Abstract
The immune systems of all mammals include populations of B cells producing antibodies with incredibly diverse specificities. Repertoire diversity has been described as the “miracle of immunology,” and it was long thought to be the result of essentially stochastic processes. Recently, however, analysis of high throughput gene sequencing data has shown that hard-wired biases in these processes result in antibody repertoires that are broadly predictable. The repertoires of mice and humans are both predictable, but they are strikingly different. In this review, features of the naïve antibody repertoires of the two species are contrasted. We show that the mouse repertoire includes a conspicuous population of public clonotypes that are shared by different individuals of an inbred strain. These clonotypes are the result of gene rearrangements that involve little gene processing. By skewing repertoire formation toward such sequences, which probably target commonly encountered pathogens, it may be that the relatively small mouse repertoire is appropriate and effective despite its size. Species like the mouse face challenges that are a direct consequence of their small body sizes and the limitations this places on the antibody arsenal—particularly early in ontogeny. We propose that it is the differences in the naïve repertoires of mice and humans, and the differences in the ways these repertoires are used, which ensure that the very different biological needs of the two species are met. The processes that contribute to repertoire formation may appear to be stochastic, but in both species, evolution has left little to chance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Species size and scaling of the immune system
All mammals have the ability to generate protective antibodies with enormously varying specificities. This miraculous diversity is made possible by multiple sets of highly similar germline genes that recombine to form functional genes encoding the immunoglobulin heavy and light chain polypeptides (Nossal 2003; Tonegawa 1983). The pairing of these chains creates the B cell receptors (BCR) that are expressed on the surface of B cells, and the secretion of these molecules forms a pool of free antibodies in the blood. In this review, the term “repertoire” is used to refer to the complete set of rearranged antibody genes that are carried by an individual, at a moment in time, as well as the complete set of BCR and free antibodies that these genes encode. The number of unique BCR/antibody sequences that are found within an individual’s repertoire is described here as the “diversity” of the repertoire.
It has been estimated that the human immune system is able to generate at least 1026 different antibody molecules (Saada et al. 2007). This estimate can serve as an approximation of the potential repertoire of any mammalian species, and it far exceeds the number of lymphocytes that is found in any species. There are only about 5 × 106 B cells in a neonatal house mouse (Mus musculus), and the number of cells only rises to about 108 in an adult (Spear et al. 1973). Even in a larger species like the human, the adult B cell population may be fewer than 1012 cells, as the total cell population of a human has been estimated to be just 3.7 × 1013 cells (Bianconi et al. 2013). The repertoires of all species are therefore small samplings of the potential repertoire. If the nature of the antibody repertoire is to be understood, we need to determine how stochastic processes and constraints that affect such processes ensure that each small individual sampling is a useful one. This is something that may vary with the size of the species.
The ways in which the sizes of species affect anatomical features and physiological processes have been a focus of interest for many centuries. The consequences for skeletal structure of the square-cube law were first described by Galileo Galilei in 1638. He noted that a bone’s strength is proportional to its cross-sectional area. Doubling the dimensions of an organism increases its weight eightfold, but bone strength only increases fourfold. Larger and heavier species therefore require broader bones (Galilei 1638). During the late nineteenth century, scaling laws were first described to explain the relationships between species’ mass and brain size (Snell 1892). In the twentieth century, the relationships between mass and other fundamental biological variables including life span and metabolic rate were reported (Haldane 1926; Hoffman 1983; Kleiber 1947). These relationships can be described by simple power laws that are multiples of one fourth (Kleiber 1947), and this has now been explained by reference to the fractal-like networks from which the circulation and respiratory systems are built (West et al. 1999).
We are aware of only three groups that have considered scaling and the immune system. Early investigations by Hsu and colleagues showed that the antibody response in Xenopus tadpoles becomes more complex after their metamorphosis into frogs (Hsu and Du Pasquier 1984; Flajnik et al. 1987). Stage-specific features of the antibody response appeared to remain after adults and tadpoles were manipulated to be of similar size. The authors concluded that features such as the higher affinity of adult antibodies were not a consequence of the larger repertoire of adult frogs, but were the result of metamorphosis-associated changes in the nature of the antibody repertoire (Hsu and Du Pasquier 1992).
Langman and Cohn argued that it would be absurd for different immune systems to have evolved to meet the differing needs of species of varying sizes (Langman and Cohn 1987). Instead, they proposed that the immune repertoire is modular, with each “protecton” being made up of about 107 cells targeting 106 different antigens. A hummingbird might carry one such modular protecton, while an elephant could carry 107 protectons.
Wiegel and Perelson proposed that many features of the immune system scale with species mass (M) (Wiegel and Perelson 2004). They assumed that a repertoire is made up of many clones of equal size, and by reference to models of lymphocyte trafficking, the metabolic rate of the lymphocyte population, and other variables, they deduced that clone size scales as M. They also concluded that the diversity of the repertoire, which they described as repertoire size, scales as ln(cM), where c is a dimensionless constant, and cM is the lifetime number of infections faced by an organism (Wiegel and Perelson 2004). The total number of B cells would therefore scale as M ln(cM). On the assumption that the lifetime total number of infections faced by a mouse is 10, they suggested that there is five times as much diversity in the human repertoire as in the mouse repertoire, and that mouse and human clone sizes are 10 and 100,000 cells, respectively.
The theoretical equations derived by Wiegel and Perelson were informed by very little experimental repertoire data, because almost no suitable data was available at the time. The situation is now changing, as a result of the development of high throughput sequencing (HTS), and its application to the study of antibody genes (Boyd et al. 2009; Boyd et al. 2010; Weinstein et al. 2009). Sets of germline genes are being reported for many different species (Corcoran et al. 2016; Guo et al. 2011; Walther et al. 2015), and repertoire studies will surely follow. For the moment, extensive repertoire datasets for mammalian species are largely confined to the C57BL/6 and BALB/c strains of inbred mouse and to the human. Although few of these studies explicitly address issues like clone size, or calculate repertoire diversity, they allow many features of the repertoire to be described. These features in turn allow the architectures of different immune repertoires to be seen. In this review, we will outline key differences between the naïve repertoires of the two mouse strains, and between the two species.
We recognize that the naive repertoire emerges from the pre-B cell repertoire as a result of negative selection against non-productive and self-reactive specificities (Gu et al. 1991; Larimore et al. 2012; Meng et al. 2011; Wardemann et al. 2003), but selection will not be a focus here. Analysis of millions of antibody gene sequences has shown that despite selection, measureable features of the expressed repertoire are relatively stable between individuals and allow predictive models to be constructed (Briney et al. 2012; Elhanati et al., 2015; Glanville et al. 2011; Greiff et al. 2017; Rubelt et al. 2016). In addition, it is the diversity of this selected repertoire and clone sizes within the repertoire that are critical determinants of how the repertoire serves the biological needs of the species. Our focus is therefore upon the expressed naïve repertoire.
The generation of antibody diversity
Functional immunoglobulin heavy chain genes, called VDJ genes, are the product of genes that recombine early in B cell development. Within each developing B lymphocyte, genomic rearrangement brings together one IGHV gene from the set of heavy chain variable (V) genes, one IGHD gene from the set of diversity (D) genes, and one IGHJ gene from the set of heavy chain joining (J) genes (Bassing et al. 2002; Lieber 2008; Tonegawa 1983). Intervening sequences are lost from the genome. Each VDJ gene that forms encodes the variable part of the immunoglobulin heavy chain polypeptide, and it is expressed in association with one gene from the set of immunoglobulin heavy chain constant region genes. These constant region genes determine the functional class or isotype of each antibody (Schroeder and Cavacini 2010). As the naïve repertoire is the focus of this review, discussion will be largely confined to the IgM isotype.
There are two classes of light chain—kappa and lambda—and these are both the product of recombining light chain V and J genes. Kappa chains are encoded by IGKV and IGKJ genes, while lambda chains are encoded by IGLV and IGLJ genes. The rearranged VJ genes are expressed in association with kappa or lambda constant region genes, but in contrast to the heavy chain, no distinct functions are associated with the different light chain isotypes.
Heavy chain genes rearrange during the pro-B cell stage of B cell development. After association of the expressed heavy chain with the surrogate light chain, these pro-B cells may undergo limited rounds of cell division. This gives rise to clones of cells with shared heavy chains that we will refer to here as pro-B cell clones. In the mouse, it has been shown that four to six cell divisions often take place (Decker et al. 1991; Hess et al. 2001), but the expansion appears to vary depending upon the expressed IGHV gene (Meng et al. 2011). This may be a consequence of variable pairing efficiencies of different IGHV genes with the surrogate light chain. The extent of pro-B cell expansion in the human is unclear, but the larger size of humans could easily allow greater clonal expansion than is seen in the mouse.
Light chain genes rearrange at the pre-B cell stage of B cell development, and heavy and light chain association then leads to the expression of native antibody on the surface of the cells. The heavy chain of a pro-B cell clone therefore comes to be expressed in association with a variety of light chains, and it may be that there is subsequently a limited homeostatic expansion of some, or all, of these clonotypes (Fig. 1). Certainly, such homeostatic expansion has been reported for T lymphocytes (Haluszczak et al. 2009; Hogan et al. 2015), and the end result is a T cell repertoire made up of clonotypes and sets of clonotypes that target different antigens with a range of frequencies (Alanio et al. 2010; Blattman et al. 2002; Lythe et al. 2016).
Development of pre-B cells from pro-B cells involves clonal expansion of particular heavy chain rearrangements. A chance sharing of a heavy chain rearrangement is shown (H1). Public clonotypes may be present at high copy number, and these can develop when cells of expanded heavy chain pro-B cell clones associate by chance with commonly expressed light chains (broad arrows). Such clonotypes may also increase in abundance through homeostatic expansion. Only a limited number of pathways to heavy and light chain pairings are shown
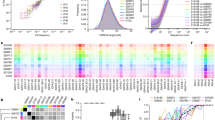
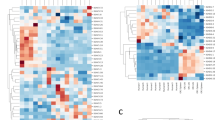
The permutations and combinations of the sets of heavy and light chain V, D, and J genes give rise to diversity that is referred to as combinatorial diversity. The diversity of the repertoire is further expanded as a result of imprecise joining at the V(D)J junctions during the recombination processes. The contribution of imprecise joining to the overall diversity of the immunoglobulin genes is referred to as junctional diversity. The processes that contribute to combinatorial and junctional diversity are often characterized as being essentially random in nature; however, this is not the case. It is now clear that the architecture or shape of the antibody repertoires of mice and humans—that is, the frequency distributions of different rearrangements within the repertoire—are pre-determined by biases in the recombination processes (Briney et al. 2012; Elhanati et al. 2015; Glanville et al. 2011; Greiff et al. 2017; Rubelt et al. 2016). A genetic basis for these biases has been strikingly demonstrated in pairs of identical human twins, for the repertoires of twins are significantly more similar than the repertoires of unrelated individuals (Glanville et al. 2011; Rubelt et al. 2016; Wang et al. 2015). An individual’s pattern of gene usage has also been shown to be stable over time (Laserson et al. 2014), and if an individual is treated with lymphocyte ablation therapy, the repertoires seen before and after lymphocyte ablation are highly similar (Glanville et al. 2011). Genetics influences both combinatorial diversity and junctional diversity and gives rise to the contrasting architectures of the human and murine repertoires.
The germline immunoglobulin genes of mice and humans
The complete human heavy chain locus was first described in 1998 (Matsuda et al. 1998), but the existence of substantial structural variation in the human locus only became apparent with more recent reports of heavy chain gene haplotypes. Most humans appear to carry 45–51 functional heavy chain germline genes (Boyd et al. 2010; Kidd et al. 2012; Watson et al. 2013), but until we have more data from disparate human populations, the extent of structural variation in the human heavy chain locus will remain uncertain (Watson and Breden 2012).
The number of functional IGHV genes in the C57BL/6 mouse strain is also unclear, despite the fact that the genome of this strain serves as the reference sequence for the species. The ImMunoGeneTics database recognizes 114 functional IGHV genes in the C57BL/6 strain, based upon an analysis of the mouse genome reference sequence and the association of these genes with intact recombination signal sequences (RSS) (Johnston et al. 2006; Lefranc et al. 2009; Riblet 2003). It is possible, however, that not all these genes are functional. Only 99 IGHV genes from the IMGT database could be identified in a set of 20,925 unique VDJ rearrangements generated from C57BL/6 splenocytes (Collins et al. 2015). Four additional IGHV genes were also seen in the VDJ dataset. IMGT considers two of these to be open reading frames of dubious functionality. The two other sequences were first reported by Johnston and colleagues (2006). One of these (musIGHV211) was recently incorporated into the IMGT database as IGHV2-9-1*01. The other (musIGHV269) is listed as a pseudogene in the VBASE2 database, because of the presence of a stop codon at codon 106 (Retter et al. 2005), but exonuclease removal of the final two nucleotides of the sequence (see below) can create an open reading frame.
The number of functional IGHV genes in other mouse strains is even less certain, but it is known that there are substantial differences between the number of IGHV genes in the C57BL/6 and BALB/c strains. In an analysis of 15,103 unique BALB/c VDJ genes, the existence of 162 distinct IGHV germline gene sequences was inferred, including 82 sequences that are absent from the IMGT database (Collins et al. 2015). Only 5 of the 162 sequences are shared with the C57BL/6 strain (Collins et al. 2015). The differences between the strains appear to be a consequence of the early breeding histories of the classical inbred mouse strains. This has resulted in the genomes of inbred mice having a mosaic structure, with different blocks of genes being derived from the three subspecies of the house mouse (Yang et al. 2011). The heavy chain locus of the BALB/c strain is derived from Mus musculus domesticus and the locus of the C57BL/6 strain is derived from Mus musculus (Fig. 2). The loci have therefore evolved in different parts of the world, under differing selection pressures, since the emergence of the three subspecies from a common ancestor 350,000 years ago (Geraldes et al. 2011).
There are a number of uncertainties regarding the human and murine IGHD loci. The human IGHD locus was first described by Corbett and colleagues and was shown to include 25 unique sequences that are found as 27 IGHD genes (Corbett et al. 1997). The IGHD4-4*01 and IGHD4-11*01 coding sequences are identical, as are IGHD5-5*01 and IGHD5-18*01. The IMGT database defines four IGHD sequences as ORFs of dubious functionality because of their association with aberrant RSS (Lefranc et al. 2009). However, two of these sequences (IGHD4-23*01 and IGHD5-24*01) are frequently identified in VDJ rearrangements, while the supposedly functional IGHV6-25*01 is not (Lee et al. 2006). There therefore appear to be 23 unique and functional IGHD sequences in the standard human germline IGHD repertoire.
Seventeen murine IGHD genes have been reported in the BALB/c strain that are recognized by IMGT as functional genes. Nineteen C57BL/6 genes are also recognized by IMGT, including 13 functional genes, 1 pseudogene, and 5 open reading frames (Lefranc et al. 2009). In an analysis of the C57BL/6 genome reference sequence, Ye identified only ten functional genes and six additional “potential D genes” (Ye 2004). In a recent analysis of murine VDJ rearrangements, we could only infer the existence of 9 functional C57BL/6 IGHD genes and 12 functional BALB/c IGHD genes (Collins et al. 2015). Some of these are duplicate sequences, so most if not all D gene diversity in the strains is the product of just eight and ten unique sequences.
Six functional IGHJ genes are found in the human and four functional IGHJ genes are found in the mouse (Lefranc et al. 2009). The mouse IGHJ1, IGHJ2, IGHJ3, and IGHJ4 genes are orthologues of human IGHJ2, IGHJ4, IGHJ5, and IGHJ6. Interestingly, the utilization frequencies of the orthologous genes are quite different. For example, musIGHJ1*03 is seen in 16% of C57BL/6 VDJ rearrangements (Collins et al. 2015), but humIGHJ2*01 is seen in just 1% of human rearrangements (Kidd et al. 2016).
The human IGK locus includes two domains spaced approximately 800 kb apart. The IGK proximal domain includes 40 IGKV genes, pseudogenes, and open reading frames (ORFs) as well as IGKJ genes and IGK constant region genes. The distal domain is essentially a duplication of the IGKV genes of the proximal domain and contains an additional 36 IGKV genes, pseudogenes, and ORFs (Zachau 1989; Zachau 1993). Overall, the kappa locus includes 63 distinct functional IGKV sequences and 10 additional ORFs. In comparison to the heavy chain locus, there are relatively few allelic variants of the light chain genes (Collins et al. 2008; Hoi and Ippolito 2013; Jackson et al. 2012), and although recent studies have highlighted previously unrecognized structural variation in the loci, the variation is much less than is seen at the heavy chain locus (Watson et al. 2015).
There is greater kappa germline gene diversity in the mouse. Genomic sequencing of the C57BL/6 mouse identified 93 functional or potentially functional IGKV genes (Thiebe et al. 1999), but a recent analysis of over 250,000 C57BL/6 VJ rearrangements inferred the existence of 101 functional IGKV genes (Aoki-Ota et al. 2012). These include ten sequences defined by IMGT as ORFs of unknown functionality. Many of these genes are also carried by the BALB/c strain, and both the kappa and lambda loci of each strain are derived from Mus musculus domesticus (Fig. 2).
The structure of the mouse lambda locus is different from all other loci of the mouse and human. It contains just two small gene clusters which each contain IGLV, IGLJ, and constant region genes (Lefranc and Lefranc 2004). Together they provide just four possible lambda VJ rearrangements (see Table 1).
The contributions of combinatorial diversity to repertoire formation
A first estimate of the contributions of combinatorial diversity to the naïve repertoires of the mouse and the human can be made by simply calculating the permutations and combinations that can be achieved given the number of unique and functional germline gene sequences in each recombining gene set. By this measure, heavy chain combinatorial diversity is very similar in the human and mouse. A lower number of IGHV genes in the human is balanced by a higher number of IGHD and IGHJ genes (see Table 1).
Combinatorial diversity is expanded in the human because IGHD genes can be read in all three reading frames. Although it is true that selection creates strong biases against certain reading frames of some human genes (Benichou et al. 2013), the human-expressed repertoire is still substantially increased by the alternative reading frames. This is not true in the mouse. There is a single very strongly preferred reading frame for each mouse IGHD gene. Although this likely reflects negative selection of particular mouse rearrangements (Schelonka et al. 2008; Zemlin et al. 2008), it may also reflect positive biases that result from the joining of murine IGHD and IGHJ genes via short homologous sequences at their gene ends (Feeney 1992b).
The contribution of IGHD genes to diversity is further reduced in the mouse, compared to the human, because of the relatively short lengths and the similarities of the mouse IGHD genes. The human IGHD genes come from seven disparate gene families, while the mouse genes come from four families. In the C57BL/6 mouse, six of the nine expressed IGHD sequences are highly similar members of the IGHD2 family, and these are all very similar to the IGHD1-1*01 gene.
Combinatorial diversity varies between outbred individuals as a result of gene deletions and gene duplications. In the human, IGHV deletion polymorphisms and gene duplications are common (Boyd et al. 2010; Chimge et al. 2005; Kidd et al. 2012; Pramanik and Li 2002; Watson et al. 2013). Substantial IGHD gene polymorphisms involving multiple contiguous genes have also been identified within the human population (Kidd et al. 2012). Data on such structural variation is unavailable for the mouse. Because the IGH loci of the BALB/c and C57BL/6 strains appear to have come from different M. musculus subspecies, direct comparisons cannot be made between their gene sets. Meaningful comparisons will only be possible when the complete locus of the BALB/c strain has been sequenced, and when the loci of other M. musculus domesticus and M. musculus musculus-derived strains can be compared to the C57BL/6 and BALB/c germline gene sets. This will likely lead to a major revision of the current murine IGHV gene nomenclature, which assumes the common evolutionary descent of all inbred mouse strains.
Combinatorial diversity is expanded by gene polymorphisms that lead to heterozygosity at some loci within an outbred individual. In the human, alternative alleles are typically seen at six to eight IGHV gene loci (Kidd et al. 2012), but individuals have been seen in whom virtually every locus is heterozygous (Scheepers et al. 2015). Such allelic variation increases combinatorial diversity, and despite the fact that allelic variants are often highly similar, the differences can be biologically significant (Liu and Lucas 2003; Avnir et al. 2016).
Calculations of combinatorial diversity say little about the contribution each heavy and light chain rearrangement makes to the expressed repertoire, for this is determined by the highly variable utilization frequencies of the different recombining genes, by the extent of pro-B cell clonal expansion, by homeostatic expansion of naïve B cells, and by the size of the naïve lymphocyte compartment. Heavy chain V, D, and J genes are used at frequencies that range through three or even four orders of magnitude (0.001–10%) in mice (Collins et al. 2015; Greiff et al. 2017) and in humans (Boyd et al. 2010; Glanville et al. 2011). The frequency of expression of most genes is highly predictable, but the utilization of a few human genes such as IGHV3-23*01 and IGHV1-69*01 appears to be more variable. This is a consequence of the presence of varying numbers of identical copies of these gene sequences within the genomes of different individuals (Sasso et al. 1996; Watson et al. 2013).
In a high throughput sequencing (HTS) study of human kappa chains, the dominance of one particular IGKV gene was extreme. The IGKV3-20*01 gene was present in 16 and 17% of rearrangements of two individuals. In two other individuals, the same sequence was present in 31 and 32% of rearrangements (Jackson et al. 2012). The higher utilization in these individuals is likely to be a consequence of gene duplication. IGKV1-5*03, IGKV1-39*01/IGKV1D-39*01, IGKV2-30*01, IGKV3-11*01, and IGKV3-15*01 also are prominent in the human repertoire (Foster et al. 1997; Jackson et al. 2012). Although no murine gene has been seen that is as highly utilized as human IGKV3-20*01, seven IGKV genes together account for over 40% of all mouse kappa rearrangements (Aoki-Ota et al. 2012).
Both in vitro and in vivo studies have shown that variation in utilization frequencies is partly a consequence of differences in the RSS that promote interaction between the recombining genes and the recombination machinery (Cowell et al. 2004; Feeney et al. 2000; Williams et al. 2001). Nevertheless, highly utilized genes and rarely utilized genes can be associated with identical RSS, as well as identical promoter sequences (Brekke and Garrard 2004).
Chromosomal location is also an important determinant of gene utilization (Choi et al. 2013), and this is particularly true during the development of the fetal repertoire (see below). A clear example of location-based bias in the expressed repertoire of the adult is the pairing bias that is seen in human heavy chain DJ recombination. We and others have reported that 3′ human IGHD genes preferentially pair with 5′ IGHJ genes, while 5′ IGHD genes tend to pair with 3′ IGHJ genes (Kidd et al. 2016; Souto-Carneiro et al. 2005; Volpe and Kepler 2008). Pairing frequencies differ in individuals with IGHD gene haplotypes that include deletion polymorphisms (Kidd et al. 2016), perhaps as a consequence of the resulting variations in distances between IGHD and IGHJ genes within the genome. Despite homology between human and murine IGHJ genes, DJ pairing bias has not been reported in the mouse.
There is evidence of positional biases in kappa VJ recombination. Kappa rearrangements in the mouse are biased toward rearrangement of the 5′ IGKJ1 gene (Yamagami et al. 1999). If this rearrangement is unproductive, additional rounds of secondary rearrangement are triggered. This process is known as receptor editing (Prak and Weigert 1995; Retter and Nemazee 1998), and its potential is maximized by the targeting of IGKJ1 in primary murine rearrangements. This targeting is a consequence of the action of the proximal IGKJ germline transcript promoter, which is one of two IGKJ promoters in the mouse genome (Vettermann et al. 2015).
The process of secondary rearrangement has complex effects upon the ultimate frequency of different productive murine VJ pairings. Many murine IGKV genes are present within the genome in the same transcriptional orientation as the IGKJ and IGKC genes. IGKV rearrangement with these IGKJ genes involves the loss of intervening genes. As these lost genes are unavailable for secondary rearrangement, the net result of large numbers of primary rearrangements of this kind, and of subsequent secondary rearrangements, should be an elevation in the frequency of 5′ IGKV and 3′ IGKJ pairings. A complication affecting the detection of this kind of VJ bias in the kappa repertoire of the mouse is that some IGKV genes have the opposite transcriptional orientation to the IGKJ and IGJC genes. Their recombination is associated with the inversion of intervening IGKV genes rather than their deletion. The mix of gene orientations may explain why preferential pairings of 5′ IGKV and 3′ IGKJ have not been reported. There are, however, other detectable biases including a biased use of murine IGKJ1 by most of the highly utilized IGKV genes (Aoki-Ota et al. 2012). In contrast, many of the most rarely utilized IGKV genes preferentially pair with IGKJ5, probably as a result of secondary rearrangements (Aoki-Ota et al. 2012).
One clear consequence of the process of secondary kappa VJ rearrangement in the mouse is that it increases the likelihood that productive kappa chain rearrangements will form. It therefore must increase the relative abundance of kappa-bearing antibodies compared to lambda-bearing antibodies. The nature and meaning of this ratio was a subject of debate for many years (Langman and Cohn 1992; Langman and Cohn 1995).
Theoretical kappa to lambda ratios can be calculated from the likelihood that any VJ rearrangement is productive, and from knowledge of the order in which the kappa and lambda loci rearrange. Functional rearrangements must maintain the required reading frame of downstream J genes and constant region genes, and consequently two thirds of random rearrangements are out-of-frame and unproductive. If a kappa locus rearrangement is unproductive, the second kappa locus can rearrange. If rearrangement of this chromosome is also unproductive, one of the two lambda gene loci can rearrange. Finally, if this rearrangement is unproductive, the second lambda gene locus offers a last chance for a productive rearrangement.
In the absence of secondary rearrangements, the ratio of kappa to lambda rearrangements should be 69:31 (1/3 + 2/3 × 1/3: 4/9 × 1/3 + 4/9 × 2/3 × 1/3). The ratio that is seen in the mouse is quite different (95:5) (Langman and Cohn 1992), and the relative abundance of kappa rearrangements is almost certainly a consequence of secondary rearrangements. We know of no formal demonstration that secondary rearrangement occurs in the human. The observed human kappa to lambda ratio is about 60:40, suggesting that the human kappa repertoire is dominated by primary rearrangements. Interestingly, in each species, the expressed kappa to lambda ratio closely matches the ratio of the number of germline kappa genes to the number of germline lambda V genes (see Table 1).
Stable expression of paired heavy and light chains on the B cell surface is an important checkpoint during the development of each cell. Early studies of human monoclonal antibodies suggested that the stability of different heavy and light chain pairs was highly variable (De Preval and Fougereau 1976). This suggested that biases in pairings could shape the antibody repertoire. The development of single-cell PCR allowed the issue to be more extensively explored (Brezinschek et al. 1998; de Wildt et al. 1999; Ghia et al. 1996), and these studies as well as more recent studies in humans using HTS have failed to identify any such biases (DeKosky et al. 2016).
If the stability of heavy and light chain pairings has the potential to be so variable, the sets of heavy and light chain genes would have to co-evolve. The fact that the heavy chain genes and light chain genes of the C57BL/6 mouse are derived from different subspecies of the house mouse (Fig. 2) raises the possibility that many pairings could be relatively unstable in this strain. This would make pairing biases readily detectable. HTS has now been applied to the study of C57BL/6 heavy and light chain pairs, but no biases have been observed (Busse et al. 2014; Tiller et al. 2009).
The contribution of junctional diversity to repertoire formation
V(D)J recombination is described as an imprecise process because during recombination, nucleotides can be both lost and gained by each of the joining gene ends. Recombination begins when RSS are recognized by the recombination activation gene proteins RAG-1 and RAG-2, which first nick one of the DNA strands between the RSS and the coding region of each of the joining genes, then introduce double-stranded breaks in the DNA (Schatz and Swanson 2011). This results in covalently sealed hairpins at each coding end. Asymmetric opening of hairpinned coding ends is then mediated by the Artemis:DNA-protein kinase catalytic subunit (DNA-PKCS) complex (Lu et al. 2007; Ma et al. 2002). This contributes to diversity because it results in inverted repeat nucleotide overhangs that can be incorporated into the junction region as palindromic (P) nucleotides. Often, these overhangs are removed by exonuclease activity, which can also remove varying numbers of nucleotides from the germline-encoded gene ends. This further expands diversity. The nucleases responsible have yet to be identified, though it is likely that the Artemis:DNA-PKCS complex is involved (Chang and Lieber 2016; Ma et al. 2002).
Prior to the joining of the recombining genes, non-template-encoded N nucleotides are added to the gene ends by the enzyme terminal deoxynucleotidyl transferase (TdT) (Basu et al. 1983). This arguably makes the greatest contribution to repertoire diversity, though diversity is constrained by a strong TdT bias for additions of Gs and Cs (Basu et al. 1983), and for the formation of homopolymer tracts (Gauss and Lieber 1996; Jackson et al. 2007; Murugan et al. 2012). Glycine, encoded by GGN codons, is the amino acid that is most frequently added to the junction by N nucleotides (Hofle et al. 2000).
The processes that generate junctional diversity are common to the human and the mouse, but the contributions to repertoire diversity are very different in the two species. In vitro studies show that the efficiency of action of the Artemis:DNA-PKCS complex varies for different human (Ezekiel et al. 1997; Lu et al. 2007) and mouse (Nadel and Feeney 1995; Nadel and Feeney 1997) gene ends, and most of the gene ends of the two species are different. The primary hairpin opening sites are also sequence dependent. In the human, the dominant IGHV opening site is 2 nts 3′ of the hairpin tip, but 3′ overhangs of variable length are generated (Lu et al. 2007). As many as seven P nucleotides can be added to a human heavy chain gene (Ohm-Laursen et al. 2006), but most human genes show little P addition (Jackson et al. 2004). In contrast, P addition is common in mouse rearrangements, though additions of more than four P nucleotides are very rare, and most additions are of just one or two nucleotides (Collins et al. 2015). P additions are particularly common at the 3′ ends of the murine IGHV and IGHD genes. P nucleotides not only contribute to the murine VDJ junctions, but they also influence repertoire development by promoting recombination at regions of short homology (see below).
Exonuclease trimming of joining gene ends has rarely been studied and is poorly understood. Studies in the mouse suggest that DNA polymerase mu may antagonize exonuclease activity and act to limit nucleotide removals during light chain gene rearrangement (Bertocci et al. 2003). The extent of removals is also affected by the gene end sequences. High G/C content seems to limit removals, while A/T-rich ends are susceptible to nucleotide loss (Gauss and Lieber 1996; Jackson et al. 2004; Nadel and Feeney 1995). It is likely that this explains why there are generally more exonuclease removals from human IGHD and IGHJ genes than from IGHV genes, and more removals from the 5′ ends of IGHD genes than from the 3′ IGHD gene ends (Souto-Carneiro et al. 2005). We are aware of no study that has compared exonuclease removals in the human with removals in the mouse.
Non-template-encoded N addition is a major driver of repertoire diversity, but it makes very different contributions to the heavy and light chain repertoires and very different contributions to the repertoires of the human and mouse. An average of 7.7 nucleotides are added to the VD junctions of the human heavy chain and 6.5 nucleotides are added to the DJ junctions (Jackson et al. 2007). On average, just 4.0 and 2.9 nucleotides are added to these regions in the C57BL/6 mouse, and there is even less addition in the BALB/c strain: 3.4 and 2.7 nucleotides, respectively (Collins et al. 2015). As no attempt was made in these studies to distinguish between P and N nucleotides, and as P nucleotides are more common in the mouse, this disparity in the number of N nucleotides in the two species is probably underestimated.
TdT is expressed in bone marrow pro-B cells prior to heavy chain variable region gene rearrangement, and its expression persists until the pre-B cell stage. It is barely detectable by the time that light chain rearrangement is taking place (Li et al. 1993). Light chain variable region genes therefore normally display minimal N addition in the mouse (Benedict et al. 2000; Bentolila et al. 1999; Richl et al. 2008) and the human. An analysis of human kappa sequences found an average of just 1.2 N additions per sequence (Jackson et al. 2012).
Few murine heavy or light chain gene sequences, and few human light chain gene sequences, contain N nucleotides that fully encode any amino acid in the polypeptide chains. The N nucleotides that are added to these sequences mainly function to complete codons that are partially encoded by germline nucleotides. Redundancy in the genetic code also means that the amino acids encoded by such codons are often entirely determined by the germline-encoded nucleotides. In particular, exonuclease removals of 3′ IGHD nucleotides very often have no consequences for the resulting amino acid sequence (Jackson et al. 2013). This can be illustrated by an analysis of human sequences that include the IGHD6-6*01 gene in its preferred reading frame. In this reading frame, the six 3′ nucleotides of IGHD6-6*01 encode two serine residues. In almost 70% of sequences, where these 3′ serine codons are affected by exonuclease removals of nucleotides, the completion of the codon by either N nucleotide additions or by germline-encoded IGHJ nucleotides results in another serine codon (unpublished).
Public clonotypes: a predictable outcome of repertoire formation
The biases in the generation of combinatorial and junctional diversity are so profound that the likelihood of the formation of different heavy and light chain pairs must vary by at least ten orders of magnitude. In a sufficiently large repertoire, certain heavy and light chain pairs may be relatively abundant, while other sequences may be so unlikely to form that they are never seen. Sequences that are more likely to form in one individual are also likely to form in other individuals. Such sequences are likely to be what can be described as public clonotypes. Public clonotypes have been an important focus of studies of the T cell receptor repertoire, and in these studies, public clonotypes are usually defined as sequences with identical amino acid CDR3 regions that are shared by different individuals (Price et al. 2009; Quigley et al. 2010). Here, we define B cell public clonotypes as V(D)J rearrangements that are shared by different individuals, which utilize identical germline V genes and J genes, and which share CDR3 amino acid sequences.
Studies of public clonotypes mostly focus on either heavy chains, or light chains, though a true public clonotype must share both chains. However, the light chain repertoire is so lacking in diversity that a heavy chain that is shared in small samplings of the repertoires of different individuals will almost certainly be a true IGH/IGL public clonotype. It will likely be found as such a clonotype when the heavy chain is associated with the more commonly expressed light chain rearrangements.
A handful of VJ combinations dominate the light chain repertoires of humans and mice, for light chains lack both combinatorial diversity and junctional diversity. The mouse kappa chain repertoire is hardly expanded at all by N additions, and expression of the polymerase Pol mu may further restrict kappa diversity by limiting exonuclease removals of the ends of the recombining genes (Bertocci et al. 2003; Bertocci et al. 2006). It has been calculated that there is only a tenfold increase in the diversity of the murine kappa chain repertoire as a result of gene processing and N addition (Aoki-Ota et al. 2012).
The human light chain repertoire has little more diversity than the mouse repertoire. We have calculated that the entire human kappa repertoire may include fewer than 104 unique amino acid sequences (Jackson et al. 2012). Within this very small repertoire, some public kappa chain clonotypes are so commonly expressed that they were repeatedly identified, by independent laboratories, even in the era of Sanger sequencing (Collins et al. 2008). HTS has shown that the lambda repertoire is similarly rich in public clonotypes (Hoi and Ippolito 2013).
Paradoxically, it is in the small antibody repertoire of the mouse, rather than in the larger repertoire of the human, that heavy chain public clonotypes are conspicuous. Analysis of hundreds of thousands of human VDJ rearrangements from four individuals found that just 1 in 4000 sequences shared CDR3s (Vollmers et al. 2013). By contrast, in a HTS study involving relatively small samplings of the repertoires of C57BL/6 and BALB/c mice, we observed that 4–6% of sequences included CDR3 sequences that were shared by two or more mice from the same strain (Collins et al. 2015). A HTS study with a much larger sampling of the expressed repertoire reported that 14% of CDR3 sequences were shared by individuals from the same strain (Greiff et al. 2017). In a re-analysis of data from our 2015 study, 2% of sequences were public clonotypes, with shared IGHV and IGHJ genes, as well as shared CDR3 regions (unpublished).
The abundance of public clonotypes in the mouse helps explain early observations of the mouse antibody response to simple chemically defined haptens. The anti-hapten responses of different mice often include the expression of shared antibodies encoded by particular V(D)J rearrangements (Cumano and Rajewsky 1985; Siekevitz et al. 1983). Some of the antibodies that were identified in these early studies are known to carry essential specificities. For example, the public T15 idiotype is protective against pathogenic streptococcal infections (Vale et al. 2013). The presence of such critical specificities in the mouse repertoire can be almost guaranteed by the biases that shape the repertoire, and the immune system has evolved to ensure that these specificities are even available to the newborn mouse (Briles et al. 1982; Feeney 1991).
The presence of public clonotypes in the mouse and human can be explained by the same biases that shape their overall repertoires. Since only a handful of heavy chain public clonotypes have been identified in the human, there is no data reporting N addition in these sequences. On the other hand, public kappa chain clonotypes are common in the human. They have an average of 0.4 N nucleotides in the VJ junction, while private clonotypes have an average of 2.5 N additions (Jackson et al. 2012).
There is less N addition in the mouse than in the human, and many of the murine sequences with the least amount of N addition seem to be public clonotypes. In a re-analysis of data from our 2015 study (Collins et al. 2015), public clonotypes had an average of 2.0 and 2.1 nucleotides in the VD and DJ junctions, respectively, while private clonotypes had 4.0 and 2.9 additions (unpublished).
The challenge of the neonatal repertoire
The neonatal B cell compartment is very small, and the formation of a suitable neonatal repertoire is therefore a special challenge. This is particularly true for the mouse, as a newborn mouse can weigh just 0.5 g. The neonatal mouse appears to meet this challenge by its reliance on a phenotypically distinct population of cells called B1 B cells (Baumgarth 2011; Baumgarth 2013), and it may be that this cell population has evolved to meet the challenges posed by very small repertoires. Controversy surrounds this cell population in the human, and the unequivocal identification of a counterpart to the murine population has proven difficult (Covens et al. 2013).
Antibodies that are generated in the neonatal mouse are overwhelmingly encoded by germline gene-derived nucleotides. Eighty-four percent of productive rearrangements lack N nucleotides as a result of a lack of TdT expression during the fetal and neonatal periods (Richl et al. 2008). This lack of N nucleotides is also an outcome of the joining of V(D)J genes at regions of short sequence homology. Eighty percent of the VD and DJ junctions of neonatal mouse heavy chains are the result of such joining (Feeney 1992a; Feeney 1992b), and this also helps ensure that recombination favors joining of the IGHD gene in the preferred reading frame.
In the mouse, VDJ joining at short homologous sequences is promoted by both germline gene ends and by germline-derived palindromic sequences. For example, all sequences of the dominant murine IGHV1 gene family end with the 3′ motif CAAGA, and these and other IGHV sequences are therefore capable of forming the P nucleotide motifs T, TC, and TCT. This promotes joining without N addition to the T-rich 5′ end of the highly expressed IGHD1-1*01 gene, as well as to the TCTAC motif that is present at the 5′ end of most members of the dominant IGHD2 gene family. Joining at short homologous sequences is equally common at the DJ junction. Most murine IGHD genes end with the 3′ motif CTAC, which is also found at the 5′ end of the IGHJ1 and IGHJ2 genes.
In the human, neonatal sequences are also more germline-oriented than sequences that are generated later in life. Human sequences can join at regions of short sequence homology, but human gene ends share fewer motifs than gene ends of the mouse, and this joining process is seen less often in the human (Bauer et al. 2007). N nucleotide addition is also limited in human fetal and newborn sequences (Feeney 1990; Feeney 1992a; Zemlin et al. 2001).
Positional biases in gene usage are evident in the neonatal repertoire of both species. Early studies of heavy chain gene rearrangements during fetal development found a bias toward the use of 3′ murine IGHV genes (Perlmutter et al. 1985; Yancopoulos et al. 1984), and the most 3′ human IGHV gene is similarly prominent in the fetal repertoire (Rogosch et al. 2012; Schroeder and Wang 1990). The most 3′ IGHD gene is also conspicuous in the fetal repertoire of both species (Rogosch et al. 2012; Schelonka et al. 2010; Shiokawa et al. 1999; Zemlin et al. 2001), and interestingly, this IGHD gene sequence is identical in both species (Corbett et al. 1997).
Repertoires in action: how the contrasting repertoires of humans and mice serve their varying biological needs
The human and the murine B cell repertoires appear to be shaped by the same biased processes, but differences in the repertoires emerge from the differing strengths of the biases in the two species. In comparison to the human repertoire, the mouse repertoire is skewed toward B cells that express receptors with more germline-encoded specificities. This is the result of the lack of N addition, as well as a greater reliance on P nucleotide addition in the generation of the murine repertoire. This limits the diversity of the CDR3 region, which is further limited by the small number of highly similar IGHD genes that are available to the mouse. The germline-focused repertoire that results is one that can deliver critical specificities, even in the tiny repertoire of the neonate. As an individual mouse matures and ages, these critical specificities will be maintained. This is partly the result of the self-renewing nature of the B-1 B cell population (Baumgarth 2013), but it is also the result of the fact that throughout life, these germline-encoded antibodies will be generated in the bone marrow at relatively high frequencies.
The relative proportions of public and private clonotypes are different in mice and humans (Fig. 3). Although the proportion of cells associated with private clonotypes remains to be determined for each species, it is clear that an ever-changing population of cells produces private clonotypes that dominate the human repertoire. Biases in the recombination process should still ensure that the human repertoire includes a core of critical public clonotypes. This core will be maintained throughout life, just as public clonotypes are maintained in the mouse.
Proposed models of the human and mouse naïve antibody repertoires, contrasting their relative proportions of public (black) and private (grey) clonotypes. The overall sizes of the repertoires shown are not to scale. The mouse repertoire has a conspicuous core of public clonotypes, but the size of the human core remains uncertain. Only the human expresses a large repertoire of private clonotypes
The repertoire of the mouse includes a conspicuous population of public clonotypes. The diversity of the repertoire of public clonotypes might even be comparable to that of the human, though suitable datasets are not yet available to allow proper comparisons. There is certainly far less private clonotype diversity in the murine repertoire than in the human repertoire. The likelihood of any particular private clonotype being generated is by definition very low, and so it is likely that in any individual mouse, private clonotypes will be carried by just a single cell. Public clonotypes, on the other hand, are not only shared by different mice, but because of the likelihood of their generation, they will often be present at high copy number in an individual mouse. As has been noted by others (Hershberg and Laning Prak 2015), this has implications for the study of B cell clonality, for standard analyses of clone trees assume that any B cells that share germline antibody gene rearrangements must be related by descent from a common ancestor (Steiman-Shimony et al. 2006; Tabibian-Keissar et al. 2008).
The presence in the naïve repertoire of multiple cells that have independently rearranged genes to express the same public clonotype has implications for the kinetics of the immune response. After invasion by a pathogen, the time to detection and clonal selection of specific B cells should be determined by the size of the population of pathogen-specific cells in the naïve repertoire. If the germline genes of the mouse have evolved to target important pathogens with public clonotypes that are present at high copy number, then invasion by these pathogens should result in a rapid response. This would be an inevitable consequence of the elevated frequency of pathogen-specific B cells in the naïve repertoire. T cell studies suggest that the resulting clonal expansion is also affected by the frequency of precursor cells that become activated (La Gruta et al. 2010; Quiel et al. 2011; De Boer and Perelson 2013).
The response to a newly emergent pathogen could be different, as suitable anti-pathogen antibodies are unlikely to be hardwired in the germline genes of the mouse. It is likely that such pathogens would trigger a slower response as a result of a greater dependence upon private rather than public clonotypes.
In both humans and mice, a response that begins with public clonotypes could be diversified over time by the recruitment of private clonotypes. It is possible that this accounts in part for observations of a shift in the repertoire of responding B cells over time (Brown et al. 2000; Shannon and Mehr 1999; Wu et al. 2010). It is also possible that in the response to a persistent infection, private clonotypes carrying advantageous specificities may not be available at the time of pathogen invasion, but may be generated over time (Zarnitsyna et al. 2013). This dynamic aspect of the private repertoire could be particularly advantageous to a species like the human, which is relatively resilient by virtue of its size and which requires prolonged survival to reproduce. Mice, on the other hand, have a high metabolic rate and lack metabolic reserves. They are vulnerable to both starvation and dehydration (Schmidt-Nielsen 1984). Because a mouse can so quickly sicken and die, the need for the rapid detection and elimination of microbial invaders may have driven the evolution of the murine repertoire.
A mouse is unlikely to benefit from a dynamic private repertoire, because there are relatively few useful specificities available in the private repertoire at any moment in time. The mouse response, in keeping with the biology of the mouse, seems to be one that is devoted to speed, and a speedy response is provided by the public repertoire. Speed may also be the factor that explains the existence of such large sets of IGHV and IGKV genes in the mouse. These genes encode the CDR1 and CDR2 regions of the heavy and light chains. The abundance of IGHV and IGKV genes may allow the mouse to quickly respond with relatively high affinity to many “expected” pathogens, without the need for suitable diversity to emerge through the process of somatic point mutation of the CDR1 and CDR2 sequences.
Somatic point mutations accumulate in murine B cells after clonal selection, but the extent of diversification is not as impressive as that which is seen in the human. In the human, clonal selection is followed by massive diversification of the expanding clones. This diversification by mutation is linked to isotype switching that in turn is linked to cell division (Tangye et al. 2002; Tangye and Hodgkin 2004). As a consequence, the human IgG response involves cells that nearly all carry highly mutated antibody genes. The mean number of mutations of human IgG-committed B cells ranges from 16.5 to 21.9, according to the IgG subclass, and very few IgG-committed B cells express unmutated antibody genes (Jackson et al. 2014). Unmutated genes are seen in just 6% of IgG3-committed cells, and fewer than 1% of cells that have switched to other IgG subclasses (Jackson et al. 2014). In contrast, few murine B cells carry more than seven or eight point mutations, and unmutated antibody genes are seen in between 27% (IgG1) and 44% (IgG2c) of sequences in IgG-committed B cells of the C57BL/6 strain (Collins et al. 2015). This disconnection between class switching and somatic point mutation suggests that the mouse may have evolved to recruit IgG-mediated effector functions as quickly as possible (Collins 2016).
The extent to which the need for a speedy response to pathogens has guided the evolution of the mouse antibody repertoire, and the mouse immune system more generally, is something that will remain uncertain until our knowledge of the immune systems of other species can inform our understanding. It is our hope that this review will stimulate repertoire studies in other species, and that eventually this should allow the relationships between species size and many aspects of the immune system to be better understood.
References
Alanio C, Lemaitre F, Law HK, Hasan M, Albert ML (2010) Enumeration of human antigen-specific naive CD8+ T cells reveals conserved precursor frequencies. Blood 115(18):3718–3725. https://doi.org/10.1182/blood-2009-10-251124
Aoki-Ota M, Torkamani A, Ota T, Schork N, Nemazee D (2012) Skewed primary Igkappa repertoire and V-J joining in C57BL/6 mice: implications for recombination accessibility and receptor editing. J Immunol 188(5):2305–2315. https://doi.org/10.4049/jimmunol.1103484
Avnir Y, Watson CT, Glanville J, Peterson EC, Tallarico AS, Bennett AS, Qin K, Fu Y, Huang CY, Beigel JH, Breden F, Zhu Q, Marasco WA (2016) IGHV1-69 polymorphism modulates anti-influenza antibody repertoires, correlates with IGHV utilization shifts and varies by ethnicity. Sci Rep 6(1):20842. https://doi.org/10.1038/srep20842
Bassing CH, Swat W, Alt FW (2002) The mechanism and regulation of chromosomal V(D)J recombination. Cell 109(Suppl):S45–S55. https://doi.org/10.1016/S0092-8674(02)00675-X
Basu M, Hegde MV et al (1983) Synthesis of compositionally unique DNA by terminal deoxynucleotidyl transferase. Biochem Biophys Res Commun 111:1105–1112
Bauer K, Hummel M, Berek C, Paar C, Rosenberger C, Kerzel S, Versmold H, Zemlin M (2007) Homology-directed recombination in IgH variable region genes from human neonates, infants and adults: implications for junctional diversity. Mol Immunol 44(11):2969–2977. https://doi.org/10.1016/j.molimm.2007.01.003
Baumgarth N (2011) The double life of a B-1 cell: self-reactivity selects for protective effector functions. Nat Rev Immunol 11(1):34–46. https://doi.org/10.1038/nri2901
Baumgarth N (2013) Innate-like B cells and their rules of engagement. Adv Exp Med Biol 785:57–66. https://doi.org/10.1007/978-1-4614-6217-0_7
Benedict CL, Gilfillan S et al (2000) Terminal deoxynucleotidyl transferase and repertoire development. Immunol Rev 175:150–157
Benichou J, Glanville J, Prak ETL, Azran R, Kuo TC, Pons J, Desmarais C, Tsaban L, Louzoun Y (2013) The restricted DH gene reading frame usage in the expressed human antibody repertoire is selected based upon its amino acid content. J Immunol 190(11):5567–5577. https://doi.org/10.4049/jimmunol.1201929
Bentolila LA, Olson S, Marshall A, Rougeon F, Paige CJ, Doyen N, Wu GE (1999) Extensive junctional diversity in Ig light chain genes from early B cell progenitors of mu MT mice. J Immunol 162(4):2123–2128
Bertocci B, De Smet A, Berek C, Weill JC, Reynaud CA (2003) Immunoglobulin kappa light chain gene rearrangement is impaired in mice deficient for DNA polymerase mu. Immunity 19(2):203–211. https://doi.org/10.1016/S1074-7613(03)00203-6
Bertocci B, De Smet A, Weill JC, Reynaud CA (2006) Nonoverlapping functions of DNA polymerases mu, lambda, and terminal deoxynucleotidyltransferase during immunoglobulin V(D)J recombination in vivo. Immunity 25(1):31–41. https://doi.org/10.1016/j.immuni.2006.04.013
Bianconi E, Piovesan A, Facchin F, Beraudi A, Casadei R, Frabetti F, Vitale L, Pelleri MC, Tassani S, Piva F, Perez-Amodio S, Strippoli P, Canaider S (2013) An estimation of the number of cells in the human body. Ann Hum Biol 40(6):463–471. https://doi.org/10.3109/03014460.2013.807878
Blattman JN, Antia R, Sourdive DJ, Wang X, Kaech SM, Murali-Krishna K, Altman JD, Ahmed R (2002) Estimating the precursor frequency of naive antigen-specific CD8 T cells. J Exp Med 195(5):657–664. https://doi.org/10.1084/jem.20001021
Boyd SD, Marshall EL et al (2009) Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Sci Transl Med 1:12ra23
Boyd SD, Gaeta BA, Jackson KJ, Fire AZ, Marshall EL, Merker JD, Maniar JM, Zhang LN, Sahaf B, Jones CD, Simen BB, Hanczaruk B, Nguyen KD, Nadeau KC, Egholm M, Miklos DB, Zehnder JL, Collins AM (2010) Individual variation in the germline Ig gene repertoire inferred from variable region gene rearrangements. J Immunol 184(12):6986–6992. https://doi.org/10.4049/jimmunol.1000445
Brekke KM, Garrard WT (2004) Assembly and analysis of the mouse immunoglobulin kappa gene sequence. Immunogenetics 56(7):490–505. https://doi.org/10.1007/s00251-004-0659-0
Brezinschek HP, Foster SJ et al (1998) Pairing of variable heavy and variable kappa chains in individual naive and memory B cells. J Immunol 160:4762–4767
Briles DE, Forman C, Hudak S, Claflin JL (1982) Anti-phosphorylcholine antibodies of the T15 idiotype are optimally protective against Streptococcus pneumoniae. J Exp Med 156(4):1177–1185. https://doi.org/10.1084/jem.156.4.1177
Briney BS, Willis JR, McKinney BA, Crowe JE (2012) High-throughput antibody sequencing reveals genetic evidence of global regulation of the naive and memory repertoires that extends across individuals. Genes Immun 13(6):469–473. https://doi.org/10.1038/gene.2012.20
Brown M, Schumacher MA et al (2000) The structural basis of repertoire shift in an immune response to phosphocholine. J Exp Med 191(12):2101–2112. https://doi.org/10.1084/jem.191.12.2101
Busse CE, Czogiel I et al (2014) Single-cell based high-throughput sequencing of full-length immunoglobulin heavy and light chain genes. European J Immunol 44:597–603
Chang HH, Lieber MR (2016) Structure-specific nuclease activities of Artemis and the Artemis: DNA-PKcs complex. Nucleic Acids Res 44(11):4991–4997. https://doi.org/10.1093/nar/gkw456
Chimge NO, Pramanik S, Hu G, Lin Y, Gao R, Shen L, Li H (2005) Determination of gene organization in the human IGHV region on single chromosomes. Genes Immun 6(3):186–193. https://doi.org/10.1038/sj.gene.6364176
Choi NM, Loguercio S, Verma-Gaur J, Degner SC, Torkamani A, Su AI, Oltz EM, Artyomov M, Feeney AJ (2013) Deep sequencing of the murine IgH repertoire reveals complex regulation of nonrandom V gene rearrangement frequencies. J Immunol 191(5):2393–2402. https://doi.org/10.4049/jimmunol.1301279
Collins AM (2016) IgG subclass co-expression brings harmony to the quartet model of murine IgG function. Immunol Cell Biol 94(10):949–954. https://doi.org/10.1038/icb.2016.65
Collins AM, Wang Y, Singh V, Yu P, Jackson KJ, Sewell WA (2008) The reported germline repertoire of human immunoglobulin kappa chain genes is relatively complete and accurate. Immunogenetics 60(11):669–676. https://doi.org/10.1007/s00251-008-0325-z
Collins AM, Wang Y, Roskin KM, Marquis CP, Jackson KJL (2015) The mouse antibody heavy chain repertoire is germline-focused and highly variable between inbred strains. Philos Trans R Soc Lond Ser B Biol Sci 370(1676):20140236. https://doi.org/10.1098/rstb.2014.0236
Corbett SJ, Tomlinson IM, Sonnhammer ELL, Buck D, Winter G (1997) Sequence of the human immunoglobulin diversity (D) segment locus: a systematic analysis provides no evidence for the use of DIR segments, inverted D segments, “minor” D segments or D-D recombination. J Mol Biol 270(4):587–597. https://doi.org/10.1006/jmbi.1997.1141
Corcoran MM, Phad GE, Bernat NV, Stahl-Hennig C, Sumida N, Persson MAA, Martin M, Hedestam GBK (2016) Production of individualized V gene databases reveals high levels of immunoglobulin genetic diversity. Nat Commun 7:13642. https://doi.org/10.1038/ncomms13642
Covens K, Verbinnen B, Geukens N, Meyts I, Schuit F, van Lommel L, Jacquemin M, Bossuyt X (2013) Characterization of proposed human B-1 cells reveals pre-plasmablast phenotype. Blood 121(26):5176–5183. https://doi.org/10.1182/blood-2012-12-471953
Cowell LG, Davila M, Ramsden D, Kelsoe G (2004) Computational tools for understanding sequence variability in recombination signals. Immunol Rev 200(1):57–69. https://doi.org/10.1111/j.0105-2896.2004.00171.x
Cumano A, Rajewsky K (1985) Structure of primary anti-(4-hydroxy-3-nitrophenyl)acetyl (NP) antibodies in normal and idiotypically suppressed C57BL/6 mice. European J Immunol 15:512–520
De Boer RJ, Perelson AS (2013) Antigen-stimulated CD4 T cell expansion can be limited by their grazing of peptide-MHC complexes. J Immunol 190:5454–5458
De Preval C, Fougereau M (1976) Specific interaction between VH and VL regions of human monoclonal immunoglobulins. J Mol Biol 102(3):657–678. https://doi.org/10.1016/0022-2836(76)90340-5
de Wildt RM, Hoet RM et al (1999) Analysis of heavy and light chain pairings indicates that receptor editing shapes the human antibody repertoire. J Mol Biol 285(3):895–901. https://doi.org/10.1006/jmbi.1998.2396
Decker DJ, Boyle NE, Koziol JA, Klinman NR (1991) The expression of the Ig H chain repertoire in developing bone marrow B lineage cells. J Immunol 146(1):350–361
DeKosky BJ, Lungu OI, Park D, Johnson EL, Charab W, Chrysostomou C, Kuroda D, Ellington AD, Ippolito GC, Gray JJ, Georgiou G (2016) Large-scale sequence and structural comparisons of human naive and antigen-experienced antibody repertoires. Proc Natl Acad Sci U S A 113(19):E2636–E2645. https://doi.org/10.1073/pnas.1525510113
Elhanati Y, Sethna Z, Marcou Q, Callan CG Jr, Mora T, Walczak AM (2015) Inferring processes underlying B-cell repertoire diversity. Philos Trans R Soc Lond Ser B Biol Sci 370(1676):20140243. https://doi.org/10.1098/rstb.2014.0243
Ezekiel UR, Sun T, Bozek G, Storb U (1997) The composition of coding joints formed in V(D)J recombination is strongly affected by the nucleotide sequence of the coding ends and their relationship to the recombination signal sequences. Mol Cell Biol 17(7):4191–4197. https://doi.org/10.1128/MCB.17.7.4191
Feeney AJ (1990) Lack of N regions in fetal and neonatal mouse immunoglobulin V-D-J junctional sequences. J Exp Med 172(5):1377–1390. https://doi.org/10.1084/jem.172.5.1377
Feeney AJ (1991) Predominance of the prototypic T15 anti-phosphorylcholine junctional sequence in neonatal pre-B cells. J Immunol 147(12):4343–4350
Feeney AJ (1992a) Comparison of junctional diversity in the neonatal and adult immunoglobulin repertoires. Int Rev Immunol 8(2-3):113–122. https://doi.org/10.3109/08830189209055567
Feeney AJ (1992b) Predominance of VH-D-JH junctions occurring at sites of short sequence homology results in limited junctional diversity in neonatal antibodies. J Immunol 149(1):222–229
Feeney AJ, Tang A et al (2000) B-cell repertoire formation: role of the recombination signal sequence in non-random V segment utilization. Immunol Rev 175:59–69
Flajnik MF, Hsu E, Kaufman JF, Du Pasquier L (1987) Changes in the immune system during metamorphosis of Xenopus. Immunol Today 8(2):58–64. https://doi.org/10.1016/0167-5699(87)90240-4
Foster SJ, Brezinschek HP et al (1997) Molecular mechanisms and selective influences that shape the kappa gene repertoire of IgM+ B cells. J Clin Invest 99:1614–1627
Galilei G. Discorsi e Dimostrazioni Matematiche intorno à due nuoue scienze Attenenti alla Mecanica & i Movimenti Locali, Lodewijk Elzevir Leiden, 1638
Gauss GH, Lieber MR (1996) Mechanistic constraints on diversity in human V(D)J recombination. Mol Cell Biol 16(1):258–269. https://doi.org/10.1128/MCB.16.1.258
Geraldes A, Basset P et al (2011) Higher differentiation among subspecies of the house mouse (Mus musculus) in genomic regions with low recombination. Mol Ecol 20(22):4722–4736. https://doi.org/10.1111/j.1365-294X.2011.05285.x
Ghia P, ten Boekel E, Sanz E, de la Hera A, Rolink A, Melchers F (1996) Ordering of human bone marrow B lymphocyte precursors by single-cell polymerase chain reaction analyses of the rearrangement status of the immunoglobulin H and L chain gene loci. J Exp Med 184(6):2217–2229. https://doi.org/10.1084/jem.184.6.2217
Glanville J, Kuo TC, von Budingen HC, Guey L, Berka J, Sundar PD, Huerta G, Mehta GR, Oksenberg JR, Hauser SL, Cox DR, Rajpal A, Pons J (2011) Naive antibody gene-segment frequencies are heritable and unaltered by chronic lymphocyte ablation. Proc Nat Acad Sci USA 108(50):20066–20071. https://doi.org/10.1073/pnas.1107498108
Greiff V, Menzel U, Miho E, Weber C, Riedel R, Cook S, Valai A, Lopes T, Radbruch A, Winkler TH, Reddy ST (2017) Systems analysis reveals high genetic and antigen-driven predetermination of antibody repertoires throughout B cell development. Cell Rep 19(7):1467–1478. https://doi.org/10.1016/j.celrep.2017.04.054
Gu H, Tarlinton D, Müller W, Rajewsky K, Förster I (1991) Most peripheral B cells in mice are ligand selected. J Exp Med 173(6):1357–1371. https://doi.org/10.1084/jem.173.6.1357
Guo Y, Bao Y, Wang H, Hu X, Zhao Z, Li N, Zhao Y (2011) A preliminary analysis of the immunoglobulin genes in the African elephant (Loxodonta africana). PLoS One 6(2):e16889. https://doi.org/10.1371/journal.pone.0016889
Haldane JBS (1926) On being the right size. In, Harper’s Magazine
Haluszczak C, Akue AD, Hamilton SE, Johnson LD, Pujanauski L, Teodorovic L, Jameson SC, Kedl RM (2009) The antigen-specific CD8+ T cell repertoire in unimmunized mice includes memory phenotype cells bearing markers of homeostatic expansion. J Exp Med 206(2):435–448. https://doi.org/10.1084/jem.20081829
Hershberg U, Luning Prak ET (2015) The analysis of clonal expansions in normal and autoimmune B cell repertoires. Philos Trans R Soc Lond Ser B Biol Sci 370(1676):20140239. https://doi.org/10.1098/rstb.2014.0239
Hess J, Werner A, Wirth T, Melchers F, Jack HM, Winkler TH (2001) Induction of pre-B cell proliferation after de novo synthesis of the pre-B cell receptor. Proc Natl Acad Sci U S A 98(4):1745–1750. https://doi.org/10.1073/pnas.98.4.1745
Hoffman MA (1983) Energy metabolism, brain size and longevity in mammals. Q Rev Biol 58(4):495–512. https://doi.org/10.1086/413544
Hofle M, Linthicum DS, Ioerger T (2000) Analysis of diversity of nucleotide and amino acid distributions in the VD and DJ joining regions in Ig heavy chains. Mol Immunol 37(14):827–835. https://doi.org/10.1016/S0161-5890(00)00110-3
Hogan T, Gossel G, Yates AJ, Seddon B (2015) Temporal fate mapping reveals age-linked heterogeneity in naive T lymphocytes in mice. Proc Natl Acad Sci U S A 112(50):E6917–E6926. https://doi.org/10.1073/pnas.1517246112
Hoi KH, Ippolito GC (2013) Intrinsic bias and public rearrangements in the human immunoglobulin Vlambda light chain repertoire. Genes Immun 14(4):271–276. https://doi.org/10.1038/gene.2013.10
Hsu E, Du Pasquier L (1984) Ontogeny of the immune system in Xenopus II. Antibody repertoire differences between larval and adults. Differentiation 28(2):116–122. https://doi.org/10.1111/j.1432-0436.1984.tb00274.x
Hsu E, Du Pasquier L (1992) Changes in the amphibian antibody repertoire are correlated with metamorphosis and not with age or size. Develop Immunol 2(1):1–6. https://doi.org/10.1155/1992/28568
Jackson KJ, Gaeta B et al (2004) Exonuclease activity and P nucleotide addition in the generation of the expressed immunoglobulin repertoire. BMC Immunol 5:19
Jackson KJ, Gaeta BA et al (2007) Identifying highly mutated IGHD genes in the junctions of rearranged human immunoglobulin heavy chain genes. J Immunol Methods 324(1-2):26–37. https://doi.org/10.1016/j.jim.2007.04.011
Jackson KJ, Kidd MJ et al (2013) The shape of the lymphocyte receptor repertoire: lessons from the B cell receptor. Front Immunol 4:263
Jackson KJ, Wang Y et al (2012) Divergent human populations show extensive shared IGK rearrangements in peripheral blood B cells. Immunogenetics 64(1):3–14. https://doi.org/10.1007/s00251-011-0559-z
Jackson KJ, Wang Y et al (2014) Human immunoglobulin classes and subclasses show variability in VDJ gene mutation levels. Immunol Cell Biol 92(8):729–733. https://doi.org/10.1038/icb.2014.44
Johnston CM, Wood AL, Bolland DJ, Corcoran AE (2006) Complete sequence assembly and characterization of the C57BL/6 mouse Ig heavy chain V region. J Immunol 176(7):4221–4234. https://doi.org/10.4049/jimmunol.176.7.4221
Kidd MJ, Chen Z, Wang Y, Jackson KJ, Zhang L, Boyd SD, Fire AZ, Tanaka MM, Gaeta BA, Collins AM (2012) The inference of phased haplotypes for the immunoglobulin H chain V region gene loci by analysis of VDJ gene rearrangements. J Immunol 188(3):1333–1340. https://doi.org/10.4049/jimmunol.1102097
Kidd MJ, Jackson KJ et al (2016) DJ pairing during VDJ recombination shows positional biases that vary among individuals with differing IGHD locus immunogenotypes. J Immunol 196(3):1158–1164. https://doi.org/10.4049/jimmunol.1501401
Kleiber M (1947) Body size and metabolic rate. Physiol Rev 27(4):511–541
La Gruta NL, Rothwell WT, Cukalac T, Swan NG, Valkenburg SA, Kedzierska K, Thomas PG, Doherty PC, Turner SJ (2010) Primary CTL response magnitude in mice is determined by the extent of naive T cell recruitment and subsequent clonal expansion. J Clin Invest 120(6):1885–1894. https://doi.org/10.1172/JCI41538
Langman RE, Cohn M (1987) The E-T (elephant-tadpole) paradox necessitates the concept of a unit of B-cell function: the protection. Mol Immunol 24(7):675–697. https://doi.org/10.1016/0161-5890(87)90050-2
Langman RE, Cohn M (1992) How might the K/lambda ratio expressed by antigen-unselected B cells be explained? Res Immunol 143(8):804–811; discussion 830-9. https://doi.org/10.1016/0923-2494(92)80095-3
Langman RE, Cohn M (1995) The proportion of B-cell subsets expressing kappa and lambda light chains changes following antigenic selection. Immunol Today 16(3):141–144. https://doi.org/10.1016/0167-5699(95)80131-6
Larimore K, McCormick MW, Robins HS, Greenberg PD (2012) Shaping of human germline IgH repertoires revealed by deep sequencing. J Immunol 189(6):3221–3230. https://doi.org/10.4049/jimmunol.1201303
Laserson U, Vigneault F, Gadala-Maria D, Yaari G, Uduman M, Vander Heiden JA, Kelton W, Taek Jung S, Liu Y, Laserson J, Chari R, Lee JH, Bachelet I, Hickey B, Lieberman-Aiden E, Hanczaruk B, Simen BB, Egholm M, Koller D, Georgiou G, Kleinstein SH, Church GM (2014) High-resolution antibody dynamics of vaccine-induced immune responses. Proc Natl Acad Sci U S A 111(13):4928–4933. https://doi.org/10.1073/pnas.1323862111
Lee CE, Gaeta B et al (2006) Reconsidering the human immunoglobulin heavy-chain locus: 1. An evaluation of the expressed human IGHD gene repertoire. Immunogenetics 57(12):917–925. https://doi.org/10.1007/s00251-005-0062-5
Lefranc M-P, Lefranc G (2004) Immunoglobulin lambda (IGL) genes of human and mouse. In: Honjo T, Alt FW, Neuberger MS (eds) Molecular biology of B cells. Academic Press, Elsevier Science, pp 37–59. https://doi.org/10.1016/B978-012053641-2/50005-8
Lefranc MP, Giudicelli V, Ginestoux C, Jabado-Michaloud J, Folch G, Bellahcene F, Wu Y, Gemrot E, Brochet X, Lane J, Regnier L, Ehrenmann F, Lefranc G, Duroux P (2009) IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res 37(Database):D1006–D1012. https://doi.org/10.1093/nar/gkn838
Li YS, Hayakawa K, Hardy RR (1993) The regulated expression of B lineage associated genes during B cell differentiation in bone marrow and fetal liver. J Exp Med 178(3):951–960. https://doi.org/10.1084/jem.178.3.951
Lieber MR (2008) The mechanism of human nonhomologous DNA end joining. J Biol Chem 283(1):1–5. https://doi.org/10.1074/jbc.R700039200
Liu L, Lucas AHIGH (2003) V3-23*01 and its allele V3-23*03 differ in their capacity to form the canonical human antibody combining site specific for the capsular polysaccharide of Haemophilus influenzae type b. Immunogenetics 55(5):336–338. https://doi.org/10.1007/s00251-003-0583-8
Lu H, Schwarz K, Lieber MR (2007) Extent to which hairpin opening by the Artemis:DNA-PKcs complex can contribute to junctional diversity in V(D)J recombination. Nucleic Acids Res 35(20):6917–6923. https://doi.org/10.1093/nar/gkm823
Lythe G, Callard RE, Hoare RL, Molina-Paris C (2016) How many TCR clonotypes does a body maintain? J Theor Biol 389:214–224
Ma Y, Pannicke U, Schwarz K, Lieber MR (2002) Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 108(6):781–794. https://doi.org/10.1016/S0092-8674(02)00671-2
Matsuda F, Ishii K, Bourvagnet P, Kuma K, Hayashida H, Miyata T, Honjo T (1998) The complete nucleotide sequence of the human immunoglobulin heavy chain variable region locus. J Exp Med 188(11):2151–2162. https://doi.org/10.1084/jem.188.11.2151
Meng W, Yunk L, Wang LS, Maganty A, Xue E, Cohen PL, Eisenberg RA, Weigert MG, Mancini SJC, Luning Prak ET (2011) Selection of individual VH genes occurs at the pro-B to pre-B cell transition. J Immunol 187(4):1835–1844. https://doi.org/10.4049/jimmunol.1100207
Murugan A, Mora T, Walczak AM, Callan CG (2012) Statistical inference of the generation probability of T-cell receptors from sequence repertoires. Proc Natl Acad Sci U S A 109(40):16161–16166. https://doi.org/10.1073/pnas.1212755109
Nadel B, Feeney AJ (1995) Influence of coding-end sequence on coding-end processing in V(D)J recombination. J Immunol 155(9):4322–4329
Nadel B, Feeney AJ (1997) Nucleotide deletion and P addition in V(D)J recombination: a determinant role of the coding-end sequence. Mol Cell Biol 17(7):3768–3778. https://doi.org/10.1128/MCB.17.7.3768
Nossal GJ (2003) The double helix and immunology. Nature 421(6921):440–444. https://doi.org/10.1038/nature01409
Ohm-Laursen L, Nielsen M et al (2006) No evidence for the use of DIR, D-D fusions, chromosome 15 open reading frames or VH replacement in the peripheral repertoire was found on application of an improved algorithm, JointML, to 6329 human immunoglobulin H rearrangements. Immunol 119:265–277
Perlmutter RM, Kearney JF, Chang SP et al (1985) Developmentally controlled expression of immunoglobulin VH genes. Science 227(4694):1597–1601
Prak EL, Weigert M (1995) Light chain replacement: a new model for antibody gene rearrangement. J Exp Med 182(2):541–548. https://doi.org/10.1084/jem.182.2.541
Pramanik S, Li H (2002) Direct detection of insertion/deletion polymorphisms in an autosomal region by analyzing high-density markers in individual spermatozoa. Am J HumG E N 71:1342–1352
Price DA, Asher TE, Wilson NA, Nason MC, Brenchley JM, Metzler IS, Venturi V, Gostick E, Chattopadhyay PK, Roederer M, Davenport MP, Watkins DI, Douek DC (2009) Public clonotype usage identifies protective Gag-specific CD8+ T cell responses in SIV infection. J Exp Med 206(4):923–936. https://doi.org/10.1084/jem.20081127
Quiel J, Caucheteux S, Laurence A, Singh NJ, Bocharov G, Ben-Sasson SZ, Grossman Z, Paul WE (2011) Antigen-stimulated CD4 T-cell expansion is inversely and log-linearly related to precursor number. Proc Natl Acad Sci U S A 108(8):3312–3317. https://doi.org/10.1073/pnas.1018525108
Quigley MF, Greenaway HY, Venturi V, Lindsay R, Quinn KM, Seder RA, Douek DC, Davenport MP, Price DA (2010) Convergent recombination shapes the clonotypic landscape of the naive T-cell repertoire. Proc Natl Acad Sci U S A 107(45):19414–19419. https://doi.org/10.1073/pnas.1010586107
Retter I, Althaus HH, Münch R, Müller W (2005) VBASE2, an integrative V gene database. Nucleic Acids Res 33(Database issue):D671–D674. https://doi.org/10.1093/nar/gki088
Retter MW, Nemazee D (1998) Receptor editing occurs frequently during normal B cell development. J Exp Med 188(7):1231–1238. https://doi.org/10.1084/jem.188.7.1231
Riblet R (2003) Immunoglobulin heavy chain genes in the mouse. In: Honjo T, Alt FW, Neuberger M (eds) Molecular biology of B cells. Elsevier Academic Press, London
Richl P, Stern U, Lipsky PE, Girschick HJ (2008) The lambda gene immunoglobulin repertoire of human neonatal B cells. Mol Immunol 45(2):320–327. https://doi.org/10.1016/j.molimm.2007.06.155
Rogosch T, Kerzel S, Hoss K, Hoersch G, Zemlin C, Heckmann M, Berek C, Schroeder HW, Maier RF, Zemlin M (2012) IgA response in preterm neonates shows little evidence of antigen-driven selection. J Immunol 189(11):5449–5456. https://doi.org/10.4049/jimmunol.1103347
Rubelt F, Bolen CR, McGuire HM, Heiden JAV, Gadala-Maria D, Levin M, M. Euskirchen G, Mamedov MR, Swan GE, Dekker CL, Cowell LG, Kleinstein SH, Davis MM (2016) Individual heritable differences result in unique cell lymphocyte receptor repertoires of naive and antigen-experienced cells. Nat Commun 7:11112. https://doi.org/10.1038/ncomms11112
Saada R, Weinberger M, Shahaf G, Mehr R (2007) Models for antigen receptor gene rearrangement: CDR3 length. Immunol Cell Biol 85(4):323–332. https://doi.org/10.1038/sj.icb.7100055
Sasso EH, Johnson T, Kipps TJ (1996) Expression of the immunoglobulin VH gene 51p1 is proportional to its germline gene copy number. J Clin Invest 97(9):2074–2080. https://doi.org/10.1172/JCI118644
Schatz DG, Swanson PC (2011) V(D)J recombination: mechanisms of initiation. Ann Rev Gen 45:167–202
Scheepers C, Shrestha RK, Lambson BE, Jackson KJL, Wright IA, Naicker D, Goosen M, Berrie L, Ismail A, Garrett N, Abdool Karim Q, Abdool Karim SS, Moore PL, Travers SA, Morris L (2015) Ability to develop broadly neutralizing HIV-1 antibodies is not restricted by the germline Ig gene repertoire. J Immunol 194(9):4371–4378. https://doi.org/10.4049/jimmunol.1500118
Schelonka RL, Szymanska E, Vale AM, Zhuang Y, Gartland GL, Schroeder HW (2010) DH and JH usage in murine fetal liver mirrors that of human fetal liver. Immunogenetics 62(10):653–666. https://doi.org/10.1007/s00251-010-0469-5
Schelonka RL, Zemlin M, Kobayashi R, Ippolito GC, Zhuang Y, Gartland GL, Szalai A, Fujihashi K, Rajewsky K, Schroeder HW (2008) Preferential use of DH reading frame 2 alters B cell development and antigen-specific antibody production. J Immunol 181(12):8409–8415. https://doi.org/10.4049/jimmunol.181.12.8409
Schmidt-Nielsen K (1984) Scaling. Why is animal size so important? Cambridge University Press, New York. https://doi.org/10.1017/CBO9781139167826
Schroeder HW Jr, Cavacini L (2010) Structure and function of immunoglobulins. J All Clin Immunol 125:S41–S52
Schroeder HW Jr, Wang JY (1990) Preferential utilization of conserved immunoglobulin heavy chain variable gene segments during human fetal life. Proc Natl Acad Sci U S A 87(16):6146–6150. https://doi.org/10.1073/pnas.87.16.6146
Shannon M, Mehr R (1999) Reconciling repertoire shift with affinity maturation: the role of deleterious mutations. J Immunol 162(7):3950–3956
Shiokawa S, Mortari F, Lima JO, Nuñez C, Bertrand FE 3rd, Kirkham PM, Zhu S, Dasanayake AP, Schroeder HW Jr (1999) IgM heavy chain complementarity-determining region 3 diversity is constrained by genetic and somatic mechanisms until two months after birth. J Immunol 162(10):6060–6070
Siekevitz M, Huang SY, Gefter ML (1983) The genetic basis of antibody production: a single heavy chain variable region gene encodes all molecules bearing the dominant anti-arsonate idiotype in the strain A mouse. Eur J Immunol 13(2):123–132. https://doi.org/10.1002/eji.1830130207
Snell O, Die Abhängigkeit d, Hirngewichtes v (1892) dem Körpergewicht und den geistigen Fähigkeiten. Archiv für Psychiatrie und Nervenkrankheiten 23(2):436–446. https://doi.org/10.1007/BF01843462
Souto-Carneiro MM, Sims GP, Girschik H, Lee J, Lipsky PE (2005) Developmental changes in the human heavy chain CDR3. J Immunol 175(11):7425–7436. https://doi.org/10.4049/jimmunol.175.11.7425
Spear PG, Wang AL, Rutishauser U, Edelman GM (1973) Characterization of splenic lymphoid cells in fetal and newborn mice. J Exp Med 138(3):557–573. https://doi.org/10.1084/jem.138.3.557
Steiman-Shimony A, Edelman H, Hutzler A, Barak M, Zuckerman NS, Shahaf G, Dunn-Walters D, Stott DI, Abraham RS, Mehr R (2006) Lineage tree analysis of immunoglobulin variable-region gene mutations in autoimmune diseases: chronic activation, normal selection. Cell Immunol 244(2):130–136. https://doi.org/10.1016/j.cellimm.2007.01.009
Tabibian-Keissar H, Zuckerman NS, Barak M, Dunn-Walters DK, Steiman-Shimony A, Chowers Y, Ofek E, Rosenblatt K, Schiby G, Mehr R, Barshack I (2008) B-cell clonal diversification and gut-lymph node trafficking in ulcerative colitis revealed using lineage tree analysis. Eur J Immunol 38(9):2600–2609. https://doi.org/10.1002/eji.200838333
Tangye SG, Ferguson A, Avery DT, Ma CS, Hodgkin PD (2002) Isotype switching by human B cells is division-associated and regulated by cytokines. J Immunol 169(8):4298–4306. https://doi.org/10.4049/jimmunol.169.8.4298
Tangye SG, Hodgkin PD (2004) Divide and conquer: the importance of cell division in regulating B-cell responses. Immunology 112(4):509–520. https://doi.org/10.1111/j.1365-2567.2004.01950.x
Thiebe R, Schable KF et al (1999) The variable genes and gene families of the mouse immunoglobulin kappa locus. Eur J Immunol 29(7):2072–2081. https://doi.org/10.1002/(SICI)1521-4141(199907)29:07<2072::AID-IMMU2072>3.0.CO;2-E
Tiller T, Busse CE, Wardemann H (2009) Cloning and expression of murine Ig genes from single B cells. J Immunol Methods 350(1-2):183–193. https://doi.org/10.1016/j.jim.2009.08.009
Tonegawa S (1983) Somatic generation of antibody diversity. Nature 302(5909):575–581. https://doi.org/10.1038/302575a0
Vale AM, Kapoor P, Skibinski GA, Elgavish A, Mahmoud TI, Zemlin C, Zemlin M, Burrows PD, Nobrega A, Kearney JF, Briles DE, Schroeder HW Jr (2013) The link between antibodies to OxLDL and natural protection against pneumococci depends on D(H) gene conservation. J Exp Med 210(5):875–890. https://doi.org/10.1084/jem.20121861
Vettermann C, Timblin GA, Lim V, Lai EC, Schlissel MS (2015) The proximal J kappa germline-transcript promoter facilitates receptor editing through control of ordered recombination. PLoS One 10(1):e0113824. https://doi.org/10.1371/journal.pone.0113824
Vollmers C, Sit RV, Weinstein JA, Dekker CL, Quake SR (2013) Genetic measurement of memory B-cell recall using antibody repertoire sequencing. Proc Natl Acad Sci U S A 110(33):13463–13468. https://doi.org/10.1073/pnas.1312146110
Volpe JM, Kepler TB (2008) Large-scale analysis of human heavy chain V(D)J recombination patterns. Immunome Res 4(1):3. https://doi.org/10.1186/1745-7580-4-3
Walther S, Rusitzka TV, Diesterbeck US, Czerny CP (2015) Equine immunoglobulins and organization of immunoglobulin genes. Dev Comp Immunol 53(2):303–319. https://doi.org/10.1016/j.dci.2015.07.017
Wang C, Liu Y, Cavanagh MM, le Saux S, Qi Q, Roskin KM, Looney TJ, Lee JY, Dixit V, Dekker CL, Swan GE, Goronzy JJ, Boyd SD (2015) B-cell repertoire responses to varicella-zoster vaccination in human identical twins. Proc Natl Acad Sci U S A 112(2):500–505. https://doi.org/10.1073/pnas.1415875112
Wang JR, de Villena FP et al (2012) Comparative analysis and visualization of multiple collinear genomes. BMC bioinformatics 13(Suppl 3):S13
Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC (2003) Predominant autoantibody production by early human B cell precursors. Science 301(5638):1374–1377. https://doi.org/10.1126/science.1086907
Watson CT, Breden F (2012) The immunoglobulin heavy chain locus: genetic variation, missing data, and implications for human disease. Genes Immun 13(5):363–373. https://doi.org/10.1038/gene.2012.12
Watson CT, Steinberg KM et al (2013) Complete haplotype sequence of the human immunoglobulin heavy-chain variable, diversity, and joining genes and characterization of allelic and copy-number variation. Am J Hum Gen 92:530–546
Watson CT, Steinberg KM, Graves TA, Warren RL, Malig M, Schein J, Wilson RK, Holt RA, Eichler EE, Breden F (2015) Sequencing of the human IG light chain loci from a hydatidiform mole BAC library reveals locus-specific signatures of genetic diversity. Genes Immun 16(1):24–34. https://doi.org/10.1038/gene.2014.56
Weinstein JA, Jiang N, White RA, Fisher DS, Quake SR (2009) High-throughput sequencing of the zebrafish antibody repertoire. Science 324(5928):807–810. https://doi.org/10.1126/science.1170020
West GB, Brown JH, Enquist BJ (1999) The fourth dimension of life: fractal geometry and allometric scaling of organisms. Science 284(5420):1677–1679. https://doi.org/10.1126/science.284.5420.1677
Wiegel FW, Perelson AS (2004) Some scaling principles for the immune system. Immunol Cell Biol 82(2):127–131. https://doi.org/10.1046/j.0818-9641.2004.01229.x
Williams GS, Martinez A, Montalbano A, Tang A, Mauhar A, Ogwaro KM, Merz D, Chevillard C, Riblet R, Feeney AJ (2001) Unequal VH gene rearrangement frequency within the large VH7183 gene family is not due to recombination signal sequence variation, and mapping of the genes shows a bias of rearrangement based on chromosomal location. J Immunol 167(1):257–263. https://doi.org/10.4049/jimmunol.167.1.257
Wu YC, Kipling D et al (2010) High-throughput immunoglobulin repertoire analysis distinguishes between human IgM memory and switched memory B-cell populations. Blood 116:1070–1078
Yamagami T, ten Boekel E et al (1999) Frequencies of multiple IgL chain gene rearrangements in single normal or kappaL chain-deficient B lineage cells. Immunity 11:317–327
Yancopoulos GD, Desiderio SV, Paskind M, Kearney JF, Baltimore D, Alt FW (1984) Preferential utilization of the most JH-proximal VH gene segments in pre-B-cell lines. Nature 311(5988):727–733. https://doi.org/10.1038/311727a0
Yang H, Wang JR, Didion JP, Buus RJ, Bell TA, Welsh CE, Bonhomme F, Yu AHT, Nachman MW, Pialek J, Tucker P, Boursot P, McMillan L, Churchill GA, de Villena FPM (2011) Subspecific origin and haplotype diversity in the laboratory mouse. Nat Genet 43(7):648–655. https://doi.org/10.1038/ng.847
Ye J (2004) The immunoglobulin IGHD gene locus in C57BL/6 mice. Immunogenetics 56(6):399–404. https://doi.org/10.1007/s00251-004-0712-z
Zachau HG (1989) Immunoglobulin genes. Immunol Today 10(8):S9–10
Zachau HG (1993) The immunoglobulin kappa locus—or—what has been learned from looking closely at one-tenth of a percent of the human genome. Gene 135(1-2):167–173. https://doi.org/10.1016/0378-1119(93)90062-8
Zarnitsyna V, Evavold B, Schoettle L, Blattman J, Antia R (2013) Estimating the diversity, completeness, and cross-reactivity of the T cell repertoire. Front Immunol 4. https://doi.org/10.3389/fimmu.2013.00485
Zemlin M, Bauer K, Hummel M, Pfeiffer S, Devers S, Zemlin C, Stein H, Versmold HT (2001) The diversity of rearranged immunoglobulin heavy chain variable region genes in peripheral blood B cells of preterm infants is restricted by short third complementarity-determining regions but not by limited gene segment usage. Blood 97(5):1511–1513. https://doi.org/10.1182/blood.V97.5.1511
Zemlin M, Schelonka RL, Ippolito GC, Zemlin C, Zhuang Y, Gartland GL, Nitschke L, Pelkonen J, Rajewsky K, Schroeder HW (2008) Regulation of repertoire development through genetic control of DH reading frame preference. J Immunol 181(12):8416–8424. https://doi.org/10.4049/jimmunol.181.12.8416
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Collins, A.M., Jackson, K.J.L. On being the right size: antibody repertoire formation in the mouse and human. Immunogenetics 70, 143–158 (2018). https://doi.org/10.1007/s00251-017-1049-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-017-1049-8