: (i) External: used to measure the goodness of a clustering structure with respect to external information; (ii) Internal: used to measure the goodness of a clustering structure without supplying any class labels; and (iii) Relative: used to compare different clusterings.
We adopt an external clustering validation perspective, using as external source of information the clusterings obtained with the EEG. There is a long list of external validation indices proposed in the literature [39, 40, 42, 43], which can be categorized as follows: (i) Counting Pairs Methods: a class of criteria based on counting the pairs of points on which two clusterings agree/disagree, Wallace [44], Fowlkes and Mallows [42], and Rand’s [45] are the most representatives of this class; (ii) Set Matching: based on set matching cardinality, \(\mathcal H\) criterion [39], and consistency index (Ci) [46, 47] are representative of this class; (iii) Information Theoretic: based on information theoretic concepts (entropy and mutual information); representatives of this class of criteria are the Variation of Information (VI) index [39] and Dom’s index [43].
In this work, we compare the partitions obtained with the ECG with the ones obtained with the EEG (taken as ground-truth), and following the idea proposed in [14]. We use indices of the three categories, to verify the consistency of the results in several perspectives, namely: Rand [45], a modified version of the Consistency Index entitled Average Cluster Consistency (ACC) [47], and VI [39]. All the three indices take values between 0 and 1. Rand’s index and the ACC take the value 1 for a perfect match between partitions, and for the VI index, 0 corresponds to a perfect match.
4 Experimental Results
Figure 6 exemplifies the clustering of the ECG templates obtained for one of the subjects, using the clustering combination (CC) method. It shows, for each line of the concentration task, the clusters to which the templates in that line belong to. The first observation to note is that the lines are not characterized by a single cluster, but rather by two or three clusters that alternate between them. However, it is possible to perceive the existence of different groups of lines. In this particular case, lines 0–2 are mainly composed by clusters 1, 2, and 3, lines 4–7 are composed by clusters 7 and 8, and the remaining lines are composed by clusters 4, 5, and 6. Another interesting note is the fact that the number of templates per line decreases throughout the completion of the task, implying that the first few lines of the task take longer to complete than the last lines. These observations are valid for the majority of the subjects, although the number of clusters and their distribution differs from subject to subject, forming different groups of lines.
Inter-subject variability is evidenced in Fig. 7, where the clustering obtained, across all subjects, with the EEG (using PLF features and ALDID clustering—Fig. 7a) is compared to the clustering obtained with the ECG (using CC clustering—Fig. 7b). Remember that, in the case of the ECG, each line is represented by the most frequent cluster in that line. It is possible to observe that the ECG produces a higher number of clusters than the EEG, where each cluster tends to form groups of contiguous lines. Contrastingly, in the ECG it is frequent to have transitions to clusters seen in previous lines.
The results of the cluster validation are shown in Tables 1, 2, and 3 for the Average Cluster Consistency (ACC), Variation of Information (VI), and Rand’s index, respectively. For the ACC criterion, the highest agreement is obtained between the ECG clustering with CC and both the EEG clustering using AL (BPF) and ALDID (PLF), with a value of 0.79. Regarding the VI measure, the strongest agreements are seen for the ECG clustering using the AL algorithm, in particular with the ALDID method applied to the PLF features from the EEG, with a value of 0.20. Concerning Rand’s index, the highest value, 0.63, is obtained between the ECG clustering through CC with the EEG clustering using WLDID (BPF).
5 Discussion
Our work addresses the following questions: (i) “Is ECG morphological analysis capable of identifying affective states throughout the realization of a task that demands a high attention span?”; (ii) “Are the obtained states related to the ones found while analyzing EEG data?”; and (iii) “What techniques can be considered to be more suitable for the analysis of the ECG?”
The validation of the partitions found using ECG, when considering the EEG partitions as ground-truth, shows that there is evidence of correlation between them, revealing that ECG can be used to infer affective states. The ECG partitions have a much higher number of partitions than the EEG ones, leading to distinct results over the various validation criteria (considering the different perspectives), associated with moderate to high matching. This was mainly due to small variations over time of the ECG heartbeats, that lead to slow time transitions between the different clusters.
The clustering technique that presents the best results varies depending on the validation index. When considering the average cluster consistency (ACC), the consensus clustering (CC) obtains partitions that lead to a best match; when using variation of information (VI) criterion, the Average Link (AL) method is the one that leads to best match; and when using the Rand’s index there is not a method which can be considered a clear winner. The situations with best results are partitions with high number of clusters, which correspond to Average linkage and Consensus Clustering.
6 Conclusions
In this work we present a methodology for attention detection based on the morphological analysis of ECG signals, using data collected during the course of a task requiring a high level of attention span. We compare the ECG morphology results with the analysis performed using the EEG. This comparison was accomplished using clustering validation indices.
The ECG analysis was divided into several steps. For the feature extraction step, the signal was first digitally filtered, segmented based on the peaks found by a modification of the Engelse and Zeelenberg algorithm, and templates were formed using means of 5 consecutive heart beats. For the clustering step, several state of the art techniques were used, since the ECG heartbeats have very small variations over time, leading to touching clusters.
Several clustering validation indices were used, trying to compare the partitions using different perspectives. Each of the validation indices showed that there is a high evidence of correlation between the partitions obtained by the ECG and the EEG. There is not a clear winner method, but Average Linkage and Consensus Clustering can be considered suitable methods for this kind of analysis.
Notes
- 1.
This is the number typically proposed in the reference literature.
References
Fairclough, S.H.: Fundamentals of physiological computing. Interact. Comput. 21(1), 133–145 (2009)
Pell, M.D., Jaywant, A., Monetta, L., Kotz, S.A.: Emotional speech processing: disentangling the effects of prosody and semantic cues. Cogn. Emot. 25(5), 834–853 (2011)
Epp, C., Lippold, M., Mandryk, R.L.: Identifying emotional states using keystroke dynamics. In: Proceedings of the 2011 Annual Conference on Human Factors in Computing Systems, pp. 715–724. ACM (2011)
Murray, I.R., Arnott, J.L.: Toward the simulation of emotion in synthetic speech: a review of the literature on human vocal emotion. J. Acoust. Soc. Am. 93, 1097 (1993)
Zheng, W., Zhou, X., Zou, C., Zhao, L.: Facial expression recognition using kernel canonical correlation analysis (kcca). IEEE Trans. Neural Networks 17(1), 233–238 (2006)
Aviezer, H., Trope, Y., Todorov, A.: Body cues, not facial expressions, discriminate between intense positive and negative emotions. Science 338(6111), 1225–1229 (2012)
Canento, F., Fred, A., Silva, H., Gamboa, H., Lourenço, A.: Multimodal biosignal sensor data handling for emotion recognition. In: Proceedings of the IEEE Sensors Conference, pp. 647–650. IEEE Press (2011)
Silva, H., Fred, A., Eusebio, S., Torrado, M., Ouakinin, S.: Feature extraction for psychophysiological load assessment in unconstrained scenarios. In: Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), pp. 4784–4787. IEEE Press (2012)
Ahern, G.L., Schwartz, G.E.: Differential lateralization for positive and negative emotion in the human brain: EEG spectral analysis. Neuropsychologia 23(6), 745–755 (1985)
Coan, J.A., Allen, J.J.: Handbook of emotion elicitation and assessment. Oxford University Press, Oxford (2007)
Mak, J.N., Wolpaw, J.R.: Clinical applications of brain-computer interfaces: current state and future prospects. IEEE Rev. Biomed. Eng. 2, 187–199 (2009)
Medina, L.: Identification of stress states from ECG signals using unsupervised learning methods. Master’s thesis, Universidade Técnica de Lisboa, Instituto Superior Técnico (2009)
Belle, A., Ji, S.Y., Ansari, S., Hakimzadeh, R., Ward, K., Najarian, K.: Frustration detection with electrocardiograph signal using wavelet transform. In: IEEE International Conference on Biosciences (BIOSCIENCESWORLD), pp. 91–94. IEEE Press (2010)
Belle, A., Hargraves, R.H., Najarian, K.: An automated optimal engagement and attention detection system using electrocardiogram. Comput. Math. Meth. Med. 2012 (2012)
Carreiras, C., Aidos, H., Silva, H., Fred, A.: Exploratory EEG analysis using clustering and phase-locking factor. In: Proceedings of the 6th International Conference on Bio-Inspired Systems and Signal Processing (BIOSIGNALS 2013), pp. 79–88. SCITEPRESS (2013)
Silva, H., Lourenço, A., Lourenço, R., Leite, P., Coutinho, D., Fred, A.: Study and evaluation of a single differential sensor design based on electro-textile electrodes for ECG biometrics applications. In: Proceedings of the IEEE Sensors Conference, pp. 1764–1767. IEEE Press (2011)
Levenson, R.W.: Autonomic nervous system differences among emotions. Psychol. Sci. 3(1), 23–27 (1992)
Malmivuo, J., Plonsey, R.: Bioelectromagnetism: Principles and Applications of Bioelectric and Biomagnetic Fields. Oxford University Press, Oxford (1995)
Gamboa, H., Silva, H., Fred, A.: HiMotion Project. Technical report, Instituto Superior Técnico, Lisbon, Portugal (2007)
Fulton, J.: Mensa Book of Total Genius. Barnes & Noble Books, Totowa (1999)
Engelse, W.A.H., Zeelenberg, C.: A single scan algorithm for QRS-detection and feature extraction. Comput. Cardiol. 6, 37–42 (1979)
Canento, F., Lourenço, A., Silva, H., Fred, A., Raposo, N.: On real time ECG algorithms for biometric applications. In: Proceedings of the 6th International Conference on Bio-Inspired Systems and Signal Processing (BIOSIGNALS 2013), pp. 228–235. SCITEPRESS (2013)
Pfurtscheller, G., Lopes da Silva, F.H.: Event-related EEG/MEG synchronization and desynchronization: basic principles. Clin. Neurophysiol. 110, 1842–1857 (1999)
Almeida, M., Bioucas-Dias, J., Vigário, R.: Source separation of phase-locked subspaces. In: Proceedings of the International Conference on Independent Component Analysis and Signal Separation, vol. 5441, pp. 203–210 (2009)
Jain, A.K., Dubes, R.C.: Algorithms for Clustering Data. Prentice-Hall Inc, Upper Saddle River (1988)
Theodoridis, S., Koutroumbas, K.: Patern Recognition. Academic Press (1999)
Fred, A., Jain, A.: Evidence accumulation clustering based on the k-means algorithm. In: Structural, Syntactic, and Statistical Pattern Recognition, pp. 303–333 (2002)
Fred, A., Leitão, J.: A new cluster isolation criterion based on dissimilarity increments. IEEE Trans. Pattern Anal. Mach. Intell. 25(8), 944–958 (2003)
Aidos, H., Fred, A.: Hierarchical clustering with high order dissimilarities. In: Proceedings of the 7th International Conference on Machine Learning and Data Mining (MLDM 2011). LNCS, vol. 6871, pp. 280–293. New York, USA (2011)
Aidos, H., Fred, A.: Statistical modeling of dissimilarity increments for d-dimensional data: application in partitional clustering. Pattern Recogn. 45(9), 3061–3071 (2012)
Ghosh, J., Acharya, A.: Cluster ensembles. WIREs Data Mining Knowled. Discovery 1(4), 305–315 (2011)
Vega-Pons, S., Ruiz-Shulcloper, J.: A survey of clustering ensemble algorithms. Int. J. Pattern Recogn. Artifical Intell. (IJPRAI) 25(3), 337–372 (2011)
Fred, A., Jain, A.K.: Combining multiple clustering using evidence accumulation. IEEE Trans. Pattern Anal. Mach. Intell. 27(6), 835–850 (2005)
Lourenço, A., Fred, A., Jain, A.K.: On the scalability of evidence accumulation clustering. In: Proceedings of the 20th International Conference on Pattern Recognition (ICPR), pp. 782–785. IEEE Press (2010)
Kuncheva, L.I., Vetrov, D.P.: Evaluation of stability of k-means cluster ensembles with respect to random initialization. IEEE Trans. Pattern Anal. Mach. Intell. 28(11), 1798–1808 (2006)
Lourenço, A., Rota Bulò, S., Rebagliati, N., Figueiredo, M., Fred, A., Pelillo, M.: Probabilistic evidence accumulation for clustering ensembles. In: Proceedings of the International Conference on Pattern Recognition Applications and Methods (ICPRAM), pp. 58–67. SCITEPRESS (2013)
Dubes, R., Jain, A.: Validity studies in clustering methodologies. Pattern Recogn. 11, 235–254 (1979)
Halkidi, M., Batistakis, Y., Vazirgiannis, M.: On clustering validation techniques. J. Intell. Inform. Syst. 17, 107–145 (2001)
Meilă, M.: Comparing clusterings–an information based distance. J. Multivar. Anal. 98(5), 873–895 (2007)
Ben-Hur, A., Elisseeff, A., Guyon, I.: A stability based method for discovering structure in clustered data. In: Pacific Symposium on Biocomputing, pp. 6–17 (2002)
Luo, P., Xiong, H., Zhan, G., Wu, J., Shi, Z.: Information-theoretic distance measures for clustering validation: generalization and normalization. IEEE Trans. Knowl. Data Eng. 21(9), 1249–1262 (2009)
Fowlkes, E.B., Mallows, C.L.: A method for comparing two hierarchical clusterings. J. Am. Statis. Assoc. 78(383), 553–569 (1983)
Dom, B.E.: An information-theoretic external cluster-validity measure. Technical Report IBM Research Report RJ 10219 (2001)
Wallace, D.L.: A method for comparing two hierarchical clusterings: comment. J. Am. Statis. Assoc. 78(383), 569–576 (1983)
Rand, W.M.: Objective criteria for the evaluation of clustering methods. J. Am. Statis. Assoc. 66(336), 846–850 (1971)
Fred, A.: Finding consistent clusters in data partitions. In: Proceedings of the Second International Workshop on Multiple Classifier Systems, pp. 309–318. Springer-Verlag (2001)
Duarte, F., Duarte, J., Fred, A., Rodrigues, M.: Average cluster consistency for cluster ensemble selection. In: Fred, A., Dietz, J., Liu, K., Filipe, J. (eds.) Knowledge Discovery, Knowlege Engineering and Knowledge Management. Communications in Computer and Information Science, vol. 128, pp. 133–148. Springer, Berlin (2011)
Acknowledgments
This work was partially funded by the Portuguese Science Foundation (FCT) under grants PTDC/EEI-SII/2312/2012, SFRH/BD/65248/2009 and SFRH/PROTEC/49512/2009, and by Área Departamental de Engenharia Electrónica e Telecomunicações e de Computadores (ISEL), whose support the authors gratefully acknowledge.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this paper
Cite this paper
Carreiras, C., Lourenço, A., Aidos, H., da Silva, H.P., Fred, A.L.N. (2016). Unsupervised Analysis of Morphological ECG Features for Attention Detection. In: Madani, K., Dourado, A., Rosa, A., Filipe, J., Kacprzyk, J. (eds) Computational Intelligence. IJCCI 2013. Studies in Computational Intelligence, vol 613. Springer, Cham. https://doi.org/10.1007/978-3-319-23392-5_24
Download citation
DOI: https://doi.org/10.1007/978-3-319-23392-5_24
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-23391-8
Online ISBN: 978-3-319-23392-5
eBook Packages: EngineeringEngineering (R0)