Abstract
Gene conversion, mediated by activation-induced cytidine deaminase (AID), has been found to contribute to generation of the primary antibody repertoire in several vertebrate species. Generation of the primary antibody repertoire by gene conversion of immunoglobulin (Ig) genes occurs primarily in gut-associated lymphoid tissues (GALT) and is best described in chicken and rabbit. Here, we discuss current knowledge of the mechanism of gene conversion as well as the contribution of the microbiota in promoting gene conversion of Ig genes. Finally, we propose that the antibody diversification strategy used in GALT species, such as chicken and rabbit, is conserved in a subset of human and mouse B cells.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
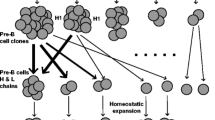
Activation-induced cytidine deaminase (AID) catalyzes three important immunological processes—somatic hypermutation, class-switch recombination, and somatic gene conversion (Muramatsu et al. 2000; Arakawa et al. 2002). Although somatic hypermutation and class-switch recombination are widely utilized among vertebrate species, somatic gene conversion has been observed, as an immunological mechanism, primarily in vertebrates that diversify their primary antibody (Ab) repertoires in gut-associated lymphoid tissues (GALT ). These species generate their primary antibody repertoires through a fundamentally different strategy than that used by mice and humans (Fig. 1). B cells of mice and humans, for example, generate a highly diverse antibody repertoire by utilizing many different V, (D), and J gene segments during V(D)J gene rearrangement of the immunoglobulin (Ig) heavy and light chain loci. In contrast, B cells of vertebrates that diversify their primary antibody repertoires in GALT use V(D)J gene recombination primarily for assembling functional Ig heavy and light chain genes, rather than for generating antibody diversity. B cells in these species preferentially utilize a small subset of V (and often (D) and J) gene segments during V(D)J gene rearrangement, and the resulting V(D)J genes serve as mutation substrates during a subsequent phase of antibody repertoire diversification in GALT. During repertoire diversification, mutations are introduced into the rearranged V(D)J genes through somatic gene conversion and somatic hypermutation. This process generates an extremely large array of different antibody specificities, collectively termed the primary antibody repertoire , which serves to anticipate and protect against a vast range of potential pathogens.
Two strategies for generating a diverse primary antibody repertoire. B cells of humans and mice (left) utilize many different V, (D), and J gene segments during V(D)J gene rearrangement of the immunoglobulin (Ig) heavy and light chain loci (Ig heavy chain rearrangement is shown). A diverse primary antibody repertoire is thus generated directly through V(D)J gene rearrangement, and the B cells exit the bone marrow and enter the periphery. In contrast, B cells of chickens, rabbits, and artiodactyls (right) preferentially utilize a small subset of V (and often (D) and J) gene segments during V(D)J gene rearrangement and thus generate much less initial antibody diversity. Upon exiting the bone marrow, B cells migrate to gut-associated lymphoid tissues (GALT), where they undergo proliferation and mutate their V(D)J genes through somatic gene conversion and somatic hypermutation. A tissue section from rabbit appendix, the major site of rabbit GALT, is shown, with proliferating B cells stained green and non-proliferating B cells stained red. V(D)J gene mutation is initiated by AID-mediated deamination of cytidines, and the resulting lesions (G/U mismatches) induce somatic hypermutation or somatic gene conversion. Somatic hypermutation introduces point mutations (arrows) throughout the V(D)J gene and into the J–C intron. Somatic gene conversion transfers nucleotide tracts (bounded by yellow lines) from upstream V donors into the rearranged V gene. A highly diverse primary antibody repertoire is generated and the B cells exit GALT and enter the periphery
2 Prevalence of AID-Mediated Antibody Repertoire Diversification Among Vertebrates
This strategy for generating a diverse primary antibody repertoire was first recognized in the chicken, which has only one functional V gene at both the Ig heavy and light chain loci and uses gene conversion to diversify its primary antibody repertoire in a gut-associated lymphoid tissue called the bursa of Fabricius (Reynaud et al. 1985, 1987). Shortly thereafter, the Ig light chain loci of the mallard duck, turkey, quail, pigeon, hawk, and cormorant were found to preferentially utilize a single VL gene segment during V–J recombination, suggesting that these avian species also use gene conversion to diversify their primary antibody repertoires (McCormack et al. 1989a). While this strategy was briefly thought unique to birds, it was subsequently found that rabbits (Knight and Becker 1990), swine (Sun et al. 1998), and cattle (Parng et al. 1996) also generate limited antibody diversity during V(D)J gene recombination due to preferential rearrangement of a few V gene segments. In these species, Ig gene diversification also occurs in GALT, primarily the appendix in rabbits and the ileal Peyer’s patch in cattle. It is not clear where Ig gene diversification occurs in swine, as it has recently been shown not to occur in the ileal Peyer’s patch (Butler et al. 2011). These tissues contain thousands of lymphoid follicles where B cells proliferate and diversify their Ig genes and are considered mammalian equivalents of the bursa of Fabricius.
3 Somatic Gene Conversion vs. Somatic Hypermutation
Although somatic hypermutation and somatic gene conversion are both mediated by AID, they are fundamentally different mutational processes. Both processes are initiated by AID-mediated deamination of cytidine residues, particularly at RGYW consensus hotspots, in V(D)J genes, but they represent two different pathways of resolving this initial DNA lesion. Somatic hypermutation introduces point mutations through error-prone repair of the resulting uracil residues (Di Noia and Neuberger 2007). While primarily known for driving affinity maturation in germinal centers during primary immune responses, somatic hypermutation is also utilized, in an antigen-independent manner, to diversify the primary antibody repertoire in some species, e.g., the rabbit. Somatic gene conversion, rather than introducing point mutations, transfers nucleotide tracts from upstream donor V gene segments into the rearranged V(D)J gene through nonreciprocal homologous recombination. The unidirectionality of the sequence exchange, leaving the donor sequence unaltered, distinguishes somatic gene conversion from double homologous recombination. During diversification of the primary antibody repertoire, a single round of gene conversion can introduce multiple nucleotide substitutions, as well as codon insertions and/or deletions, and multiple rounds of gene conversion can occur within a given V(D)J gene.
4 Gene Conversion-Mediated Antibody Repertoire Diversification in the Chicken
Somatic gene conversion was first recognized as a mechanism for antibody repertoire diversification in the chicken by Reynaud et al. (1987). The chicken Ig heavy and light chain loci each contain only one functional V gene segment (V H 1 and V L 1, respectively), as well as a single functional J gene segment (J H and J L ) (Reynaud et al. 1985, 1989). In addition, only limited junctional diversity is introduced by V(D)J gene rearrangement, which does not continue throughout life, but solely during a brief period of early embryonic development (Weill et al. 1986; McCormack et al. 1989b). Thus, unlike humans and mice, little antibody diversity is generated during rearrangement of the chicken Ig loci. The majority of repertoire diversity is generated subsequently in the bursa of Fabricius , beginning between days 15 and 18 of embryonic development, by gene conversion-mediated replacement of nucleotide tracts in the rearranged V H 1 and V L 1 gene segments (Reynaud et al. 1987; Thompson and Neiman 1987).
The chicken Ig light chain locus contains 25 V L pseudogene (ψV L ) segments in the 19 kb region upstream of the V L 1 and J L gene segments (Reynaud et al. 1987, 1989). Reynaud et al. (1985, 1987) first observed that tracts of nucleotide substitutions within diversified V L 1 cDNA sequences corresponded to nucleotide sequences present in the ψV L gene segments. The chicken Ig heavy chain locus contains around 80 ψV H gene segments, similar to V–D joints, in a 60–80 kb region upstream of V H 1, about 15 D H gene segments and a single J H gene segment (Reynaud et al. 1989). Reynaud et al. (1989) similarly found tracts of nucleotide substitutions in diversified V H 1 cDNA sequences that corresponded to nucleotide sequences in ψV H gene segments (Reynaud et al. 1989). All ψV L and ψV H gene segments lack a promoter, leader exon, or V(D)J recombination signals. Few of them contain stop codons or frameshift mutations, but many are truncated at their 5′ or 3′ ends. Carlson et al. (1990) demonstrated that the diversifying nucleotide tracts in the chicken Ig light chain gene result from intrachromosomal gene conversion. By restriction mapping donor ψV L gene segments and recipient V L 1 genes in a panel of v-rel-transformed chicken B cell lines and sequencing donor ψV L gene segments used in V L 1 nucleotide substitutions, these authors showed that donor ψV L gene segments were not modified during the sequence exchange. The authors further showed, by means of allelic V L 1 and ψV L gene segment polymorphisms in the SC chicken strain (an F1 cross between the G4 and S3 inbred strains), that gene conversion uses donor and recipient V L gene segments from the same allele and thus from the same chromosome.
5 Gene Conversion-Mediated Antibody Repertoire Diversification in the Rabbit
The recognition of preferential V H gene segment usage in the rabbit, first reported by Knight and Becker (1990), provided a solution to the long-standing problem of allelically inherited rabbit VH allotypes (Knight and Becker 1990). The presence in rabbits of VH allotypic markers, inherited in a simple Mendelian fashion, on 80–90 % of serum Ig molecules had been difficult to explain under the assumption that multiple V H gene segments were utilized during rearrangement of the heavy chain locus. This problem was resolved with the recognition that rabbit B cells, like those of chickens, preferentially utilize the 3′-most V H gene segment during V(D)J gene rearrangement and generate antibody repertoire diversity through subsequent mutation of the rearranged IgH genes (Knight and Becker 1990; Becker and Knight 1990).
While rabbits and chickens use the same strategy to generate a diverse primary antibody repertoire, there are some interesting differences between the two species. Rabbits, for example, generate a more diverse range of Ig genes during the initial V(D)J gene rearrangements. Unlike the chicken Ig heavy chain locus, many of the upstream rabbit V H gene segments are potentially functional, and a small number of them are used in 10–20 % of VDJ gene rearrangements (Friedman et al. 1994). In contrast to the single chicken J H gene segment, the rabbit heavy chain locus contains five functional J H gene segments, although J H 4 is preferentially used in VDJ gene rearrangements (Becker et al. 1989; Lavinder et al. 2014). Also unlike chickens, multiple Igκ gene segments are utilized during rearrangement of the rabbit Ig light chain locus, thus contributing significant diversity to the initial antibody repertoire (Sehgal et al. 1999; Lavinder et al. 2014). Although the rabbit light chain locus contains three functional J L gene segments, one of these, IGKJ1_2, is used nearly exclusively in Ig light chain gene rearrangements (Lavinder et al. 2014). Interestingly, in both chickens and rabbits, gene conversion also contributes to Ig gene mutation in germinal centers during antigen-specific immune responses, in addition to its role in diversifying the primary antibody repertoire (Winstead et al. 1999; Arakawa et al. 1996).
While chickens rely exclusively on gene conversion to diversify the primary antibody repertoire, rabbits use both gene conversion and somatic hypermutation (Becker and Knight 1990; Weinstein et al. 1994). As in chickens, rabbit V(D)J genes are diversified in GALT , primarily the appendix in rabbits (Weinstein et al. 1994; Vajdy et al. 1998). In chickens, B cells seed the bursa of Fabricius and begin diversifying their V(D)J genes shortly before hatch (Reynaud et al. 1994). Ig gene diversification in the chicken thus begins as a developmentally programmed event that occurs in the absence of exogenous antigen, as well as in experimentally manipulated B cells expressing truncated B cell receptors that lack the antigen-binding region (Sayegh et al. 1999). In contrast, rabbits begin diversifying their primary antibody repertoire shortly after birth. The differential timing of the onset of repertoire diversification in rabbits and chickens might reflect their differing rates of embryonic development. While newly hatched chicks are well-developed precocial offspring, rabbit pups are altricial offspring requiring maternal care for the first 2–3 weeks of life.
Rabbits further differ from chickens in requiring select members of the intestinal microbiota to initiate the diversification process (Zhai and Lanning 2013; Rhee et al. 2004). Rhee et al. (2004) demonstrated that maintaining the rabbit appendix as a germ-free tissue by ligating it at birth to prevent microbial colonization prevented V(D)J gene diversification. These authors further found that V(D)J gene diversification could be induced by colonizing germ-free ligated appendix with colony-purified isolates from the rabbit intestinal microbiota. Surprisingly, only co-colonization with Bacillus subtilis and Bacteroides fragilis induced V(D)J gene diversification, despite each of several tested isolates attaining similar colonization densities (Rhee et al. 2004). Rabbit B cells thus require select members of the intestinal microbiota to initiate V(D)J gene diversification in GALT.
As in chickens, repertoire diversification in rabbits is initiated in an antigen-independent manner. The diversification patterns in appendix B cell Ig genes, for example, differ strikingly from those driven by an immunizing antigen in splenic germinal centers (Sehgal et al. 2002). Severson et al. (2010) reported evidence that bacterial superantigen-like molecules drive repertoire diversification in rabbit GALT. These authors found that spores from Bacillus subtilis and other Bacillus species bind a superantigen-like binding site on rabbit IgM and that surface molecules from B. anthracis stimulate B cell proliferation in rabbit GALT. Furthermore, Rhee et al. (2005) identified a putative superantigen-like binding site on rabbit VHa allotype Ig by comparing the amino acid sequences of the VHa and VHn allotypes. V H 1, the preferentially utilized V H gene segment, encodes the VHa allotype, while the less frequently utilized V H gene segments primarily encode the VHn allotype. Mutant ali/ali rabbits lack V H 1, and, as a result, VHn B cells comprise the vast majority of peripheral and GALT B cells during the first 6 weeks of life (Pospisil et al. 1995). Between 6 and 11 weeks of age, however, the small VHa B cell population expands rapidly and becomes the dominant B cell type. Rhee et al. (2005) demonstrated that this shift from VHn to VHa B cell dominance is driven by the intestinal microbiota in GALT, because it did not occur when the appendix was ligated to prevent microbial colonization and all other organized GALT was removed from newborn ali/ali pups. Taken together, these data suggest that bacterial superantigen-like molecules induce repertoire diversification in rabbit GALT by polyclonally activating B cells that express VHa allotype B cell receptors.
6 Determinants of Gene Conversion Donor Usage
The chicken Ig light chain locus has proven useful for determining some of the molecular requirements for gene conversion, because it is compact and the nucleotide sequences of all 25 of its ψV L gene segments are known. Efficient intrachromosomal gene conversion in mammalian cells requires at least 200–300 bp of homologous nucleotide sequence and an overall nucleotide sequence identity of >80 % (Liskay et al. 1987; Waldman and Liskay 1987). In the chicken IgL locus, the frequency with which ψV L gene segments are used as gene conversion donors is similarly influenced by the extent of their homology with V L 1 (McCormack and Thompson 1990). This is dependent on both their percent nucleotide sequence identity with V L 1 and their length. Truncated ψV L gene segments, for example, are used less frequently than full-length ψV L gene segments (McCormack and Thompson 1990). Proximity to the rearranged V L 1–J L gene also influences the frequency of ψV L gene segment use, with those located most proximal utilized more frequently than those located more distally (Reynaud et al. 1987; McCormack and Thompson 1990). A third determinant of ψV L gene segment usage is relative orientation with respect to the V L 1–J L gene, those in inverted or antisense orientation being preferentially utilized. Gene conversion tracts are not found in the 5′ region of V L 1, despite the presence of ψV L gene segments with homology to this region (Reynaud et al. 1987), but are found throughout the remainder of V L 1, with particularly high frequency in CDR1, the FR2/CDR2 boundary, and CDR3 (McCormack and Thompson 1990). Gene conversion tracts range in length from 8 bp to around 200 bp (McCormack and Thompson 1990). Although the 5′ ends of gene conversion tracts always begin in regions of homology between the ψV L gene segment and V L 1, the 3′ ends can occur in nonhomologous regions and frequently contain nucleotide insertions or deletions, suggesting that the gene conversion mechanism operates with a 5′ to 3′ polarity (McCormack and Thompson 1990).
7 The Chicken DT40 Cell Line: A Model for Studying Ig Gene Conversion
The avian leukemia virus (ALV)-induced chicken B cell lymphoma line DT40 has provided additional insights into the molecular mechanism of gene conversion (Arakawa and Buerstedde 2004). DT40 appears to be arrested at the stage of bursal B cells and continues to undergo gene conversion of the Ig light chain gene during in vitro culture (Buerstedde et al. 1990; Kim et al. 1990). It also integrates transfected gene constructs highly efficiently into essentially any gene locus, regardless of transcriptional activity (Buerstedde and Takeda 1991). Although such highly efficient targeted integration is not seen in human or murine B cell lines or chicken non-B cell lines, it is seen in other chicken B cell lines that do not undergo Ig gene conversion (Buerstedde and Takeda 1991). This suggests that targeted integration in chicken B cell lines is mediated by a homologous recombination activity that supports, but is not sufficient for, gene conversion. Studies of DT40 attribute this supportive role to the RAD52 pathway.
8 The Role of the RAD52 Pathway in Gene Conversion
The RAD52 pathway mediates double-strand break (DSB) repair by homologous recombination and is required for both gene conversion and targeted integration in the yeast Saccharomyces cerevisiae. It is comprised of proteins that recognize double-strand breaks (RAD50, MRE11, and XRS2) and proteins that promote homology searches and strand invasion (RAD51, RAD52, RAD54, RAD55, and RAD57). Homologues of the genes encoding several of these yeast proteins have been cloned from chicken bursal cells, and their roles in targeted integration and gene conversion have been studied by disrupting them in DT40 cells (Arakawa and Buerstedde 2004). RAD54-deficient DT40 cells, for example, are highly sensitive to DNA damage and exhibit a 100-fold decrease in targeted integration efficiency, as well as reduced Ig light chain gene conversion activity (Bezzubova et al. 1997). DT40 cells deficient in NBS1, the vertebrate homologue of the yeast XRS2 gene, exhibit a similar phenotype (Tauchi et al. 2002). Five vertebrate genes (RAD51B, RAD51C, RAD51D, XRCC2, and XRCC3) are considered Rad51 paralogs on the basis of sequence similarity with the yeast RAD51 gene. The RAD51 proteins are structural homologues of the bacterial DNA repair protein, recA (Kawabata et al. 2005). Disruption of each of these individually in DT40 cells results in reduced targeted integration efficiency and deficiencies in DSB repair, while disruption of all five loci is lethal (Takata et al. 2000, 2001; Sonoda et al. 1998). Although S. cerevisiae RAD52 mutants exhibit severe recombination and DNA repair defects, DNA repair is normal and homologous recombination only moderately reduced in RAD52-deficient DT40 cells. The function of RAD52, however, appears to partially overlap that of XRCC3 in chicken B cells because, while disruption of either results in comparatively mild deficiencies, disruption of both leads to chromosome instability and cell death (Fujimori et al. 2001).
These studies suggest that gene conversion is dependent on the more general processes of homologous gene targeting and recombinational repair mediated by the RAD52 pathway. Indeed, co-localization of RAD51D and XRCC2 has been directly observed in diversifying Ig λL genes in DT40 cells, and ectopic expression of either accelerates the rate of gene conversion and influences the length of gene conversion tracts (Ordinario et al. 2009). RAD51 is also highly expressed in the rabbit appendix, which, like the chicken bursa, is a site of gene conversion-mediated antibody diversification (Barrington et al. 1999; Schiaffella et al. 1998). While establishing that the homologous recombination activity of the RAD52 pathway is necessary for efficient Ig gene conversion, these studies do not explain how gene conversion is initiated and specifically targeted to the Ig loci. The resolution of this question came with the surprising discovery that gene conversion of Ig genes requires activation-induced cytidine deaminase (AID), a protein previously identified as essential for both somatic hypermutation and class-switch recombination.
9 AID Is Required for Gene Conversion of Ig Genes
AID was first identified as a protein specifically upregulated in germinal center B cells that shares homology with the apolipoprotein B mRNA-editing enzyme, APOBEC-1 (Muramatsu et al. 1999). It was subsequently found to be required for both somatic hypermutation and class-switch recombination of Ig genes (Muramatsu et al. 1999, 2000; Revy et al. 2000). Shortly thereafter, disruption of the AID locus in chicken DT40 cells was found to cause complete loss of Ig gene conversion activity (Arakawa et al. 2002; Harris et al. 2002). Thus, remarkably, AID was identified as a master regulator of all three B cell-specific modifications of rearranged Ig genes. It is currently thought that all three modifications are initiated by AID-mediated deamination of cytidines and that the manner in which the resulting lesions (G/U mismatches) are resolved determines whether somatic hypermutation, gene conversion, or class-switch recombination follows. Inhibition of uracil-DNA glycosylase , for example, reduces gene conversion and activates somatic hypermutation in DT40 cells (Di Noia and Neuberger 2002). This observation demonstrates that gene conversion is favored by uracil excision of AID-induced G/U mismatches, rather than by their recognition by the mismatch repair complex. It further suggests that gene conversion might be facilitated by a DNA strand break generated by an apyrimidinic endonuclease acting on an abasic site (Di Noia and Neuberger 2002).
Impairment of homologous recombination in DT40 cells can also reduce gene conversion activity and activate somatic hypermutation of Ig genes. Disrupting the RAD51 paralogs RAD51B, XRCC2, or XRCC3, for example, shifts the pattern of Ig V region mutations from gene conversion tracts to frequent point mutations localized preferentially at G/C base pairs, especially at RGYW motifs, known as somatic hypermutation hotspots (Sale et al. 2001; Rogozin and Kolchanov 1992). This observation supports the idea that gene conversion and somatic hypermutation represent distinct pathways for resolving a common AID-mediated lesion in the Ig V gene and suggests that recombination-mediated repair influences the choice of pathway. Furthermore, preventing Ig gene conversion by deleting the upstream ψV L gene segments in DT40 cells induces somatic hypermutation of the IgL gene (Arakawa et al. 2004). As in the RAD51 paralog mutants, point mutations are found primarily at G/C base pairs in RGYW hotspots. The mutations occur between 150 and 500 bp downstream of the Ig light chain promoter and are dependent on AID expression. The point mutations observed in DT40 RAD51- and ψV L -deletion mutants are thus similar to those observed in human and mouse germinal center B cells in their distribution with respect to the Ig light chain promoter, dependence on AID, preference for RGYW hotspots, and restriction to the Ig locus. They do differ, however, in preferentially occurring at G/C base pairs and consisting primarily of G to C and C to G transversions, whereas somatic hypermutation in germinal center B cells targets G/C and A/T base pairs with similar frequency and exhibits a slight bias for transitions.
10 A Model for the Regulation of Gene Conversion and Somatic Hypermutation
Arakawa and Buerstedde (2009) incorporated many of the insights gained from DT40 studies into a model explaining the initiation and regulation of somatic hypermutation and gene conversion of Ig genes. In their model, both are initiated by a common DNA lesion, AID-mediated cytidine deamination, within the Ig gene. In the absence of nearby homologous donors or high homologous recombination activity, the resulting G/U mismatch is resolved by error-prone repair pathways that introduce point mutations characteristic of somatic hypermutation. The availability of homologous donors and high homologous recombination activity, on the other hand, favors resolution by gene conversion. Irreversible commitment to the gene conversion pathway, however, only occurs during strand exchange, and prior to this step, a shift to somatic hypermutation is still possible. Thus, deletion of individual RAD51 paralogs, which participate in steps preceding strand exchange, decreases Ig gene conversion and induces Ig somatic hypermutation, while deletion of RAD52 members that are active after strand exchange, such as RAD54, only decreases Ig gene conversion (Sale et al. 2001; Bezzubova et al. 1997). This model also predicts that Ig gene conversion will only occur if homologous donors and high homologous recombination activity are both available, potentially explaining why B cells in some species utilize Ig gene conversion, while those in other species do not.
11 Antibody Repertoire Diversification in GALT in Humans and Mice
Although mice and humans use combinatorial rearrangement of multiple V, (D), and J gene segments as a strategy for generating a diverse primary antibody repertoire, might the strategy employed by chickens, rabbits, and artiodactyls be conserved in some human and mouse B cell populations? A number of observations suggest that this might indeed be the case. Casola et al. (2004), for example, found that B cell receptor-deficient transgenic mice spontaneously develop germinal centers (GCs) in Peyer’s patches. This antigen-independent B cell activation in mouse GALT, presumably induced by signals acquired from the intestinal microbiota, is reminiscent of the antigen-independent B cell activation stimulated by intestinal commensals in rabbit GALT that drives diversification of the primary antibody repertoire. Furthermore, Shimomura et al. (2008) identified an intestinal B cell population that develops independently of B cell receptor specificity, Lyn or Btk signaling, or T-cell help. A proportion of these B cells expressed mutated V(D)J genes, suggesting they had undergone antigen-independent diversification by somatic hypermutation.
A B cell population with similar characteristics has been identified in humans with common variable immune disorders caused by mutations in genes encoding proteins required for T-cell help and GC formation. A circulating population of IgM+IgD+ B cells has been identified in these patients that expresses the post-GC memory B cell marker CD27 and has mutated V(D)J genes (Weller et al. 2001). These B cells are probably not generated during T-cell-independent (TI), antigen-specific responses to bacterial capsules or cell wall components because they are also found in human infants too young to respond to TI antigens (less than 2 years of age) (Weller et al. 2008). Instead, Weill et al. (2004) suggest that human CD27+IgM+IgD+ B cells diversify their V(D)J genes independently of antigen, in a manner similar to that seen in “GALT species” (Weill et al. 2004). Interestingly, the CD27+IgM+IgD+ B cell population is markedly reduced in children deficient in MyD88 or IRAK-4 (interleukin-1 receptor-associated kinase 4), molecules required for MyD88-dependent Toll-like receptor (TLR) signaling (Maglione et al. 2014). These children suffer from life-threatening, often recurrent, pyogenic bacterial infections, particularly invasive pneumococcal disease (Picard et al. 2003; von Bernuth et al. 2008). They are, however, otherwise healthy and mount protective immune responses against other bacterial infections. Moreover, their clinical status improves with age, becoming essentially normal by 10 years of age. These observations suggest that human CD27+IgM+IgD+ B cells provide important, nonredundant immune protection against pyogenic bacteria early in life. The CD27+IgM+IgD+ phenotype is characteristic of marginal zone B cells, which provide a first line of defense because they localize in marginal sinuses where the blood first enters the spleen and produce low affinity, polyreactive “innate IgM” that effectively recognizes repetitive epitopes, such as microbial glycans and carbohydrates. Significantly, Maglione et al. (2014) found that serum IgM of children with MyD88 or IRAK-4 deficiency recognizes a greatly reduced range of microbial glycans, compared to that of normal children. Taken together, these data suggest that human CD27+IgM+IgD+ B cells generate a diversified antibody repertoire in a TLR-dependent, antigen-independent manner and are important for protection against pyogenic bacteria early in life. These similarities to the antibody diversification strategy used by the “GALT species” suggest that this strategy might be conserved in human and mouse B cell populations serving specialized functions.
12 Concluding Statement
The generation of a diverse primary antibody repertoire in vertebrates is fundamentally important for protection against pathogens. The emergence, during vertebrate evolution, of primary antibody repertoire diversification by AID-mediated gene conversion provided a highly effective solution to this problem. This strategy is likely more widely utilized among vertebrates than is currently appreciated and might be conserved in specialized B cell populations in those that generate their primary antibody repertoires by combinatorial rearrangement of multiple V, (D), and J gene segments.
References
Arakawa H, Buerstedde JM (2004) Immunoglobulin gene conversion: insights from bursal B cells and the DT40 cell line. Dev Dyn 229:458–464
Arakawa H, Buerstedde JM (2009) Activation-induced cytidine deaminase-mediated hypermutation in the DT40 cell line. Philos Trans R Soc Lond B Biol Sci 364:639–644
Arakawa H, Furusawa S, Ekino S, Yamagishi H (1996) Immunoglobulin gene hyperconversion ongoing in chicken splenic germinal centers. EMBO J 15(10):2540–2546
Arakawa H, Hauschild J, Buerstedde JM (2002) Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science 295:1301–1306
Arakawa H, Saribasak H, Buerstedde JM (2004) Activation-induced cytidine deaminase initiates immunoglobulin gene conversion and hypermutation by a common intermediate. PLoS Biol 2, E179
Barrington RA, Fasullo M, Knight KL (1999) A role for RAD51 in the generation of immunoglobulin gene diversity in rabbits. J Immunol 162:911–919
Becker RS, Knight KL (1990) Somatic diversification of immunoglobulin heavy chain VDJ genes: evidence for somatic gene conversion in rabbits. Cell 63:987–997
Becker RS, Zhai SK, Currier SJ, Knight KL (1989) Ig VH, DH and JH germ-line gene segments linked by overlapping cosmid clones of rabbit DNA. J Immunol 142:1351–1355
Bezzubova O, Silbergleit A, Yamaguchi-Iwai Y, Takeda S, Buerstedde J-M (1997) Reduced X-ray resistance and homologous recombination frequencies in a RAD54-/- mutant of the chicken DT40 cell line. Cell 89:185–193
Buerstedde JM, Takeda S (1991) Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell 67:179–188
Buerstedde JM, Reynaud CA, Humphries HE, Olson W, Ewert DL, Weill JC (1990) Light chain gene conversion continues at high rate in an ALV-induced cell line. EMBO J 9:921–927
Butler JE, Santiago-Mateo K, Sun XZ, Wertz N, Sinkora M, Francis DH (2011) Antibody repertoire development in fetal and neonatal piglets. XX. B cell lymphogenesis is absent in the ileal Peyer’s patches, their repertoire development is antigen dependent, and they are not required for B cell maintenance. J Immunol 187:5141–5149
Carlson LM, Mccormack WT, Postema CE, Humphries EH, Thompson CB (1990) Templated insertions in the rearranged chicken IgL V gene segment arise by intrachromosomal gene conversion. Genes Dev 4:536–547
Casola S, Otipoby KL, Alimzhanov M, Humme S, Uyttersprot N, Kutok JL, Carroll MC, Rajewsky K (2004) B cell receptor signal strength determines B cell fate. Nat Immunol 5:317–327
Di Noia J, Neuberger MS (2002) Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature 419:43–48
Di Noia JM, Neuberger MS (2007) Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem 76:1–22
Friedman ML, Tunyaplin C, Zhai SK, Knight KL (1994) Neonatal VH, D and JH gene usage in rabbit B-lineage cells. J Immunol 152:632–641
Fujimori A, Tachiiri S, Sonoda E, Thompson LH, Dhar PK, Hiraoka M, Takeda S, Zhang Y, Reth M, Takata M (2001) Rad52 partially substitutes for the Rad51 paralog XRCC3 in maintaining chromosomal integrity in vertebrate cells. EMBO J 20:5513–5520
Harris RS, Sale JE, Petersen-Mahrt SK, Neuberger MS (2002) AID is essential for immunoglobulin V gene conversion in a cultured B cell line. Curr Biol 12:435–438
Kawabata M, Kawabata T, Nishibori M (2005) Role of recA/RAD51 family proteins in mammals. Acta Med Okayama 59:1–9
Kim S, Humphries EH, Tjoelker L, Carlson L, Thompson CB (1990) Ongoing diversification of the rearranged immunoglobulin light-chain gene in a bursal lymphoma cell line. Mol Cell Biol 10:3224–3231
Knight KL, Becker RS (1990) Molecular basis of the allelic inheritance of rabbit immunoglobulin VH allotypes: implications for the generation of antibody diversity. Cell 60:963–970
Lavinder JJ, Hoi KH, Reddy ST, Wine Y, Georgiou G (2014) Systematic characterization and comparative analysis of the rabbit immunoglobulin repertoire. PLoS One 9, e101322
Liskay RM, Letsou A, Stachelek JL (1987) Homology requirement for efficient gene conversion between duplicated chromosomal sequences in mammalian cells. Genetics 115:161–167
Maglione PJ, Simchoni N, Black S, Radigan L, Overbey JR, Bagiella E, Bussel JB, Bossuyt X, Casanova JL, Meyts I, Cerutti A, Picard C, Cunningham-Rundles C (2014) IRAK-4 and MyD88 deficiencies impair IgM responses against T-independent bacterial antigens. Blood 124:3561–3571
Mccormack WT, Thompson CB (1990) Chicken IgL variable region gene conversions display pseudogene donor preference and 5/ and 3/ polarity. Genes Dev 4:548–558
Mccormack WT, Carlson LM, Tjoelker LW, Thompson CB (1989a) Evolutionary comparison of the avian IgL locus: combinatorial diversity plays a role in the generation of the antibody repertoire in some avian species. Int Immunol 1:332–341
Mccormack WT, Tjoelker LW, Barth CF, Carlson LM, Petryniak B, Humphries EH, Thompson CB (1989b) Selection for B cells with productive IgL gene rearrangements occurs in the bursa of Fabricius during chicken embryonic development. Genes Dev 3:838–847
Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T (1999) Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem 274:18470–18476
Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T (2000) Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102:553–563
Ordinario EC, Yabuki M, Handa P, Cummings WJ, Maizels N (2009) RAD51 paralogs promote homology-directed repair at diversifying immunoglobulin V regions. BMC Mol Biol 10:98
Parng C-L, Hansal S, Goldsby RA, Osborne BA (1996) Gene conversion contributes to Ig light chain diversity in cattle. J Immunol 157:5478–5486
Picard C, Puel A, Bonnet M, Ku CL, Bustamante J, Yang K, Soudais C, Dupuis S, Feinberg J, Fieschi C, Elbim C, Hitchcock R, Lammas D, Davies G, Al-Ghonaium A, Al-Rayes H, Al-Jumaah S, Al-Hajjar S, Al-Mohsen IZ, Frayha HH, Rucker R, Hawn TR, Aderem A, Tufenkeji H, Haraguchi S, Day NK, Good RA, Gougerot-Pocidalo MA, Ozinsky A, Casanova JL (2003) Pyogenic bacterial infections in humans with IRAK-4 deficiency. Science 299:2076–2079
Pospisil R, Young-Cooper GO, Mage RG (1995) Preferential expansion and survival of B lymphocytes based on VH framework 1 and framework 3 expression: “Positive” selection in appendix of normal and VH-mutant rabbits. Proc Natl Acad Sci USA 92:6961–6965
Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, Tezcan I, Ersoy F, Kayserili H, Ugazio AG, Brousse N, Muramatsu M, Notarangelo LD, Kinoshita K, Honjo T, Fischer A, Durandy A (2000) Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell 102:565–575
Reynaud C-A, Anquez V, Dahan A, Weill J-C (1985) A single rearrangement event generates most of the chicken immunoglobulin light chain diversity. Cell 40:283–291
Reynaud C-A, Anquez V, Grimal H, Weill J-C (1987) A hyperconversion mechanism generates the chicken light chain preimmune repertoire. Cell 48:379–388
Reynaud C-A, Dahan A, Anquez V, Weill J-C (1989) Somatic hyperconversion diversifies the single VH gene of the chicken with a high incidence in the D region. Cell 59:171–183
Reynaud C-A, Bertocci B, Dahan A, Weill J-C (1994) Formation of the chicken B-cell repertoire: Ontogenesis, regulation of Ig gene rearrangement, and diversification by gene conversion. Adv Immunol 57:353–378
Rhee KJ, Sethupathi P, Driks A, Lanning DK, Knight KL (2004) Role of commensal bacteria in development of gut-associated lymphoid tissues and preimmune antibody repertoire. J Immunol 172:1118–1124
Rhee KJ, Jasper PJ, Sethupathi P, Shanmugam M, Lanning D, Knight KL (2005) Positive selection of the peripheral B cell repertoire in gut-associated lymphoid tissues. J Exp Med 201:55–62
Rogozin IB, Kolchanov NA (1992) Somatic hypermutagenesis in immunoglobulin genes. II. Influence of neighbouring base sequences on mutagenesis. Biochim Biophys Acta 1171:11–18
Sale JE, Calandrini DM, Takata M, Takeda S, Neuberger MS (2001) Ablation of XRCC2/3 transforms immunoglobulin V gene conversion into somatic hypermutation. Nature 412:921–926
Sayegh CE, Demaries SL, Iacampo S, Ratcliffe MJ (1999) Development of B cells expressing surface immunoglobulin molecules that lack V(D)J-encoded determinants in the avian embryo bursa of fabricius. Proc Natl Acad Sci USA 96:10806–10811
Schiaffella E, Fuschiotti P, Bensinger SJ, Mage RG (1998) High RAD51 mRNA levels in young rabbit appendix. A role in B-cell gene conversion? Immunogenetics 48:108–115
Sehgal D, Johnson G, Wu TT, Mage RG (1999) Generation of the primary antibody repertoire in rabbits: expression of a diverse set of Igk-V genes may compensate for limited combinatorial diversity at the heavy chain locus. Immunogenetics 50:31–42
Sehgal D, Obiakor H, Mage RG (2002) Distinct clonal Ig diversification patterns in young appendix compared to antigen-specific splenic clones. J Immunol 168:5424–5433
Severson KM, Mallozzi M, Driks A, Knight KL (2010) B cell development in GALT: role of bacterial superantigen-like molecules. J Immunol 184:6782–6789
Shimomura Y, Ogawa A, Kawada M, Sugimoto K, Mizoguchi E, Shi HN, Pillai S, Bhan AK, Mizoguchi A (2008) A unique B2 B cell subset in the intestine. J Exp Med 205:1343–1355
Sonoda E, Sasaki MS, Buerstedde JM, Bezzubova O, Shinohara A, Ogawa H, Takata M, Yamaguchi-Iwai Y, Takeda S (1998) Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J 17:598–608
Sun J, Hayward C, Shinde R, Christenson R, Ford SP, Butler JE (1998) Antibody repertoire development in fetal and neonatal piglets. I. Four VH genes account for 80 percent of VH usage during 84 days of fetal life. J Immunol 161:5070–5078
Takata M, Sasaki MS, Sonoda E, Fukushima T, Morrison C, Albala JS, Swagemakers SM, Kanaar R, Thompson LH, Takeda S (2000) The Rad51 paralog Rad51B promotes homologous recombinational repair. Mol Cell Biol 20:6476–6482
Takata M, Sasaki MS, Tachiiri S, Fukushima T, Sonoda E, Schild D, Thompson LH, Takeda S (2001) Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol Cell Biol 21:2858–2866
Tauchi H, Kobayashi J, Morishima K, Van Gent DC, Shiraishi T, Verkaik NS, Vanheems D, Ito E, Nakamura A, Sonoda E, Takata M, Takeda S, Matsuura S, Komatsu K (2002) Nbs1 is essential for DNA repair by homologous recombination in higher vertebrate cells. Nature 420:93–98
Thompson CB, Neiman PE (1987) Somatic diversification of the chicken immunoglobulin light chain gene is limited to the rearranged variable gene segment. Cell 48:369–378
Vajdy M, Sethupathi P, Knight KL (1998) Dependence of antibody somatic diversification on gut-associated lymphoid tissue in rabbit. J Immunol 160:2725–2729
Von Bernuth H, Picard C, Jin Z, Pankla R, Xiao H, Ku CL, Chrabieh M, Mustapha IB, Ghandil P, Camcioglu Y, Vasconcelos J, Sirvent N, Guedes M, Vitor AB, Herrero-Mata MJ, Arostegui JI, Rodrigo C, Alsina L, Ruiz-Ortiz E, Juan M, Fortuny C, Yague J, Anton J, Pascal M, Chang HH, Janniere L, Rose Y, Garty BZ, Chapel H, Issekutz A, Marodi L, Rodriguez-Gallego C, Banchereau J, Abel L, Li X, Chaussabel D, Puel A, Casanova JL (2008) Pyogenic bacterial infections in humans with MyD88 deficiency. Science 321:691–6
Waldman AS, Liskay RM (1987) Differential effects of base-pair mismatch on intrachromosomal versus extrachromosomal recombination in mouse cells. Proc Natl Acad Sci USA 84:5340–5344
Weill JC, Reynaud CA, Lassila O, Pink JR (1986) Rearrangement of chicken immunoglobulin genes is not an ongoing process in the embryonic bursa of Fabricius. Proc Natl Acad Sci USA 83:3336–3340
Weill JC, Weller S, Reynaud CA (2004) A bird’s eye view on human B cells. Semin Immunol 16:277–281
Weinstein PD, Anderson AO, Mage RG (1994) Rabbit IgH sequences in appendix germinal centers: VH diversification by gene conversion-like and hypermutation mechanisms. Immunity 1:647–659
Weller S, Faili A, Garcia C, Braun MC, Le Deist FF, de Saint Basile GG, Hermine O, Fischer A, Reynaud CA, Weill JC (2001) CD40-CD40L independent Ig gene hypermutation suggests a second B cell diversification pathway in humans. Proc Natl Acad Sci USA 98:1166–1170
Weller S, Mamani-Matsuda M, Picard C, Cordier C, Lecoeuche D, Gauthier F, Weill JC, Reynaud CA (2008) Somatic diversification in the absence of antigen-driven responses is the hallmark of the IgM+ IgD+ CD27+ B cell repertoire in infants. J Exp Med 205:1331–1342
Winstead CR, Zhai SK, Sethupathi P, Knight KL (1999) Antigen-induced somatic diversification of rabbit IgH genes: gene conversion and point mutation. J Immunol 162:6602–6612
Zhai SK, Lanning DK (2013) Diversification of the primary antibody repertoire begins during early follicle development in the rabbit appendix. Mol Immunol 54:140–147
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Lanning, D.K., Knight, K.L. (2015). Diversification of the Primary Antibody Repertoire by AID-Mediated Gene Conversion. In: Hsu, E., Du Pasquier, L. (eds) Pathogen-Host Interactions: Antigenic Variation v. Somatic Adaptations. Results and Problems in Cell Differentiation, vol 57. Springer, Cham. https://doi.org/10.1007/978-3-319-20819-0_12
Download citation
DOI: https://doi.org/10.1007/978-3-319-20819-0_12
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-20818-3
Online ISBN: 978-3-319-20819-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)