Abstract
The ensemble average diffusion propagator (EAP) obtained from diffusion MRI (dMRI) data captures important structural properties of the underlying tissue. As such, it is imperative to derive accurate estimate of the EAP from the acquired diffusion data. Taking inspiration from the theory of radial basis functions, we propose a method for estimating the EAP by representing the diffusion signal as a linear combination of 3D anisotropic Gaussian basis functions centered at the sample points in the q-space. This is in contrast to other methods, that always center the Gaussians at the origin in q-space. We also derive analytical expressions for the estimated diffusion orientation distribution function (ODF), the return-to-the-origin probability (RTOP) and the mean-squared-displacement (MSD). We validate our method on data obtained from a physical phantom with known crossing angle and on in-vivo human brain data. The performance is compared with the 3D-SHORE method of [4, 9] and radial basis function based method of [15].
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- Diffusion Propagator
- Mean Square Displacements (MSD)
- Diffusion Signal
- Radial Basis Function Method
- Normalized Mean Square Error (NMSE)
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
A classical method in dMRI is Diffusion tensor imaging (DTI) [3] which assumes the EAP to be a single Gaussian centered at the origin. However, this over-simplified assumption has limitations in voxels where there are more complicated structures. To resolve this issue, Diffusion spectrum imaging (DSI) was proposed in [17]. However, a large number of measurements and a long acquisition time makes it impractical to use DSI in clinical setting. To this end, many imaging methods have been proposed, which reduce the number of measurements by using suitable signal models or by using compressed sensing techniques. For example, Hybrid diffusion imaging (HYDI) [18], Diffusion Propagator Imaging (DPI) [5], the SHORE basis [9, 11], the Bessel Fourier basis [6], the Spherical Polar Fourier (SPF) basis [2, 4] and the spherical ridgelet basis [14] extend the spherical representation of the signal on a single shell to multiple shells with a continuous radial term. On the other hand, MAP-MRI [12] represents the diffusion signal using Hermite polynomials. Finally, the method of [19] (NODDI) estimates axonal dispersion while the CHARMED model [1] uses very high b-value to estimate the axon diameter distribution.
2 Our Contributions
In this work, we use 3D Gaussian functions for representing the diffusion signal and computing the EAP. The diffusion signal is expressed as a linear combination of Gaussian basis functions centered at several locations in the q-space at which measurements are available. This is in contrast to other mixture models, which typically center the basis functions at the origin in q-space [7, 13]. The present work is a generalization of the radial basis functions method in [15], incorporating directional anisotropic (non-radial) Gaussians for continuous representation of the diffusion signal and the propagator. Since the Fourier transform of a Gaussian is another Gaussian, one obtains simple analytical expressions for the EAP, the ODF, the return-to-the-origin probability (RTOP) and the mean-squared-displacement (MSD). We validate our method on a physical phantom data set with known fiber crossing and on in-vivo human brain data set. We also compare our method to the one using radial basis functions (RBF) [15], 3D-SHORE [9], and show that adding constraints helps in improving the performance of 3D-SHORE.
3 Signal Representation
Accurate reconstruction of high dimensional continuous functions from finite number of samples can be achieved using as a linear combination of radial basis functions centered around the given data points [8]. We use a similar methodology to represent the dMRI signal continuously in the q-space. Given a sampling of diffusion signal \(E(\boldsymbol{q})\) at N data locations \(\{\boldsymbol{q}_{1},\ldots,\boldsymbol{q}_{N}\}\) in q-space, we consider its reconstruction using
\(\boldsymbol{q}_{0} = 0\) and \(\phi _{0}(\boldsymbol{q}) = e^{-\boldsymbol{q}^{T}D_{ 0}\boldsymbol{q}}\). The tensor D is assumed to have a cylindrical shape with eigenvalues \(\sigma _{0},\sigma _{1},\sigma _{2}\) such that σ 1 = σ 2. The interpolation weights w n’s can then be computed by solving the following linear system: \(E(\boldsymbol{q}_{i}) =\sum _{ n=0}^{N}w_{n}\phi _{n}(\boldsymbol{q}_{i} -\boldsymbol{q}_{n})\text{for}i = 0,\ldots,N\) with E(0) = 1. Due to antipodal symmetry, we enforce equal coefficients for \(\phi _{n}(\boldsymbol{q} -\boldsymbol{q}_{n})\) and \(\phi _{n}(\boldsymbol{q} + \boldsymbol{q}_{n})\).
3.1 Closed Form Expressions for EAP and ODF
The EAP is given by the Fourier transform of \(\hat{E}(\boldsymbol{q})\). Since the constructed \(\hat{E}(\boldsymbol{q})\) is a linear combination of Gaussian functions, its Fourier transform is given by a linear combination of the Fourier transforms of the individual basis functions. Hence, the estimated EAP is of the form \(P(\boldsymbol{r}) = \mathcal{F}(\hat{E}(\boldsymbol{q})) =\sum _{ n=0}^{N}w_{n}\varPhi _{n}(\boldsymbol{r})\) with \(\varPhi _{0} = \mathcal{F}(\phi _{0}(\boldsymbol{q}))\) and \(\varPhi _{n} = \mathcal{F}(\phi _{n}(\boldsymbol{q} -\boldsymbol{q}_{n}) +\phi _{n}(\boldsymbol{q} + \boldsymbol{q}_{n}))\). In particular, Φ 0 is given by
A translation of a basis function leads to a phase shift of its Fourier transform, i.e. \(\mathcal{F}(\phi _{n}(\boldsymbol{q} + \boldsymbol{q}_{n})) = e^{i2\pi \boldsymbol{r}^{T}\boldsymbol{q}_{ n}}\mathcal{F}(\phi _{n}(\boldsymbol{q}))\). Hence, for n ≥ 1,
The ODF is computed from the EAP by evaluating the integral \(\varPsi (\boldsymbol{u}) =\int _{ 0}^{\infty }P(r\boldsymbol{u})r^{2}\mathit{dr}\) [17], where \(\boldsymbol{u}\) is a unit vector and r is the radial co-ordinate. From the propagator \(P(\boldsymbol{r})\), \(\varPsi (\boldsymbol{u})\) is given analytically as \(\varPsi (\boldsymbol{u}) =\sum _{ n=0}^{N}w_{n}\varPsi _{n}(\boldsymbol{u})\) with
and
3.2 Expressions for RTOP and MSD
Similarly, closed form expressions for RTOP and MSD can be computed. RTOP, which is given by \(\int _{\mathbb{R}^{3}}\hat{E}(\boldsymbol{q})d\boldsymbol{q}\), is simply \(P(0) =\pi ^{\frac{3} {2} }(w_{0}\vert D_{0}\vert ^{-\frac{1} {2} } + 2\vert D\vert ^{-\frac{1} {2} }\sum _{n=1}^{N}w_{n})\). Similarly, the MSD, which is given by \(\int _{\mathbb{R}^{2}}P(\boldsymbol{r})\|\boldsymbol{r}\|^{2}d\boldsymbol{r}\), can be computed from \(\int _{\mathbb{R}^{2}}\varPhi _{0}(\boldsymbol{r})\|\boldsymbol{r}\|^{2}d\boldsymbol{r} = \frac{1} {2\pi ^{2}} \mathop{trace}\nolimits (D_{0})\), and
3.3 The Relation to Radial Basis Functions Method
The radial basis functions method [15] is a special case of the proposed framework where radially symmetric Gaussian functions centered at the sample locations are used to represent diffusion measurement. The basis function is of the form \(\phi _{n}^{\mathrm{rbf}} = e^{-\sigma \|\boldsymbol{q}-\boldsymbol{q}_{n}\|^{2} }\) which corresponds to an isotropic tensor D = σ I 3×3. The corresponding EAP is expressed as \(P(r) =\sum _{ n=1}^{N}w_{n}\varPhi _{n}^{\mathrm{rbf}}(\boldsymbol{r})\) with
The ODF is given by \(\varPsi (\boldsymbol{u}) =\sum _{ n=1}^{N}w_{n}\varPsi _{n}^{\mathrm{rbf}}(\boldsymbol{u})\) with
3.4 Estimation Procedure
We discuss different methods for estimating the coefficients w n. From the samples at N locations in the q-space, we obtain an N + 1 dimensional vector e with the first entry being the measurement at the origin. The tensor D 0 is computed as in standard DTI [3], while the tensor D is chosen to have the same eigenvectors as D 0. One constructs an \((N + 1) \times (N + 1)\) dimensional matrix A with
for j ≥ 1 and \(i = 1,\ldots,N + 1\). We denote by w, an N + 1 vector whose entries are the coefficients w n to be estimated. A simple method to estimate w is \(w = A^{-1}e\). However, to avoid ill conditioned matrices A, a Tikhonov regularized solution is given by \(w_{\ell_{2}} = (A^{T}A +\lambda I)^{-1}A^{T}e\) where λ > 0 is a regularization parameter.
This method, however, does not account for the fact that the diffusion measurements are monotonically decreasing with increasing b-values. Moreover, the diffusion propagator should be positive and the value of the measurement at the origin is known to be one. Thus, one could numerically enforce these constraints while estimating the vector of weight w. The corresponding function then becomes: \(\min _{w}\|Aw - e\|^{2} +\lambda \| w\|^{2},\text{s.t.}Bw \geq 0,c^{T}w = 1.\) The matrix B is of the form B = [B 1 T, B 2 T]T with each column of B 1 being the difference of a Gaussian basis functions ϕ n along a given set of gradient directions at several b-value shells and each column of B 2 being the evaluation of Φ n at given set of locations for \(\boldsymbol{r}\). The vector c contains the values of the basis functions at the origin. We note that these constraints are always feasible and a trivially feasible element is given by w = [1, 0, …, 0]T.
4 Experiments
We tested our method on a data set acquired from a spherical physical phantom with a crossing angle of 45 degree [10]. We acquired ten separate scans of the phantom with the 5 different b-values \(b =\{ 1,000,2,000,3,000,4,000,5,000\}\,\mathrm{s}/\mathrm{mm}^{2}\) and each b-value shell consisted of 81 gradient directions. The ten scans were averaged to obtain the “gold-standard” data. The test data set was acquired as follows: For each of the following number of gradient directions K = { 16, 20, 24, 26, 30, 36, 42, 60, 81} we acquired the diffusion measurement over 2 b-shells corresponding to \(b =\{ 1,000,3,000\}\,\mathrm{s}/\mathrm{mm}^{2}\). Further, five repetitions were acquired for each of these data samples to test the effect of noise on signal reconstruction quality. Each acquisition had an average SNR of about 8.5.
4.1 Comparison Metrics
We used several quantitative metrics to test the ability of the proposed algorithm. Let \(\hat{E}_{\boldsymbol{x}}\) denote the reconstructed signal in the voxel at location \(\boldsymbol{x}\) and \(E_{\boldsymbol{x},\mathrm{gold}}\) be the “gold-standard” signal. The Normalized mean squared error (NMSE) in signal reconstruction was computed as
where Ω denotes the set of locations for all voxels. The estimated angle between the two principal diffusion directions (in case of crossing) was computed as the average estimated angle (EA) given by
where \(\boldsymbol{u}_{\boldsymbol{x},1}\) and \(\boldsymbol{u}_{\boldsymbol{x},2}\) denote the direction of the two peaks in the voxel at location \(\boldsymbol{x}\) and Ω 2 denote the set of locations for voxels that have two peaks. The estimated angle was computed only in the voxels where only two peaks are detected. In order to know if the recovered signal missed or exaggerated the number of peaks, we computed the percentage of false peaks (PF) given as
where Ω gold denotes the set of locations for voxels that have n gold peaks in gold standard data which equals to 2 in this case, \(\mathcal{B}\) is an indicator function whose value is 1 if \(n_{\boldsymbol{x}} = n_{\mathrm{gold}}\) and 0 otherwise.
We used all of these metrics to quantify the reconstruction quality of the data using Gaussian basis function. We used two methods to estimate the vector \(w\). For the first one, w was chosen as \(w_{\ell_{2}}\) where the ℓ 2 regularization coefficient λ is chosen such that the condition number of A T A +λ I is bounded by 107. The second estimate of w was computed by solving a constrained quadratic programming problem. We compared the proposed methods with the ℓ 1 3D-SHORE method [4], the ℓ 2 3D-SHORE method [9], the ℓ 2 3D-SHORE method with constraint of monotonic decrease of the signal along gradient directions and the radial basis functions (RBF) method [15].
4.2 Phantom Results
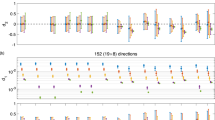
Results on the “gold standard” data (where the actual angle = 45∘) with different methods are summarized in Table 1. For the test data set, the estimated angle (averaged over the five acquisitions) with different number of gradient directions are shown in Fig. 1. The percentage of false peaks and NMSE are shown in Fig. 1b, c respectively. The parameters σ 0 = 0. 002 and σ 1 = 0. 001 were used for all the Gaussian basis functions and for all experiments done on the test data set. The estimated ODF’s with 42 gradient directions (two b-values, so a total of 84 measurements) with constrained Gaussians, constrained ℓ 2 3D-SHORE, ℓ 1 3D-SHORE and radial basis functions are shown in Fig. 2a–d, respectively.
Note that, the proposed method produces much sharper ODF’s compared to the 3D-SHORE method. Further, from the error metrics shown in Fig. 1, it becomes clear that the proposed method, while having a slightly higher error in terms of the estimated angle, is yet very successful in detecting the two peaks (i.e. significantly lower percentage of false negatives) compared to 3D-SHORE (see Fig. 1b). Further, the method of 3D-SHORE itself does much better if constraints are added, which was not done in the method presented in [4, 9].
4.3 In Vivo Results
We tested our method on in-vivo human brain data with scan parameters: b-values of {900, 2, 000, 3, 600, 5, 600} s∕mm2 with each b-value shell having 60 gradient directions. This data set was considered as the “gold-standard” data. To obtain the test data, we used two subsets of this data. The first set consisted of data with b-values \(b =\{ 900,3,600\}\,\mathrm{s}/\mathrm{mm}^{2}\) and 60 gradient directions on each shell, while the second set had the same b-values but 30 gradient directions per shell. For the rectangular region (white box) shown in Fig. 3c, the NMSE for these two sets compared to the “gold standard” are given in Table 2. The estimated ODF for the data set with 2 b-value shells and 60 gradient directions using the constrained Gaussians (proposed) and the constrained ℓ 2 3D-SHORE are shown in Fig. 3a and b.
5 Conclusion
In this work, we proposed a novel method of using anisotropic Gaussian functions centered at several locations in q-space to represent the diffusion signal and derive analytical expressions for the EAP, the MSD and RTOP. By using the same set of parameters, we showed the robustness of the proposed method to different number of gradient directions on a physical phantom data with very high noise (SNR = 8.5). We also showed that the 3D-SHORE method works better if it is constrained to ensure monotonic decrease of the signal with increasing b-value. Quantitatively, the proposed method seems to have lower error in detecting the crossing peaks compared to 3D-SHORE and RBF. A limitation of the current method is that the user has to choose the eigenvalues of the tensor for the Gaussian basis functions, which can be also be chosen by minimizing the fitting error in a leave-one-out cross-validation scheme [16].
References
Assaf, Y., Freidlin, R.Z., Rohde, G.K., Basser, P.J.: New modeling and experimental framework to characterize hindered and restricted water diffusion in brain white matter. Magn. Reson. Med. 52(5), 965–978 (2004)
Assemlal, H.E., Tschumperlé, D., Brun, L.: Efficient and robust computation of PDF features from diffusion MR signal. Med. Image Anal. 13(5), 715–729 (2009)
Basser, P., Mattiello, J., LeBihan, D.: Estimation of the effective self-diffusion tensor from the NMR spin echo. J. Magn. Reson., Ser. B 103(3), 247–254 (1994)
Cheng, J., Ghosh, A., Jiang, T., Deriche, R.: Model-free and analytical EAP reconstruction via spherical polar Fourier diffusion MRI. In: Medical Image Computing and Computer-Assisted Intervention–MICCAI 2010, pp. 590–597. Springer (2010)
Descoteaux, M., Deriche, R., Le Bihan, D., Mangin, J.F., Poupon, C.: Multiple q-shell diffusion propagator imaging. Med. Image Anal. 15(4), 603–621 (2011)
Hosseinbor, A.P., Chung, M.K., Wu, Y.C., Alexander, A.L.: Bessel fourier orientation reconstruction (bfor): an analytical diffusion propagator reconstruction for hybrid diffusion imaging and computation of q-space indices. NeuroImage 64, 650–670 (2013)
Jian, B., Vemuri, B.C.: A unified computational framework for deconvolution to reconstruct multiple fibers from diffusion weighted MRI. IEEE Trans. Med. Imaging 26(11), 1464–1471 (2007)
Lanczos, C.: Applied Analysis. Courier Dover Publications (1988)
Merlet, S.L., Deriche, R.: Continuous diffusion signal, EAP and ODF estimation via compressive sensing in diffusion MRI. Med. Image Anal. 17(5), 556–572 (2013)
Moussavi-Biugui, A., Stieltjes, B., Fritzsche, K., Semmler, W., Laun, F.B.: Novel spherical phantoms for Q-ball imaging under in vivo conditions. Magn. Reson. Med. 65(1), 190–194 (2011)
Özarslan, E., Koay, C., Shepherd, T., Blackb, S., Basser, P.: Simple harmonic oscillator based reconstruction and estimation for three-dimensional q-space MRI. In: ISMRM 17th Annual Meeting and Exhibition, Honolulu, p. 1396 (2009)
Özarslan, E., Koay, C.G., Shepherd, T.M., Komlosh, M.E., İrfanoğlu, M.O., Pierpaoli, C., Basser, P.J.: Mean apparent propagator (MAP) MRI: A novel diffusion imaging method for mapping tissue microstructure. NeuroImage 78, 16–32 (2013)
Rathi, Y., Gagoski, B., Setsompop, K., Michailovich, O., Grant, P.E., Westin, C.F.: Diffusion propagator estimation from sparse measurements in a tractography framework. In: Medical Image Computing and Computer-Assisted Intervention–MICCAI 2013, pp. 510–517. Springer (2013)
Rathi, Y., Michailovich, O., Setsompop, K., Bouix, S., Shenton, M.E., Westin, C.F.: Sparse multi-shell diffusion imaging. In: Medical Image Computing and Computer-Assisted Intervention–MICCAI 2011, pp. 58–65. Springer (2011)
Rathi, Y., Niethammer, M., Laun, F., Setsompop, K., Michailovich, O., Grant, P.E., Westin, C.F.: Diffusion propagator estimation using radial basis functions. In: Computational Diffusion MRI and Brain Connectivity, pp. 57–66. Springer (2014)
Rippa, S.: An algorithm for selecting a good value for the parameter c in radial basis function interpolation. Adv. Comput. Math. 11(2–3), 193–210 (1999)
Wedeen, V.J., Hagmann, P., Tseng, W.Y.I., Reese, T.G., Weisskoff, R.M.: Mapping complex tissue architecture with diffusion spectrum magnetic resonance imaging. Magn. Reson. Med. 54(6), 1377–1386 (2005)
Wu, Y., Alexander, A.: Hybrid diffusion imaging. NeuroImage 36(3), 617–629 (2007)
Zhang, H., Schneider, T., Wheeler-Kingshott, C.A., Alexander, D.C.: NODDI: Practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage 61(4), 1000–1016 (2012)
Acknowledgements
This work has been supported by NIH grants: R01MH097979 (Rathi), R01MH074794 (Westin), P41RR013218, P41EB015902 (Kikinis, Core PI: Westin), and Swedish research grant VR 2012-3682 (Westin).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer International Publishing Switzerland
About this paper
Cite this paper
Ning, L., Michailovich, O., Westin, CF., Rathi, Y. (2014). Diffusion Propagator Estimation Using Gaussians Scattered in q-Space. In: O'Donnell, L., Nedjati-Gilani, G., Rathi, Y., Reisert, M., Schneider, T. (eds) Computational Diffusion MRI. Mathematics and Visualization. Springer, Cham. https://doi.org/10.1007/978-3-319-11182-7_13
Download citation
DOI: https://doi.org/10.1007/978-3-319-11182-7_13
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-11181-0
Online ISBN: 978-3-319-11182-7
eBook Packages: Mathematics and StatisticsMathematics and Statistics (R0)