Abstract
Public health is a critical issue, therefore we can find a great research interest to find faster and more accurate methods to detect diseases. In the particular case of cancer, the use of mass spectrometry data has become very popular but some problems arise due to that the number of mass-to-charge ratios exceed by a huge margin the number of patients in the samples. In order to deal with the high dimensionality of the data, most works agree with the necessity to use pre-processing. In this work we propose an algorithm called Heat Map Based Feature Selection (HmbFS) that can work with huge data without the need of pre-processing, thanks to a built-in compression mechanism based on color quantization. Results shows that our proposal is very competitive against some of the most popular algorithms and succeeds where other methodologies may fail due to the high dimensionality of the data.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Diseases that are detected at an advanced stage, usually lead to high death rate, hence early detection of conditions such as cancer are critical for public health [1]. With the development of new technologies for data analysis such as microarrays and mass spectrometry (MS) it has been possible to understand diseases much better than before [2]. For several diseases, but in the particular case of cancer, the use of MS to profile high resolution complex protein and compare the results of cancer vs normal samples has become very popular and early diagnosis has been made possible [3, 4]. The main idea is to find measurable indicators for a biological condition, these indicators are known as biomarkers and help in a considerable way for the early detection of diseases [5].
To gather MS data a mass spectrometer device is required, and the most common current techniques are [2]: (1) Matrix-Assisted Laser Desorption and Ionization Time-Of-Flight, known as MALDI-TOF which allows the analysis of biomolecules (such as DNA, proteins, peptides and sugars) and (2) Surface-Enhanced Laser Desorption and Ionization Time-Of-Flight, known as SELDI-TOF that it is used mainly for the analysis of protein mixtures in tissues samples such as blood, urine and other clinical samples [6].
In this research we have used data that was collected using SELDI approach. Using this technology is possible to generate protein features information that has been successfully used in the detection of several diseases using serum and other complex biological specimens [7, 8]. When analyzing the MS data, the value (height) for each feature represents the ion abundance at a specific mass-to-charge (m/z) interval. The combination of multiple m/z values produces a pattern that represents a condition or state, such representation is known as the fingerprint for such condition. Hence if we have different fingerprints for some conditions, e.g. for cancer and non-cancer samples, it would be possible to identify the disease presence or absence.
The use of high resolution MS data to search proteins that help to detect the presence of a particular disease has greatly accelerated the biomarkers discovery phase [9], however it has been found that very few biomarkers reappear in new test data, which leads to the conclusion that there is a classifier overfitting problem caused by learning from very few samples with lots of variables [10]. When dealing with a large number of variables, techniques such as Support Vector Machines (SVM) are used as they can work with very high dimensional data, however even with such approaches, it has been found that improvement is possible after variable reduction [11, 12].
The task of selecting just a part of the spectra, i.e. a subset of features, is a complex process that can be divided in two main approaches [13, 14]: (1) Feature extraction: where the key idea is to transform the high dimensional features into a whole new space of lower dimensionality e.g. Principal Component Analysis (PCA) [15], and (2) Feature selection: which main goal is to find the smallest number of features that still describes the data in a reasonable way [16]. This is the approach used in this work.
When dealing with MS data, feature selection (FS) becomes a very fundamental part of the learning process as the number-of-features to number-of-samples ratio is quite unbalanced. In our particular research we used the high-resolution ovarian dataset provided by the National Cancer Institute (NCI): Center for Cancer Research [17] which has 370,575 features for each sample and includes a total of 216 patients; such amount of data gives us a matrix size of over 80,000,000 that most works prefer to avoid due to the complexity it represents.
As part of this research we have developed an algorithm called Heat Map Based Feature Selection (HmbFS) which is domain independent (i.e. not specifically designed for MS data) and it is able to compute a large amount of data with limited memory resources. Experiments were carried out using an on-line machine learning framework known as ML Comp that allows objectively algorithm comparison and provides external researchers with the ability to reproduce all our experiments, something that is usually hard to achieve on most works of this kind [18].
The remaining of this paper is structured as follows: In Sect. 2, some related works are presented. In Sect. 3 we present the formal proposal and algorithm description. In Sect. 4 we describe our experiments and results. In Sect. 5 we present the final conclusions and discuss future work.
2 Related Works
In the past decade we have seen many works dealing with MS data and trying to reduce its inherent high dimensionality by several different techniques, in this section we present some of the works that are more closely related with ours.
In 2005, Yu, et al. [1] used a Bayesian neural networks to identify ovarian cancer from the high-resolution NCI dataset. Using binning as pre-processing it was possible to removed almost 97 % of the data, leaving only 11,301 features (m/z ratios). According to their results they managed to get an overall accuracy of 98.49 % using a leave-one-out cross validation approach.
In 2006, Susmita et al. [2] used the NCI ovarian cancer dataset but in low-resolution form (15,154 features). The FS methods Bonferroni [19] and Westfall and Young [20] were employed to reduce the original data to 1700 (11.21 %) and 1912 (12.61 %) respectively. Best result in their experiments is achieved by a combination of Westfall and Young with SVM with an accuracy of 96.5 %.
In 2009, Liu [21] proposes a new application of wavelet feature selection extraction that in conjunction with SVM was used to do classification on the same ovarian cancer dataset that we are using. A pre-processing step to resample the data was employed, resulting dataset was almost 96 % smaller and using SVM they got a classification accuracy of 98 % although using an unknown dataset partition.
In 2010, Jiqing et al. [22], used NCI low-resolution ovarian dataset and proposed a sparse representation based FS for MS data. Authors managed to remove around 68 % of the data by limiting the m/z range from 1,500 to 10,000; this decision is based on the rationale that points lower than 1,500 are distorted by those of energy-absorbing molecules (EAM). Further reduction is achieved by baseline correction using local linear regression [23] and normalization. Using a wrapper FS approach guided by SVM, authors achieved an accuracy of 98.3 %.
In 2013, Soha et al. [24], presented a genetic programming (GP) approach for FS in MS data. The data was pre-processed reducing it from the 370,575 features to 15,000. The key idea in this work is to combine two well known FS algorithms Information-Gain (IG) [25] and Relief-F [26] and use their outputs as terminals for the GP method. However the process is very exhaustive as the GP method is designed in a wrapper approach. When using a SVM approach for classification they got an accuracy of 93.15 %.
As a summary, we can see a common pattern in all related works, due to constrains such as computer resources and algorithms complexity, dealing with raw MS data is reported as unmanageable. In this work we present an efficient algorithm that is capable of dealing with huge number of features with limited resources.
3 Formal Proposal
The context of this work is focused on supervised machine learning, in order to formally express the place that FS takes in the learning process, we first must define the supervised learning process itself.
In order to build a model to solve a problem, for supervised learning it is required to have a set of data known as training set, this data is composed by a set of labeled (e.g. cancer vs normal) instances which are features vectors containing the information that is required to learn.
An instance is therefore formalized as an assignment of values V = (v 1 , …, v n ) to a set of features F = (f 1 , …, f n ), where each instance is labeled with at least one of the possible c1, …, c n classes of C. In the case of MS data, each possible value in the m/z spectrum is a feature, and the specific m/z concentration is the value assigned to that feature; later each instance is labeled with the condition they represent.
Since the training set is the input to build a classifier, it is clear that its performance is inherently dependent of the values of such features. And as pointed out by Yu [27], in theory, the more features we have, the more power to discriminate between classes we would have, however, in practice with a limited number of instances (as is the case of MS data) the excessive number of features not only causes the learning process to be slow but there is a high risk of overfitting the data, as irrelevant or noisy features may confuse the learning algorithm. To handle this problem is where FS takes place.
To formalize the concept of FS, let R be a reduced (subset) version of F and V R the value vector for R. So in general, FS can be defined [28] as finding the minimum subset R such as P(C | R = V R ) is equal or close to P(C | F = V), or in other words, if the probability distribution of different classes given the reduced subset is equal or close to the original distribution for the whole features in F.
Since our goal is to reduce the original space, it is required to identify such features that are relevant to the learning process and discard the rest. John et al. [29] propose a relevance categorization that is shown below.
Let F be the full set of features, F i a particular feature and S i = F – {F i }. Now we can identify three types of features, strongly relevant, weakly relevant and irrelevant given the following conditions:
A feature F i is strongly relevant iff: P(C | F i , S i ) ≠ P(C | S i )
A feature F i is weakly relevant iff: P(C | Fi, Si) = P(C | Si) and \( \exists \; {S^{\prime}}_{i} \subset S_{i} \;{\text{such}}\;{\text{as}}\;P(C \, | \, F_{i} , \, {S^{\prime}}_{i} ) \ne P(C \, | \, {S^{\prime}}_{i} ). \)
A feature F i is irrelevant iff: \( \forall \; {S^{\prime}}_{i} \subseteq S_{i} , \, P(C \, | \, F_{i} , \, {S^{\prime}}_{i} ) = P(C \, | \, {S^{\prime}}_{i} ) \)
Given these definitions, we have that a feature is strongly relevant if it is always required in order to keep (or improve) the original conditional class distribution, while a weakly relevant feature may or may not be required to keep the probability. Then an irrelevant feature is simply not required and its removal causes very minimal or not impact at all in class probability.
Our proposal hence consist in finding such relevant features in an automatic way employing our algorithm called Heat Map Based Feature Selection (HmbFS) which has been designed to work with very high dimensional datasets (such as MS data) with very low memory footprint thanks to a data compression mechanism. The FS process is divided in two stages: (1) Compression and (2) Selection, which are described below.
Stage 1 - Compression: in order to handle the analysis of hundred thousands of features with efficiency, a key factor that takes our proposal apart from others is that we do not perform single feature analysis but instead we join features in “groups”, hence reducing the number of required iterations to compare the usefulness of features. Therefore our relevance definition had a slight change as we do not analyze F i anymore but instead a group G i that is defined as G i = {F r , F g , F b }, these features stands for Feature Red , Feature Green and Feature Blue and their corresponding values represent color intensities, but together (i.e., as a group) the relation between them creates a true color.
In order to produce these groups G, we employ a lossy compression technique that builds a graphical representation of the data, the result of such transformation is known as Heatmap. The first step for compression is the normalization of each isolated feature at a time, i.e. each feature is treated independently, to formalize this, let I be the full set of instances (e.g. samples in MS data) and I x a particular sample for the whole dataset D, each value for F i that belongs to a instance I x (I x F i ) is then normalized to a 0–255 interval as shown:
Where min(IF i ) and max(IF i ) represents the minimum and maximum value respectively that a feature gets across all the instances of the dataset. This process is repeated for every feature F i across all instances I x . Once the features have been normalized, each of the packages containing F r , F g and F b can be mapped to a true color expressed in red/green/blue (RGB) format, together the three features can represent up to 16,777,216 different colors or patterns. However, since the idea is to build a generalization of the original data, we apply a technique called Color Quantization that allows the mapping of a true color to a lower depth color scale, in this case we have reduced to a 4-Bit 16-Colors. The idea is to discard small data variations or uniqueness, and produce a new set of data which is more general and consistent, the process of quantization is performed by finding the minimum euclidean distance between the true color and the 16 reference colors, ranging from R j=1 to R j=16 , the RGB values for each of those colors are the standard values defined in the HTML 4.01 specification.
To formalize the information quantization, let G be the set of all the built groups and G i a particular group (which is composed by F r , F g and F b ) that belongs to a instance I x , each of those I x G i combinations are then compared against all the reference colors R j in set R to produce the new single-value compressed feature F i that will represent the old 3-feature as shown below:
Once all the distances between true color and reference color have been calculated, we select the reference color R j with the minimum euclidean distance, and this process gives origin to a new dataset that is purely built in reference colors, or in other words, it is a compressed lossy version of the original. Dataset reduction is possible due to the fact that the reference color is represented by a single value (e.g.: red, which is composed by RGB values of 255,0,0) instead of three different features, this compression makes possible to reduce the original 370,575 feature dataset to only 123,525. It is important to notice that such reduction is only performed for selection, the original data is untouched.
Stage 2 - Feature Selection: after compression is completed (Stage 1), selecting relevant features is based on the rationale that different classes should look different, hence their associated quantized colors should look different as well. Since FS occurs in the new compressed dataset, the process see regular features F i although we know they represent a group in the original space. The mode is calculated for each feature-class (FC) relation i.e. a mode for feature 1 and class 1 (F 1 C 1 ) and another mode for F 1 C 2 , and so on.
The conditional probability that a given value for mode belongs to a class or another is compared and if such probability exceeds other classes then we define the feature as useful. A threshold Th can be applied to produce more strict comparisons. In order to formalize the criteria, let P(C j | Mo(F i )) be the probability that a given mode in Fi be associated to a class Cj, then the feature F i usefulness is defined as follows:
After all useful features have been identified, we need to restore the original data, however such process is very efficient as each group is mapped to exactly 3 features, e.g. if FS process selected (compressed groups) F i = {2}, {5}, {7} and {10}, we know they are mapped to the original space to F i = {4, 5, 6}, {13, 14, 15}, {19, 20, 21} and {28, 29, 30}. Below we present the algorithm’s pseudo-code:

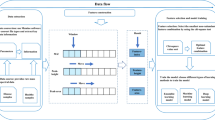
As it can be seen from pseudo-code, our proposal is very programming friendly. The compression stage allows to reduce problem complexity and evaluate features in a smaller space without losing the original data because, at the end of the process we map our results to it. Figure 1 summarizes the whole process:
In the next section we present our experiments setup and results obtained after processing the NCI High-resolution MS Ovarian Cancer dataset, which is commonly used as we reviewed in related works.
4 Experiments and Results
As correctly pointed out by Zhang [18], reproduce experiments in works such as this one is sometimes hard to achieve, in our approach we want to fill that gap by allowing readers to easily replicate our experiments and open the possibility to objectively compare any future work with ours. In order to achieve our goal we have performed our experiments in the Machine Learning (ML) Comp framework which can be accessed on-line for free in order to review our experiments or create new ones, this allows researchers to work in the same conditions than we did, with the exact same data, train/test partitions and hardware resources.
To test HmbFS we selected the NCI High-resolution ovarian cancer dataset as several works can be found using this dataset. Unlike most works, we wanted to deal with raw MS-data in order to push the limits of FS algorithms. In Table 1 we present our dataset setup compared with related works.
As it can be seen from Table 1, we performed all experiments with raw MS-data, no pre-processing of any kind was performed, the only modification to the data was a transformation to the SVM-light format that ML Comp requires. The summary of performed experiments is reviewed below:
Exp. 1 – Learning without FS: Our first goal was to find out how important FS can be when dealing with such big amount of data, for that task, we selected 5 well known multi-class algorithms and run the learning process without any FS aid.
Exp. 2 – FS in high-dimensional space: In a previous work [12] we evaluate the top 10 feature selection algorithms. In this work we used the top performers, Chi2 [30], Fcbf [31] and Relief-F [32] and coupled them with top classification algorithms to evaluate results.
Exp. 3 – Learning from HmbFS reduced datasets: Since experiments 1 & 2 proved that most current techniques are not efficient enough to handle such amount of data with limited resources, we decided that in order to have a reference point to compare HmbFS we had to reduce the datasets to allow other FS algorithms to handle the problem. However, HmbFS handled the full 370,575 features without any issue as can be seen in http://mlcomp.org/runs/36467.
The summary of classification error for our experiments is presented in Table 2, multiple partitions of data using stratified random subsampling can be found on MLComp as well as the experiments ID (reported in parenthesis) to validate our results. Two main threshold values are evaluated, using Th = 2, the number of selected features was 86,010 but for more aggressive reduction a Th = 12 was used to reduce the original data to only 2,802, over 99.4 % reduction compared with original data. The first 4 rows represent experiment 1, rows 5–8 are for experiment 2, and last 4 for experiment 3.
From Table 2, we can clearly see the problem under investigation by reviewing the first 4 rows, where most of the values are Out-Of-Memory (OOM). Current FS algorithms cannot easily handle the amount of data that raw MS represents, learning algorithms suffer from similar problems, since only Adagrad was able to process the entire dataset without any FS aid. Our proposal, HmbFS was the only FS algorithm capable of reducing the entire dataset, but in order to have a reference for comparison, we reduce the data in ~23 % (using Th=2) to make feasible the FS for competing algorithms. Best result was obtained by a combination of HmbFSTh=12 and Chi2, together they achieved an accuracy of 96.9 % with only ~0.7 % of the original data.
5 Conclusions and Future Work
From the related works we reviewed, the ones that used the high resolution NCI ovarian cancer dataset had to use different techniques for pre-processing, our experiments confirmed the necessity for such approach, as dealing with the raw MS data its very complex because at least two reasons: (1) There is a lot of noise and (2) Complexity to process such big data. After pre-processing the data however, we can notice a FS step is still performed, hence reducing the data even more. Since a FS algorithm main goal is to remove useless data (e.g. noise) and produce a small subset of feature that inherently reduce processing complexity, it would be ideal to employ a FS on MS data without the need for pre-processing, however it is hard to find FS algorithms capable to reduce such big datasets, in our experiments, none of the reviewed top algorithms was able to handle the data, all of their tests failed due to OOM errors.
Our proposal, HmbFS aims to be memory efficient in order to manage very high dimensional datasets. Experiments proves that our algorithm was able to process the almost 56 million elements (370,575 features by 151 train samples) where others failed, we were forced to help competing algorithms with a pre-filtering by HmbFS in order to have a reference point to compare with. Based on the way our algorithm works (grouping features), it can be used as a first-pass followed by a second-pass filtering by a classical FS algorithm that evaluates individual features, however this may not necessary lead to better results as shown in Table 2.
Our best result overall was achieved by using HmbfsTh=12 and Chi2 as a second-pass FS, with an accuracy of 96.9 % (run id 36502), such result however was achieved without any pre-processing, that excludes any alignment of the MS data as well, comparing with other works, we believe HmbFS can lead to very competitive results as very similar accuracies were obtained, even without pre-processing the data. Another remarkable detail of our experiments, is that all of them can be (1) replicated and (2) objectively compared, thanks to the ML Comp framework.
As far a future work it can be divided in three sections right now:
-
(1)
General dataset comparison: we want to perform more experiments of HmbFS with very different datasets in order to prove its usefulness in other areas other than MS data.
-
(2)
Automatic threshold: right now we need to set a manual threshold Th in order to decide “how much we want to reduce”, while the Th is the only required parameter and its quite intuitive, it is interesting to get rid of it by an automatic approach.
-
(3)
Human interaction in feature selection: since the theory behind HmbFS involves color generation, we would like to experiment with human-in-the-loop (HITL) to find patterns in such colors that may lead to improved selection.
References
Yu, J., Chen, X.: Bayesian neural network approaches to ovarian cancer identification from high-resolution mass spectrometry data. Bioinformatics 21, i487–i494 (2005)
Datta, S., DePadilla, L.M.: Feature selection and machine learning with mass spectrometry data for distinguishing cancer and non-cancer samples. Stat. Methodol. 3(1), 79–92 (2006). ISSN 1572-3127
Liotta, L.A., Ferrari, M., Petricoin, E.: Clinical proteomics: written in blood. Nature 425, 905 (2003)
Wulfkuhle, J.D., Loitta, L.A., Petricoin, E.F.: Proteomic applications for the early dectection of cancer. Nature 3, 267–275 (2003)
Srinivas, P.R., Verma, M., Zhao, Y., Srivastava, S.: Proteomics for cancer biomarker discovery. Clin. Chem. 48, 1160–1169 (2002)
Tang, N., Tornatore, P., Weinberger, S.R.: Current developments in SELDI affinity technology. Mass Spectrom. Rev. 23, 34–44 (2004)
Herrmann, P.C., Liotta, L.A., Petricoin, E.F.: Cancer proteomics: the state of the art. Dis. Markers 17, 49–57 (2001)
Vlahou, A., Schellhammer, P.E., Mendrinos, S., Patel, K., Kondylis, F.L., Gong, L., Nazim, S., Wright, G.L., Jr.: Development of a novel proteomic approach for the detection of transitional cell carcinoma of the bladder in urine. Am. J. Pathol. 158, 1491–1520 (2001)
Kuschner, K., Malyarenko, D., Cooke, W., Cazares, L., Semmes, O., Tracy, E.: A Bayesian network approach to feature selection in mass spectrometry data. BMC Bioinform. 11, 177 (2010)
Malyarenko, D., Cooke, W.E., Adam, B.L., Malik, G., Chen, H., Tracy, E.R., Trosset, M.W., Sasinowski, M., Semmes, O.J., Manos, D.M.: Enhancement of sensitivity and resolution of surface-enhanced laser desorption/ionization time-of-flight mass spectrometric records for serum peptides using time-series analysis techniques. Clin. Chem. 51, 65–74 (2005)
Guyon, I., Weston, J., Barnhill, S., Vapnik, V.: Gene selection for cancer classification using support vector machines. Mach. Learn. 46(1), 389–422 (2002)
Huertas, C., Juarez-Ramirez, R.: Filter feature selection performance comparison in high-dimensional data: a theoretical and empirical analysis of most popular algorithms. In: 17th Information Fusion (FUSION) Conference (2014)
Dittmann, B., Nitz, S.: Strategies for the development of reliable QA/QC methods when working with mass spectrometry-based chemosensory systems. Sens. Actuators, B 69, 253–257 (2000)
Depczynski, U., Frost, V., Molt, K.: Genetic algorithms applied to the selection of factors in principal component regression. Anal. Chim. Acta 420, 217–227 (2000)
Suganthy, M., Ramamoorthy, P.: Principal component analysis based feature extraction, morphological edge detection and localization for fast iris recognition. J. Comput. Sci. 8, 1428–1433 (2012)
Dash, M., Liu, H.: Feature selection for classification. Intell. Data Anal. 1, 131–156 (1997)
Petricoin, E., Ardekani, A.M., Hitt, B.A., Levine, P.J., Fusaro, V.A., Steinberg, S.M., Mills, G.B., Simone, C., Fishman, D.A., Kohn, E.C., Liotta, L.A.: Use of proteomic patterns in serum to identify ovarian cancer. Lancet 359, 572–577 (2002)
Zhang, X., Lu, X., Shi, Q., Xu, X., Leung, H., Harris, L., Iglehart, J., Miron, A., Liu, J., Wong, W.: Recursive SVM feature selection and sample classification for mass-spectrometry and microarray data. BMC Bioinform. 7(1), 197 (2006)
Bonferroni, C.E.: Il calcolo delle assicurazioni su gruppi di teste. In: Studi in Onore del Professore Salvatore Ortu Carboni, Rome, pp. 13–60 (1935)
Westfall, P., Young, S.: Resampling-Based Multiple Testing, Examples and Methods For p-Value Adjustment. Wiley, New York (1993)
Liu, Y.: Feature extraction and dimensionality reduction for mass spectrometry data. Comput. Biol. Med. 39(9), 818–823 (2009)
Jiqing, K., Lei, Z., Bin, H., Qi, D., Yaojia, W., Lihua, L., Shenhua, X., Hanzhou, M., Zhiguo, Z.: Sparse representation based feature selection for mass spectrometry data. In: Bioinformatics and Biomedicine Workshops (BIBMW), pp. 57–62 (2010)
Wu, B., Abbott, T., Fishman, D., McMurray, W., Mor, G., Stone, K., Ward, D., Williams, K., Zhao, H.: Comparison of statistical methods for classification of ovarian cancer using mass spectrometry data. Bioinformatics 19(13), 1636 (2003)
Ahmed, S., Zhang, M., Peng, L.: Feature selection and classification of high dimensional mass spectrometry data: a genetic programming approach. In: Vanneschi, L., Bush, W.S., Giacobini, M. (eds.) EvoBIO 2013. LNCS, vol. 7833, pp. 43–55. Springer, Heidelberg (2013)
Sebastiani, F., Ricerche, C.N.D.: Machine learning in automated text categorization. ACM Comput. Surv. 34, 1–47 (2002)
Sun, Y., Wu, D.: A relief based feature extraction algorithm. In: SDM, pp. 188–195 (2008)
Yu, L., Liu, H.: Efficient feature selection via analysis of relevance and redundancy. J. Mach. Learn. Res. 5, 1205–1224 (2004)
Koller, D., Sahami, M.: Toward optimal feature selection. In: Proceedings of the Thirteenth International Conference on Machine Learning, pp. 284–292 (1996)
John, G., Kohavi, R., Pfleger, K.: Irrelevant feature and the subset selection problem. In: Proceedings of the Eleventh International Conference on Machine Learning, pp. 121–129 (1994)
Liu, H., Setiono, R.: Chi2: feature selection and discretization of numeric attributes. In: Proceedings of the Seventh IEEE International Conference on Tools with Artificial Intelligence, pp. 388–391. IEEE Computer Society (1995)
Yu, L., Liu, H.: Feature selection for high-dimensional data: a fast correlation-based filter solution. In: Proceedings of the 20th International Conference on Machine Learning (ICML 2003), pp. 856–863 (2003)
Kira, K., Rendell, L.: A practical approach to feature selection. In: Proceedings of the Ninth International Conference on Machine Learning (ICML 1992), pp. 249–256 (1992)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this paper
Cite this paper
Huertas, C., Juárez-Ramírez, R. (2015). Heat Map Based Feature Selection: A Case Study for Ovarian Cancer. In: Mora, A., Squillero, G. (eds) Applications of Evolutionary Computation. EvoApplications 2015. Lecture Notes in Computer Science(), vol 9028. Springer, Cham. https://doi.org/10.1007/978-3-319-16549-3_1
Download citation
DOI: https://doi.org/10.1007/978-3-319-16549-3_1
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-16548-6
Online ISBN: 978-3-319-16549-3
eBook Packages: Computer ScienceComputer Science (R0)