Abstract
Clustering is regarded as one of the most difficult tasks due to the large search space that must be explored. Feature selection aims to reduce the dimensionality of data, thereby contributing to further processing. The feature subset achieved by any feature selection method should enhance classification accuracy by removing redundant features. To this end, this paper proposes a new model, called Best Clustering Normalized Mutual Information Quantile (BC-NMIQ), to rank the best features using the square root threshold. Finally, the proposed BC-NMIQ is improved with the optimal set of features selected automatically using the Incremental Association Markov Blanket (IAMB) feature selection method. The measurement criteria are applied to BC-NMIQ-IAMB as the main proposed method and to BC-NMIQ as a subsidiary proposed method. In fact, the hybrid BC-NMIQ-IAMB is the combination of the proposed filter method (BC-NMIQ) and the existing automatic filter feature selection approach (IAMB). To test the performance of the proposed BC-NMIQ-IAMB algorithm, its performance is compared with that of some other algorithms recently proposed in the literature. The results of the experiments, which were conducted on ten benchmark high-dimensional medical datasets (including binary and multi-class), confirmed that BC-NMIQ-IAMB increases the average accuracy of existing binary and multi-class algorithms to 0.92 and 0.94, respectively.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Feature selection is one of the dimensionality reduction methods, which helps to choose the most relevant features before applying the learning algorithm [39]. The goal of this method is to get a subset of all available features that are much more useful for the next trend. In other words, feature selection is aimed at eliminating irrelevant and redundant features that may lead to undesirable correlations in the next process (learning) [26]. Feature selection approaches have brought significant improvements to both the learning performance and the computational efficiency of the classification algorithms. Such improvements are particularly observed when feature selection approaches are wrapped around evolutionary computation (EC) techniques that are known to be powerful global search methods [50]. The performance of the feature selection method is usually evaluated by the machine learning model. Deep learning (a sub-branch of machine learning) algorithms have been popular for automatic recognition of digits and characters of different languages. Deep networks can be trained in a supervised mode requiring labels, or in an unsupervised way without the need for labels [5, 27]. The commonly-used machine learning models include Naïve Bayes, KNN, C4.5, SVM, BPNN, RBF-NN, K-means, Hierarchical clustering, and Density based clustering [38]. A good feature selection method should have high learning accuracy, but less computational overhead (time and space complexity). Although there have been solid reviews on feature selection [45], they mainly focus on specific research fields in feature selection. Therefore, it is still worth comprehensively surveying recent advances in feature selection and discussing some future challenges.
Feature selection has been the subject of much research in the field of supervised and unsupervised data engineering in recent decades [31], and existing methods for supervised classification are mainly divided into three categories: the filter methods [22], the wrapper methods [33], and the embedded methods [54]. The wrapper methods follow innovative guidelines for selecting a subset of features with the best predictive performance. Since the number of subsets may be extremely large and new classifiers need to be created for each updated subset, the wrapper methods are usually computationally expensive, which makes them difficult to be applied to datasets with a large number of instances or feature candidates [13, 24, 51]. Embedded methods, despite their higher speed than the wrapper methods, are still computationally heavy, and their feature selection results depend on the learning machine [13]. On the other hand, the filter feature selection methods score and rank the feature candidates according to a certain criterion, and extract one feature at a time to form a subset with a predefined dimension. They are also computationally cheaper and do not rely on particular predictors [13].
Cluster analysis is an unsupervised learning method aiming to group a set of unlabeled objects into clusters in such a way that each cluster could contain items that are more similar to the rest of the items in the same cluster than those in the other clusters [52]. Clustering can help scientists analyze data and solve practical problems; thus, it has been widely used in disparate fields, including statistical analysis [6, 7], pattern recognition [30], information retrieval [17], and bioinformatics [16]. The traditional clustering algorithms can be divided into different categories such as partition methods [11], hierarchical algorithms [32], density-based algorithms [9], intelligent traffic prediction [10], and graph-based methods [14].
Many feature selection methods use meta-heuristics, evolutionary, and swarm intelligence-based algorithms to avoid increasing computational complexity in the high-dimensional medical datasets. Meta-heuristic algorithms have been very successful in tackling many optimization problems such as data mining, machine learning, engineering design, production tasks, and Feature Selection (FS) [48]. Meta-heuristic algorithms are general-purpose stochastic methods that can find a near-optimal solution within a reasonable time. Lately, various Swarm Intelligence (SI)-based meta-heuristics have been developed and proved efficient in handling FS tasks in different fields [35, 41]. SI-based algorithms generally consist of a simple population of artificial agents. This concept is typically inspired by nature, and each agent performs an easy job, but local interactions and partly-random interactions between these agents lead to the emergence of “intelligent” global behavior, which is unknown to individual agents [39].
In the field of Machine Learning and Data Mining, many learning algorithms have been proposed, which are primarily applied to handling discrete features. However, data in real world are often continuous in nature. Hence, discretization is a commonly used data preprocessing procedure that transforms continuous features into discrete features [19]. It is the process of partitioning continuous variables into categories. Unfortunately, the number of ways to discretize a continuous attribute is infinite. Discretization is a potential time-consuming bottleneck since the number of possible discretization is exponential in the number of interval threshold candidates within the domain. Discretization techniques are often used by the classification algorithms, genetic algorithms, and a wide range of learning algorithms. The use of discrete values has a number of advantages such as:
-
Discrete features require less memory space.
-
Discrete features are often closer to a knowledge-level representation.
-
Data can be reduced and simplified through discretization, which becomes easier to understand, use, and explain.
-
Learning will be more accurate and faster using the discrete features.
The focus of this paper is on proposing a hybrid filter feature selection method including the concept of clustering, NMI, and IAMB, based on MRMR. Three feature clustering methods are used in this study, i.e., ward method, equal width discretization, and k-means. The selection of clustering methods was based on the following criteria. The ward method is noise robust and biased towards globular clusters. Equal width is straightforward and simple to apply. It works solely by having the smallest and largest value of each feature as well as the number of samples. k-means is very fast compared to many other clustering algorithms. In addition, feature selection based on learning the IAMB does not depend on a specific classifier. Moreover, IAMB does not need to know the number of features prior to applying feature selection. It is also globally optimal and less prone to over-fitting than other existing methods. The contributions of the proposed method are summarized as follows:
-
To convert features with continuous values to the respective features with discrete values using k-means, complete link, and equal width discretization clustering algorithm and to find the number of clusters of each feature by normalize mutual information based on the labels.
-
To select the most informative features using the concept of Maximum Relevance Minimum Redundancy (MRMR) within a filter method via predefined thresholds.
-
To apply Incremental Association Markov Blanket Feature Selection (IAMB) to automatically select the features.
In the rest of the paper, we will introduce the evaluation measure for feature selection. Section 2 introduces some recent studies on feature selection approaches, including wrapper, embedded, and filter approaches. Section 3 explains the proposed methodology. Section 4 presents an introduction to experimental data and setup and provides the summery of experimental results and related discussions. Finally, in Section 5, conclusion and trends for future research are presented.
2 Related work
In this section, recent studies conducted on the feature selection methods are reviewed. Sheng et al. introduced a niching memetic approach (NMA-CFS) to clustering and feature selection. In NMA-CFS, a type of variable-string length genetic algorithm was used as an optimizer, and a kind of within-cluster scatter was utilized as an evaluation criterion to find the correct number of clusters. NMA-CFS could find the exact number of clusters using a smaller number of features, but the datasets used in their experiments mostly involved a small number of clusters and features [46].
Lensen et al. put forward a comparative study of medoid-based and centroid-based encoding schemes on the PSO framework for clustering and feature selection. Although the medoid-based encoding scheme performed better than the centroid-based one, in most cases, it could not obtain the optimal number of clusters [28].
Lensen et al. proposed a three-stage PSO-based clustering and feature selection approach. In the first stage, an initial number of clusters was utilized using the Silhouette index. Then, in the second stage, the evolutionary process was carried out with the help of the initial cluster number. Finally, the best solution previously represented by the medoid-based encoding was converted to centroid-based encoding to find the optimal cluster. Like the previous ones, this approach also could not properly estimate the optimal number of clusters in most datasets [29].
Rostami et al. proposed a multi objective feature selection method on PSO-based. Their proposed method involves three main steps. At the first step, the main features are shown as a graph representation model. At the next step, feature centralities for all nodes in the chart are calculated, and finally, at the third step, an improved PSO-based search process is applied to the final feature selection. The novel approach employed in their study evaluates a feature subset by the combination of feature separability index, similarity, and feature subset size. Although the results on five medical datasets indicated that the proposed method improves previous related methods in terms of efficiency and effectiveness, several users are needed to specify the parameters used in the proposed methods. Thus, their corresponding values should be determined by users. Given that the accuracy of the learning model must be calculated to evaluate each combination of parameter values, this approach will not be applicable to situations where the construction of the learning model has a high computational complexity [39].
Rostami et al., in another research, proposed the Community Detection-based Genetic Algorithm for Feature Selection (CDGAFS) operating in three stages. The similarities of the features are calculated at the first step. Then, the features are classified by community detection algorithms into clusters at the second step. Finally, at the third step, features are selected by a genetic algorithm with a new community-based repair operation. Although the proposed method gives higher efficiency, faster convergence, and search efficiency compared to other feature selection methods, there are several user-specified parameters used in the developed feature selection methods. Therefore, their corresponding values should be determined by users. As a result, choosing the best values for the parameters is an optimization problem [42].
In another study, Rostami et al. proposed a novel pairwise constraints-based method for feature selection. In the proposed method, the similarity between the pair constraints is calculated and an uncertainty region is created based on it. Then, in an iterative process, most informative pairs are selected. The proposed method was compared to different supervised and unsupervised feature selection approaches, including LS, GCNC, FJUFS, FS, FAST, FJMI, and PCA. The reported findings indicated that, in most cases, their proposed approach was more accurate and selected fewer features [40]. Alirezanejad et al. proposed two heuristic filter methods for gene selection, namely Xvariance against Mutual Congestion (MC). Xvariance depends on internal features such as variance and mean, while the Mutual Congestion is based on the frequency of features. Experimental results showed that Mutual Congestion increased the accuracy of basic classifiers in subsequent datasets, whereas Xvariance had significant results in standard datasets. The comparisons of the results with those of the state-of-the-art methods confirmed that their methods performed better than the existing ones. Even though Xvariance and Mutual Congestion achieved significant results, both methods selected 10 best features in the data set as the final feature subset. In fact, Alirezanejad et al. used a fixed threshold in their work [8].
Abbasabadi et al. suggested that the ensemble feature selection method was adopted on the premise that combining a number of feature selection methods yields more reliable results than using just one feature selection method in order to combine rankings of features from various algorithms into a single rank for each feature, a combinational method should be used when performing ensemble feature selection. Additionally, a threshold must be established in order to acquire a functional subset of features. This study proposes Automatic Thresholding Feature Selection (ATFS), a three-step ensemble feature selection method. Diversity generation is the first step, in which various rankers are applied to each dataset to produce various feature rankings. The proposed ensemble is given automatic thresholding capabilities by fast non-dominated sorting, which is used to combine the output rankings of individual selectors. This is the second step in order to create the best feature set, feature sets are generated, which is the third step. In addition, Sorted Label Interference (SLI), a brand-new filter technique built on the interference between class labels, is suggested. Binary datasets can be used with SLI and ATFS [1].
Nematzadeh et al. introduced Whale-Mutual Congestion (WMC) as a hybrid filter feature selection method for medical datasets. It was shown that Mutual Congestion (MC) can well predict class labels. The authors also demonstrated how the whale algorithm could improve the Mutual Congestion performance by removing half of the trivial features. The accuracy, sensitivity, specificity, and MC of the proposed method were calculated in the top 10 subsets of features using a majority of votes. To further evaluate the proposed method, the whale effectiveness analysis, box diagram analysis, and whale convergence analysis were performed. The results revealed that the proposed method can achieve superior results in most cases in terms of accuracy and size of the subset. Similar to the Xvariance and Mutual Congestion proposed by Alirezanejad et al., WMC had a fixed threshold 𝜏 = 10 and could not automatically select the final feature subset [34].
Abbasabadi et al also proposed a hybrid method using a proposed filter feature selection (SLI-γ) and a wrapper GA-based feature selection approach known as GArank&rand. In the first phase, SLI-γ was used to eliminate 99% of unnecessary features. The first phase solutions were; then, optimized by GA using the SLI-γ most calculated pertinent features. For the evaluation of the measurement criteria, this paper used 11 well-known datasets, including 4 standard datasets and 7 high-dimensional datasets. The experimental findings demonstrated that SLI-γ was able to outperform the other rankers (taken into account in this study) across all datasets. Additionally, SLI-γ had a big impact on GA’s performance, in terms of, classification precision and the number of features chosen. Furthermore, when 1% of the highest ranked features were chosen for the GA population generation, the execution time of GArank&rand was significantly reduced. [2].
Al-Batah et al. applied the Correlation-based Feature Selection (CFS) algorithm to the feature selection process to reduce the dimensionality of data and find a set of discriminatory genes. Then, the Decision Table, JRip, and OneR were employed for the classification process. They indicated that the feature selection by CFS improves not only the efficiency, but also the accuracy of the classification process. However, the number of selected genes using CFS had much more features within the data set as a final feature subset [4].
Sadeghian et al. first, proposed the Information Gain Binary Butterfly Optimization Algorithm (IG-bBOA) to circumvent the S-bBOA constraints. In addition to improving classification accuracy, IG-bBOA maximized the mean of the mutual information between features and class labels. Additionally, the three-stage process of the Ensemble Information Theory-based Binary Butterfly Optimization Algorithm used IG-bBOA to decrease the number of selected features (EIT-bBOA). The first phase employs the Minimal Redundancy-Maximum New Classification Information (MRMNCI) feature selection technique to remove 80% of unnecessary and redundant features. In the subsequent phase, IG-bBOA is used to select the best subset of features. Utilizing a ranking system based on similarity, the ultimate feature subset is selected. [43].
Brankovic et al. introduced a novel feature selection approach, which employs the distance correlation (dCor) as a criterion for evaluating the dependence of the class on a given feature subset (D2CORFS). The dCorindex provides a reliable dependence measure among random vectors of arbitrary dimension, without any assumption on their distribution. The dCorindex appears to be a particularly robust criterion with respect to overfitting and redundancy issues, which are common with multivariate filter methods. The distributed combinatorial optimization scheme was used to handle the severe asymmetry of microarray datasets by dividing the feature set into several feature bins and running independently the FS algorithm on each of them [12].
The mutual information and three-dimensional mutual information (TDMI) between the features and the class label are the foundation of many feature selection algorithms. The performance of feature selection can be affected because these algorithms do not take TDMI into account when considering features. Xiangyuan et al. suggested researching feature selection based on TDMI among features in the light of the issue. The joint mutual information between the class label and feature set is used to describe relevance in accordance with the maximal relevance minimal redundancy criterion, and mutual information between feature sets is utilized to describe redundancy. The mutual information between feature sets as well as that between the class label and feature set is then separated. TDMI, among other features, is taken into account during the decomposition process in order to produce an objective function. Lastly, a feature selection algorithm based on conditional mutual information for maximal relevance with minimal redundancy (CMI-MRMR) was proposed. [23]. A novel relevancy-redundancy measurement based on distance is presented by Hallajian et al. They use an unsupervised method and the mRMR criteria concept. In addition, a supervised approach in which the features are ranked according to the separation between each pair of samples in various feature vector classes. To select the most pertinent feature subset, an ensemble of the suggested supervised and unsupervised methods was used. The effectiveness of the suggested feature selection methods was examined, investigated, and compared using the effects of 24 distance measures drawn from five major families of distance functions. The highest-ranked characteristics are chosen based on an empirically determined criterion. Three classifiers, Decision Tree, Support Vector Machine, and Naive Bayes, were used on biomedical datasets representing binary issues from the UCI data repository to assess the selected features. [25]. Thejas et al. proposed a hybrid approach with two distinguished stages, namely feature ranking and feature selection. The proposed method was independent of any number of class labels and used Random Forest as a classifier. The main idea in the feature ranking stage was to cluster the features using mini-batch k-means, which performed clustering by using a batch of data. Each cluster was scored in [0, 1] using Normalized Mutual Information (NMI). The higher the score is, the better the candidate feature is for classification. The feature ranking list was constructed through calculating cluster scores separately for all features. The features were sorted and the ranking list was created. The feature selection stage comprised two approaches: Feature Inclusion and Least Ranked Feature Exclusion. The Feature Inclusion exploited a process called Mini Batch k-means Normalized Mutual Information Feature Inclusion (KNFI). Likewise, the Least Ranked Feature Exclusion used a process called Mini-Batch k-means Normalized Mutual Information least ranked Feature Exclusion (KNFE). KNFE achieved better results when there was little relationship among features, whereas KNFI had acceptable results in the majority of the data sets. There were no evidence that their proposed method could be applied to high dimensional data sets [49]. Table 1 illustrates the existing works in Tabular format.
Unlike WOA-MC [34], Xvariance [8], and D-PSO Scaled [29], which calculated the final feature subset manually (with a pre-defined threshold), this paper proposes a hybrid filter method that automatically selects the optimal feature subset. Although the feature selection methods proposed by D-PSO Scaled [29] and KNFI-KNFE [49] had acceptable results, the applicability of the methods decreases when the number of features is considerably high (the number of features is greater than the number of samples). This paper investigates feature selection in high dimensional medical datasets with any number of class labels. The majority of works in Table 1 have moderate (MODE-CFS [26], PSO(4–2) [50], DHSTNet [6], NMA-CFS [46], Dynamic Medoid PSO [28], D-PSO Scaled [29], Xvariance [8], WOA-MC [34], IG-bBOA [43],) or high selected subset length at least in some datasets (MPSONC [39], MHBPSO1 [24], CDGAFS [42],ATFS [1], GArank&rand [2], CFS [4], CMI-MRMR [23], MRmMC [25], KNFI-KNFE [49]) The subset length calculated in this research is considerably less than existing works.
The data are, first, clustered using k-means, complete link, and equal width discretization to generate different clustering within an unsupervised approach. Next, the number of clusters of each feature is found by Normalize Mutual Information (NMI) based on the labels; then, the maximum amount of calculation is selected for each feature. Finally, the optimal set of features is selected automatically through Incremental Association Markov Blanket feature selection (IAMB).
3 Preliminaries
Considering that our method applies three clustering methods namely, Ward method, k-means, Equal Width Discretization, and subsequently NMI, we discuss these concepts below.
The objective of the Ward method, also known as Minimum Variance Method (MVM), is to reduce the sum of squared errors among individual clusters. It measures the distance among the cluster pair in two ways. First, it estimates the distance between two individual clusters (xi, xj) with single data object based on the Squared Euclidean and is defined as expressed in Eq. (1):
Second, it calculates the distance between merged cluster (xi ∪ xj) and new cluster xk based on the lance-William method and is defined as follows:
where xi, j denotes the newly-merged cluster, and Ni, Nj and Nk represent the size of the ith, jth and kth clusters, respectively [47].
Agglomerative hierarchical methods include, but are not limited to, Ward’s Link, Average Link, Complete Link, or Single Link. In this work, we focus on the Ward’s Link. For more information on clustering algorithms, we recommend Chapter 14 of Rencher’s Multivariate Analysis reference. The Ward’s clustering algorithm optimizes the “within sum of squares” (WSS) for each cluster, which is estimated for each pairwise combination of clusters, and the merging of clusters that provide the smallest contribution to the WSS measure is implemented. The Ward method is robust against outliers and provides even clusters across the data set, making it a reliable algorithm for exploratory work such as the study in [20].
The k-means algorithm is a simple iterative clustering algorithm. Using the distance (typically the Euclidean distance) as the metric and given the K number of clusters, the k-means algorithm calculates the centroids and specifies the cluster members. Assuming an arbitrary dataset with n multi-dimensional data points, K predefined number of clusters, and the Euclidean distance as the similarity index, k-means attempts to minimize the within-cluster sum of squares using Eq. (3) [56]:
where uk represents the kth center, and xi represents the ith point in the data set. The solution to the centroid uk is as expressed in Eq. (4):
If Eq. (3) is zero, then \( {u}_k=\frac{1}{n}\sum \limits_{i=1}^n{x}_i \).
The basic idea of the k-means algorithm is to randomly extract K data points from the sample set containing n multi-dimensional data points as the centroids of the initial clusters. The distance of the rest of the samples (data points) within a dataset are calculated with respect to the centroids, and the cluster members are specified. The new centroids are derived from existing clusters and the members of each cluster are updated. The k-means algorithm repeats the process until the centroids of the clusters remain unchanged. The result of the k-means algorithm directly depends on the initial centroids [37].
The mutual information (MI) is used to assess how arbitrary random variables are dependent. MI returns 1 for maximum dependency between two random variables of X and Y. On the contrary, MI = 0 shows the two random variables are completely independent. Let x ∈ X and y ∈ Y be two random variables, P (x, y ) be the joint probability density function of X and Y, and P (x) and P (y ) be the corresponding marginal probability density functions, then the mutual information X and Y can be expressed as follows in Eq. (5) [44].
Using Eq. (1) and entropy H(X) = − ∑x ∈ Xp(x)logP(x), the Normalized Mutual Information (NMI) is calculated using Eq. (6) as follows.
In statistics, the quantile function plays a crucial role in prescribing the probability distributions. It is indispensable in determining the location and spread of any given distribution, especially the median that is resistant to extreme values or outliers. The quantile function is used extensively in the simulation of non-uniform random variables and details of the use of the quantile function in modeling, statistical, reliability, and survival analysis can be found in [36]. A quantile defines a particular part of a data set; it determines how many values in a distribution are above or below a certain limit. Special quantiles are the quartile (quarter), the quintile (fifth), and the percentiles (hundredth).
The probability density function of the chi-square distribution and the cumulative distribution function are given by Eqs. (7) and (8).
where γ (., .) = incomplete gamma function and P(., .) = regularized gamma function.
The quantile (Q) approach was used to obtain the second order nonlinear differential eq. Q is applied to distributions whose Cumulative Distribution Function (CDF) is monotonously increasing and absolutely continuous. The Chi-square distribution is one of such distributions, which is expressed in Eq. (9):
where the function F−1(p) is the composition inverse of the CDF. Suppose the Probability Density Function (PDF) f(x) is known and the differentiation exists. The first-order quantile equation is obtained from the differentiation of Eq. (9) to obtain Eq. (10):
Maximum Relevance and Minimum Redundancy (MRMR) is an efficient variable selection method with confirmed successful results on biological datasets. MRMR finds the most informative features based on the correlation with class label with minimum redundancy among features. In other words, in the MRMR method, each feature is ranked based on not only its relevance to the target variable, but also its redundancy in the feature set [3].
Assuming fi is the ith feature and the class label is c, the Maximum-Relevance method selects the top m features relevant to the class label in Eq. (11).
To eliminate the redundancy among features, a Minimum-Redundancy criterion was defined in Eq. (12).
A sequential incremental algorithm was used to solve the simultaneous optimizations of Eqs. (7) and (5) in Eq. (9). Assuming G is the set of all features and Sm-1 number of features are already selected, then the task is to select the mth feature from the G − (Sm-1 ) so that it could maximize the single-variable relevance subtracted from the redundancy function in Eq. (13).
4 Proposed method
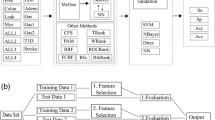
The general steps of the method (BC-NMIQ-IAMB) proposed in the current paper are shown in Fig. 1. In this method, first, the diversity of clustering is generated (using Equal Width Discretization, k-means, and Ward Link) for each feature of dataset D. Then, the Best Clustering (BC) for each feature is selected with normalized mutual information with respect to class labels (L). Next, the features are ranked based on the concept of MRMR and the initial best features are selected using a predefined threshold that is the square root of the number of features. Finally, the proposed BC-NMIQ is improved with IAMB. The measurement criteria were applied to BC-NMIQ-IAMB as the main proposed method and also to BC-NMIQ as the subsidiary proposed method. Table 2 shows the notations used in the paper.
First, each feature of a dataset (Fi) is clustered using k-means, Ward Link, and Equal Width Discretization cluster algorithm so that k could be equal to main cluster datasets to achieve equal clusters for each feature (Fi). Then, the clustering with maximum NMI with respect to the response variable (L) is selected for each Fi to construct \( {C}_i^{\ast } \). Therefore, Fi with continuous feature values was changed to the corresponding clusters with discrete values with different diversities in \( {C}_i^{\ast } \) as shown in Fig. 2.
Algorithm 2, first, clusters the features based on k-means-Ward Linkage- Equal Width Discretization and then selects the optimal clustering using NMI among all features.
The Best Clustering (BC) for each feature is selected using Maximum NMI.
Next, using the concept of normalized mutual information quantile (NMIQ), the most informative features are selected based on a filter approach in Algorithm 2, which uses an incremental search method to find the optimal features. The normalized MI between fi and fs, NMI(fi; fs) is calculated using Eq. (15).
In this paper, we propose to use the average normalized MI as a measure of redundancy between the ith feature and the subset of selected features S = {fs}, for s = 1, …, |S|, i. e. , in Eq. (16).
where ∣S∣ is the cardinality of set S. Equation (16) is a kind of correlation measure that is symmetric and takes values in [0,1]. The value of 0 indicates that feature fi and the subset S of the selected features are independent. The value of 1 indicates that feature fi is highly correlated with all features in the subset S.
The selection criterion used in NMIFS selects the feature that maximizes the measure G in Eq. (17).
The right-hand side of Eq. (17) is an adaptive redundancy penalization term, which corresponds to the average normalized MI between the candidate feature and the set of selected features [21].
The selection criterion used in NMI selects the feature that maximizes the measure. The complete NMI algorithm is in Algorithm 3.
Algorithm 3 shows how the feature subset (S) is incrementally selected using the BC-NMIQ algorithm. Finally, the Incremental Association Markov Blanket Feature Selection (IAMB) uses the subset of features (S) calculated from Algorithm 3 as an input to generate the final feature subset automatically in Algorithm 4.
Assuming the dataset with n instances and m features, Algorithms 2, 3, and 4 have the time complexity of \( O\left({n}^{\frac{3}{2}}\times logn\times m\right) \), O(\( {m}^{\frac{3}{2}}\times nlogn\Big) \), and O(m2), respectively.
5 Experimental results
In this work, the hybrid filter method was used to increase the accuracy of the machine learning model. Ten benchmark medical datasets (including binary and multi-labeled) were used to evaluate the proposed method. The evaluations are in terms of the number of selected features and the classification accuracy. Prior to introducing the datasets in Section 5.2, the classifiers used to calculate the measurement criteria are described in Section 5.1. Finally, the implementation results and the comparative results are given in Section 5.3.
5.1 Experimental setup
The experiments in this study were carried out using a standard PC with Intel Core i5, CPU 2.7 GHz, and GeForce GTX 1080 GPU, with 8 GB memory. All the experiments were performed in MATLAB using the MATLAB–R2018 libraries.
In this paper, the performance evaluation was done by randomly partitioning the original datasets into training and test sets using stratified 10-fold cross validation. The system was tuned in the validation phase so that the hyper parameters, including the depth of the decision tree, the number of trees in random forest, distance in the ward linkage, and the value of k in KNN could be cross validated for each dataset. Cross validation prevents the model from being too complex for possible over-fitting or too simple for possible under-fitting.
Decision tree (DT)
The family of decision tree algorithms is generally classified by creating a tree-like pattern in a descending way. The leaves of the decision tree correspond to the classifications. DT deals with both numerical and symbolic data.
Random Forest (RF)
RF is an ensemble technique in multiple decision trees that uses majority voting. This technique has wide applicability to pattern recognition for high-dimensional and complex problems. RF tries to decrease the high variance of decision trees.
K-nearest neighbors (KNN)
KNN is one of the machine learning algorithms that has acceptable classification performance when data are not easily separable. KNN scans through all past experiences and looks for the K closest experiences (data points), which are called the K nearest neighbors.
5.2 Experimental data
In this research, ten medical benchmark datasets were used to investigate the proposed method (BC-NMIQ-IAMBFS), as presented in Table 1. The information in Table 3 includes the number of genes, number of instances, and number of class labels. The total number of genes ranges from 2000 to 15,154.
When analyzing the performance of the partitional approaches, the information presented in Table 2 shows that k-means and Ward Linkage have outperformed the Equal Width Discretization algorithm, and the performance of Equal Width Discretization, especially in terms of the external validation, may remarkably deteriorate in some datasets (e.g., CNS and Colon). Table 4 shows that k-means with DT has competitively the best accuracy on the average. Table 5 shows that BC-NMIQ-IAMB achieves the highest average accuracies with less number of selected features than those of BC-NMIQ and BC (recalling that BC uses the entire set of features). Table 6 compares BC-NMIQ-IAMB with four existing approaches, the most accurate of which is the method we suggest.
Figure 3 shows the Receiver Operating Characteristics (ROC) curve of binary datasets, which is one of the major metrics used to check the performance of classification models. In other words, the ROC curve demonstrates to what extent the classification model is able to distinguish between classes. The ROC curve with greater Area Under Curve (AUC) shows a better separability of classes. The optimal situation is when AUC equals 1. The ROC curves in Fig. 4 are based on the DT classifier. The ROC curve is plotted with True Positive Rate (TPR) or recall in y-axis (\( Recall=\frac{TP}{FN+ TP} \)) and False Positive Rate (FPR) in x-axis (\( FPR=\frac{FP}{TN+ FP} \)). (The blue curve indicates cancer genes, and the red curve shows normal genes).
The overall goal of this paper is to select a smaller number of genes and achieve similar or better classification accuracy than using all genes. The tests of the proposed BC-NMIQ-IAMB on ten datasets using random forest classifier shows that our algorithm is better than all other compared algorithms in terms of the classification accuracy and dimension reduction. In addition, with using different evaluation measures, the proposed algorithms are found highly efficient offering better solutions than other algorithms considered in this paper, i.e., CFS [4], MC, WOA-MC [34], and D2CORFS [12]. The lower accuracies of CFS and MC compared to WOA-MC and BC-NMIQ-IAMB is predictable because CFS and MC are simple filter methods (especially CFS in which the classification accuracy decreases when the number of features exceeds in the dataset). On the other hand, D2CORFS achieves higher accuracy than CFS, MC, and WOA-MC. The proposed hybrid method is successful in reaching the best accuracy and finding the smallest subset of features, which is due to the high capability of BC-NMIQ-IAMB to model the complex problem of high dimensional data. All in all, BC-NMIQ-IAMB increases the average accuracy of similar algorithms in this domain on binary and multi-class datasets so that this increase is considerable in comparison with CFS (in both binary and multi-class datasets) as well as MC and WOA-MC (in binary datasets).
6 Conclusion
This paper proposes a feature selection approach, i.e., the hybrid BC-NMIQ-IAMB method comprising two filter methods, namely BC-NMIQ and IAMB, to find the most informative genes for the cancer classification and diagnosis. The main objective of the proposed approach is to find the smallest subset of biomarkers, which can signify the disease efficiently. The contribution of the proposed method is that it can effectively model the interactions in complex systems and is applicable to the problem of feature selection on high dimensional data. The proposed approach is evaluated on 10 popular microarray datasets and compared with some of the most recent approaches. The experimental results obtained on different datasets shows that despite the selection of a minimal subset of features, the selected genes have high influence on separating different classes. The results of the experiments demonstrate that the proposed approach achieved a high accuracy rate even with 2 or 4 highly-informative genes. There is also much work to be done in the future, for example, investigating how to improve the efficiency of algorithms; how to combine the findings of this paper with some of the current, advanced feature selection algorithms such as semi-supervised manifold learning methods and sparse classification algorithms; and/or how to apply the proposed algorithms to automatically determining the number of clusters and feature clustering with incorporating the clinical biological information.
Data availability
The datasets analyzed during the current study and the related implementation are available from the corresponding author on reasonable request.
References
Abasabadi S, Nematzadeh H, Motameni H, Akbari E (2021) Automatic ensemble feature selection using fast non-dominated sorting. Inf Syst 100:101760
Abasabadi S et al (2022) Hybrid feature selection based on SLI and genetic algorithm for microarray datasets. J Supercomput 78:19725–19753
Ahmed YA, Koçer B, Huda S, Saleh al-rimy BA, Hassan MM (2020) A system call refinement-based enhanced minimum redundancy maximum relevance method for ransomware early detection. J Netw Comput Appl 167:102753
Al-Batah M et al (2019) Gene Microarray Cancer Classification using Correlation Based Feature Selection Algorithm and Rules Classifiers. Int J Online Biomed Eng 15(8):62
Ali H, Tran SN, Benetos E, d’Avila Garcez AS (2018) Speaker recognition with hybrid features from a deep belief network. Neural Comput & Applic 29(6):13–19
Ali A et al (2019) Leveraging spatio-temporal patterns for predicting citywide traffic crowd flows using deep hybrid neural networks. In 2019 IEEE 25th international conference on parallel and distributed systems (ICPADS). IEEE
Ali A, Zhu Y, Zakarya M (2021) A data aggregation based approach to exploit dynamic spatio-temporal correlations for citywide crowd flows prediction in fog computing. Multimed Tools Appl:1–33
Alirezanejad M, Enayatifar R, Motameni H, Nematzadeh H (2020) Heuristic filter feature selection methods for medical datasets. Genomics 112(2):1173–1181
Ankerst M, Breunig MM, Kriegel HP, Sander J (1999) OPTICS: ordering points to identify the clustering structure. ACM SIGMOD Rec 28(2):49–60
Awan N, Ali A, Khan F, Zakarya M, Alturki R, Kundi M, Alshehri MD, Haleem M (2021) Modeling dynamic Spatio-temporal correlations for urban traffic flows prediction. IEEE Access 9:26502–26511
Blömer J et al (2016) Theoretical analysis of the k-means algorithm–a survey. In: Algorithm Engineering. Springer, pp 81–116
Brankovic A, Hosseini M, Piroddi L (2018) A distributed feature selection algorithm based on distance correlation with an application to microarrays. IEEE/ACM Trans Comput Biol Bioinform 16(6):1802–1815
Chandrashekar G, Sahin F (2014) A survey on feature selection methods. Comput Electr Eng 40(1):16–28
Chang H, Yeung D-Y (2008) Robust path-based spectral clustering. Pattern Recogn 41(1):191–203
Chaudhuri A, Sahu TP (2021) A hybrid feature selection method based on binary Jaya algorithm for micro-array data classification. Comput Electr Eng 90:106963
Chowdhary CL, Acharjya D (2016) A hybrid scheme for breast cancer detection using intuitionistic fuzzy rough set technique. Int J Healthc Inf Syst Inform (IJHISI) 11(2):38–61
Chowdhary CL, Acharjya D (2018) Segmentation of mammograms using a novel intuitionistic possibilistic fuzzy c-mean clustering algorithm. In: Nature Inspired Computing. Springer, pp 75–82
Debata PP, Mohapatra P (2022) Identification of significant bio-markers from high-dimensional cancerous data employing a modified multi-objective meta-heuristic algorithm. J King Saud Univ-Comput Inform Sci 34(8):4743–4755
Dimić G et al (2019) Descriptive statistical analysis in the process of educational data mining. In 2019 14th international conference on advanced technologies, systems and Services in Telecommunications (TELSIKS). IEEE
Ehlert KM, Orr MK (2019) Comparing grouping results between cluster analysis and Q-methodology. In: 2019 IEEE Frontiers in education conference (FIE). IEEE, pp 1–3
Estévez PA et al (2009) Normalized mutual information feature selection. IEEE Trans Neural Netw 20(2):189–201
Fleuret F (2004) Fast binary feature selection with conditional mutual information. J Mach Learn Res 5(9):1531–1555
Gu X, Guo J, Xiao L, Li C (2022) Conditional mutual information-based feature selection algorithm for maximal relevance minimal redundancy. Appl Intell 52(2):1436–1447
Gunasundari S, Janakiraman S, Meenambal S (2018) Multiswarm heterogeneous binary PSO using win-win approach for improved feature selection in liver and kidney disease diagnosis. Comput Med Imaging Graph 70:135–154
Hallajian B, Motameni H, Akbari E (2022) Ensemble feature selection using distance-based supervised and unsupervised methods in binary classification. Elsevier Expert Syst Appl 200:1–18
Hancer E (2020) A new multi-objective differential evolution approach for simultaneous clustering and feature selection. Eng Appl Artif Intell 87:103307
Iqbal T, Ali H (2018) Generative adversarial network for medical images (MI-GAN). J Med Syst 42(11):1–11
Lensen A, Xue B, Zhang M (2016) Particle swarm optimisation representations for simultaneous clustering and feature selection. In 2016 IEEE symposium series on computational intelligence (SSCI). IEEE
Lensen A, Xue B, Zhang M (2017) Using particle swarm optimisation and the silhouette metric to estimate the number of clusters, select features, and perform clustering. In European conference on the applications of evolutionary computation. Springer
Li J, Huang G, Zhou Y (2020) A sentiment classification approach of sentences clustering in webcast barrages. J Inf Process Syst 16(3):718–732
Mitra P, Murthy C, Pal SK (2002) Unsupervised feature selection using feature similarity. IEEE Trans Pattern Anal Mach Intell 24(3):301–312
Murtagh F, Contreras P (2012) Algorithms for hierarchical clustering: an overview. Wiley Interdiscip Rev Data Min Knowl Discov 2(1):86–97
Nakariyakul S, Casasent DP (2009) An improvement on floating search algorithms for feature subset selection. Pattern Recogn 42(9):1932–1940
Nematzadeh H, Enayatifar R, Mahmud M, Akbari E (2019) Frequency based feature selection method using whale algorithm. Genomics 111(6):1946–1955
Nguyen BH, Xue B, Zhang M (2020) A survey on swarm intelligence approaches to feature selection in data mining. Swarm Evol Comput 54:100663
Okagbue HI, Adamu MO, Anake TA (2017) Quantile approximation of the chi–square distribution using the quantile mechanics
Rathod RR, Garg RD (2017) Design of electricity tariff plans using gap statistic for K-means clustering based on consumers monthly electricity consumption data. Int J Energy Sect Manag 11:295–310
Rodriguez A, Laio A (2014) Clustering by fast search and find of density peaks. Science 344(6191):1492–1496
Rostami M, Forouzandeh S, Berahmand K, Soltani M (2020) Integration of multi-objective PSO based feature selection and node centrality for medical datasets. Genomics 112(6):4370–4384
Rostami M, Berahmand K, Forouzandeh S (2020) A novel method of constrained feature selection by the measurement of pairwise constraints uncertainty. J Big Data 7(1):1–21
Rostami M, Berahmand K, Nasiri E, Forouzandeh S (2021) Review of swarm intelligence-based feature selection methods. Eng Appl Artif Intell 100:104210
Rostami M, Berahmand K, Forouzandeh S (2021) A novel community detection based genetic algorithm for feature selection. J Big Data 8(1):1–27
Sadeghian Z, Akbari E, Nematzadeh H (2021) A hybrid feature selection method based on information theory and binary butterfly optimization algorithm. Eng Appl Artif Intell 97:104079
Sanchez EH, Serrurier M, Ortner M. (2020) Learning disentangled representations via mutual information estimation. In European conference on computer vision. Springer
Sheikhpour R, Sarram MA, Gharaghani S, Chahooki MAZ (2017) A survey on semi-supervised feature selection methods. Pattern Recogn 64:141–158
Sheng W, Liu X, Fairhurst M (2008) A niching memetic algorithm for simultaneous clustering and feature selection. IEEE Trans Knowl Data Eng 20(7):868–879
Sreedhar Kumar S et al (2019) A brief survey of unsupervised agglomerative hierarchical clustering schemes. Int J Eng Technol 8(1):29–37
Talbi E-G (2009) Metaheuristics: from design to implementation, vol 74. John Wiley & Sons
Thejas G et al (2019) Mini-batch normalized mutual information: a hybrid feature selection method. IEEE Access 7:116875–116885
Xue B, Zhang M, Browne WN (2012) Particle swarm optimization for feature selection in classification: a multi-objective approach. IEEE transactions on cybernetics 43(6):1656–1671
Yan C, Liang J, Zhao M, Zhang X, Zhang T, Li H (2019) A novel hybrid feature selection strategy in quantitative analysis of laser-induced breakdown spectroscopy. Anal Chim Acta 1080:35–42
Yang J, Ma Y, Zhang X, Li S, Zhang Y (2017) An initialization method based on hybrid distance for k-means algorithm. Neural Comput 29(11):3094–3117
Zhong W, Chen X, Nie F, Huang JZ (2021) Adaptive discriminant analysis for semi-supervised feature selection. Inf Sci 566:178–194
Zhou Y, Jin R, Hoi SCH (2010) Exclusive lasso for multi-task feature selection. In Proceedings of the thirteenth international conference on artificial intelligence and statistics. JMLR Workshop and Conference Proceedings
Zhu Z, Ong Y-S, Dash M (2007) Markov blanket-embedded genetic algorithm for gene selection. Pattern Recogn 40(11):3236–3248
Zhu J, Jang-Jaccard J, Liu T, Zhou J (2021) Joint spectral clustering based on optimal graph and feature selection. Neural Process Lett 53(1):257–273
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Asghari, S., Nematzadeh, H., Akbari, E. et al. Mutual information-based filter hybrid feature selection method for medical datasets using feature clustering. Multimed Tools Appl 82, 42617–42639 (2023). https://doi.org/10.1007/s11042-023-15143-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-15143-0