Abstract
Objective: To explore the efficacy of machine learning techniques in identifying Metabolic Syndrome (MetS) and examine the performance of models when applied to target populations with different distributions. Methods: This study utilizes data from the National Health and Nutrition Examination Survey (NHANES) and local physical examinations, where MetS is diagnosed based on the International Diabetes Federation (IDF) standards. We first employ demographic and blood test data from NHANES and predicted MetS using machine learning models (including MLP, Logistic Regression, Random Forest, XGBoost, Catboost, and Multi-layer Perceptron), and then test these predictions on different population data. Results: Models employing 59 features demonstrate commendable performance in the NHANES test set (same population testing), with the MLP model exhibiting the best performance (AUROC= 0.93). Models constructed with 32 features (excluding height, weight, and certain blood test information) still show promising results (MLP AUROC = 0.89). However, when the models are tested on the local physical examination dataset (cross-population testing), there is a substantial decline in performance (MLP AUROC = 0.71). Conclusion: Machine learning techniques can predict MetS on the NHANES dataset with high accuracy. Due to the distribution shift, examined machine learning models perform better in the setting with same population distribution.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Metabolic syndrome (MetS) is an early subclinical syndrome characterized by the aggregation of multiple risk factors of metabolic diseases. The main indicators of MetS include abdominal obesity, elevated blood pressure, hyperglycemia, hyperlipidemia, and low high-density lipoprotein cholesterol. Existing researches have shown that MetS is closely associated with the risk of numerous chronic non-communicable diseases, such as coronary heart diseases, stroke, diabetes, and hypertension [1]. As a result, it has become a significant disease burden in the field of public health and a focal issue in recent population health management (PHM) [2, 3].
Machine learning (ML) technology has been increasingly utilized in clinical areas [4]. The timely and accurately detection and diagnosis of diseases through machine learning remains a consistent and prominent topic in scientific research. Concurrently, the widespread application of AI technology has fostered the rapid growth of PHM, which aims to enable all individuals within a specific population to maintain and enhance their health. With the evolution and implementation of advanced machine learning models, population health research can model multidimensional health data from large cohorts to extract valuable insights, further applying these findings to PHM [5].
The diagnosis of metabolic syndrome (MetS), according to the diagnostic criteria in the joint statement by the International Diabetes Federation (IDF), involves multiple physical meOSasurements and laboratory tests, such as waist circumference, blood pressure, blood sugar, triglycerides, and high-density lipoprotein cholesterol, which obviously presents certain limitations in the context of PHM or individual health management. Among the others, waist circumference is the indicator that is fairly stable and can be conveniently measured. The blood pressure and blood sugar require multiple measures to confirm the reliability. The triglycerides, and high-density lipoprotein cholesterol need to be tested in hospital with lab analyser. The gold standards for the blood sugar, riglycerides, and high-density lipoprotein cholesterol are tested intrusively. In this case, out-of-hospital diagnosis of MetS has become challenging, thus, the exploiting of other valuable indicators and the use of ML approaches to assist the remote monitoring and detection in the early stage is immensely valuable.
In clinical scenarios, if a patient is suspected to have MetS, endocrinologists will be involved in the diagnosis and treatment pipeline. Patients will receive advice from endocrinologists on lifestyle management. However, this process can be time-consuming and labor-intensive, and there is also a certain risk of missed or misdiagnosis.
Given these challenges, this research aims achieve early identification of MetS using ML technologies with population health data. Better classification of MetS will assist the timely detection of MetS and thus improve personal health management.
2 Datasets and Methods
2.1 Datasets
Data Sources. In this study, we utilized the public database from the National Health and Nutrition Examination Survey (NHANES), which is a national survey program conducted by the U.S. National Center for Health Statistics. Initiated in 1960, this program periodically evaluates the health and nutritional status of the American population, collecting relevant clinical, demographic, and nutritional data. NHANES stands as one of the largest ongoing health surveys in the US population, offering a comprehensive dataset on population health, which can be employed to research various health issues, such as MetS, diabetes, and cardiovascular diseases. The NHANES survey adopts multistage sampling techniques, including random sampling, stratified sampling, and cluster sampling. Participants undergo questionnaire interviews, physical examinations, and biological sample collections. The comprehensive NHANES dataset encompasses numerous physiological indicators, biochemical markers, nutritional indices, disease diagnoses, medication usage, and health behaviors [6, 7].
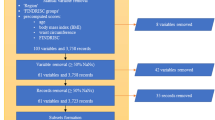
We used the NHANES data from 1999–2018, encompassing 101,316 individuals, and a local health examination dataset, encompassing 10,446 individuals, forming the original datasets for this study.
Diagnosis of Metabolic Syndrome. In this study, we diagnosed MetS based on the standards and definitions set forth in the joint statement by the IDF [8]. According to this criterion, MetS is diagnosed when three or more of the following five components are present:
-
1)
Increased waist circumference (\(\ge \)88cm for females, \(\ge \)102cm for males);
-
2)
Elevated triglycerides (TG) (\(\ge \)150mg/dL) or currently undergoing treatment for hyperlipidemia;
-
3)
Reduced high-density lipoprotein cholesterol (HDL-c) (<40mg/dL for males, <50mg/dL for females) or currently undergoing treatment for low HDL-c;
-
4)
Elevated blood pressure (systolic blood pressure (SBP) \(\ge \)130mmHg or diastolic blood pressure (DBP) \(\ge \)85mmHg or both) or currently on antihypertensive treatment or with a history of hypertension;
-
5)
Elevated fasting blood glucose (FBG) (\(\ge \)100mg/dL) or currently undergoing treatment for hyperglycemia.
Using the questionnaire information and biochemical data of participants provided by NHANES, such as blood glucose, triglycerides, and HDL-C, we diagnosed each component [9]. NHANES did not collect laboratory test information for HDL-C during the survey years from 1999 to 2004. Thus, we substituted this with medication treatment information reported in the questionnaire. Ultimately, 55,684 participants with sufficient information were diagnosed, of which 19,530 (35.1%) were categorized as MetS.
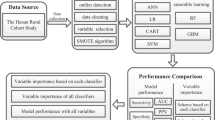
Features. As is a specialized, large-scale cross-sectional study, NHANES data encompasses various features ranging from demographic, anthropometric, to blood test variables. Given the distinct feature sets in NHANES and local health examination datasets, we utilized the overlapping feature subset in this study. We primarily based our choices on the features available in the local health examination dataset. Firstly, after excluding indicators for MetS diagnosis, we identified common demographic, anthropometric, and blood test features present in both the local health examination dataset and the NHANES dataset (a total of 59 features). Due to the issue of missing data in the local physical examination dataset, we ultimately established two feature settings encompassing 59 features and 32 features respectively, and three different experiments (Table 1). Specifically, Experiment 1 involved modeling using 59 variables in the NHANES dataset and testing in an independent validation set selected from the NHANES population. Experiments 2 and 3 utilized the same NHANES training data, with an input of 32 features, and were tested on the independent validation set from NHANES and the local dataset, respectively. Table 2 illustrates the full names and corresponding abbreviations of the features.
2.2 Methods
Data Preprocessing. The preprocessing of the data commenced with the identification of outliers in each column, those values falling outside the range (<Q1-1.5\(\times \)IQR, >Q3+1.5\(\times \)IQR) were regarded as outliers. Adopting a stratified approach based on varied age and gender demographics, any missing value in each feature was substituted with the mean value of that particular feature derived from specific age-gender subgroup. For categorical variables, missing values were imputed with a distinct value and subsequently transformed into dummy variables.
The NHANES dataset was splitted into a training set (38,978 samples) and a test set (16,706 samples) randomly with the “train_test_split” function in the “scikit-learn” package, while the whole local dataset was employed as an independent validation test set (10,446 samples). Subsequently, using the “QuantileTransformer” function in the “scikit-learn” package (Python, version 0.23.2) to normalise the datasets. This non-linear and independent transformation technique converts raw values into uniform distribution values sampled from the estimated cumulative distribution function of the feature. To mitigate the potential adverse effects of sample imbalance on training efficacy, the Synthetic Minority Over-sampling Technique (SMOTE) was used to oversample the MetS positive population within the training set. Table 3 presents the sample size for each dataset.
Models. We employed various machine learning models including Logistic Regression (LR), Random Forest (RF), XGBoost, Catboost, as well as the Multilayer Perceptron (MLP). We used 10-fold cross-validation and optimised each model, subsequently training the models with these optimal settings [10]. For the MLP model, the hyper-parameters and model architectures were optimised by a random search, with the model then being trained using this configuration.
Evaluation. Accuracy, Precision, Recall, F1-score, and the Area Under the ROC curve (AUROC) are used to evaluating the performance of the models. Accuracy measures the proportion of samples correctly classified out of the total, representing the model’s ability to classify correctly. Precision indicates the proportion of positive predictions that were actually positive, reflecting the precision of the model’s classification. Recall represents the proportion of actual positive samples that the model correctly predicted as positive, indicating the model’s coverage of positive samples. The F1-score is the harmonic mean of Precision and Recall, serving as an integrated metric to assess the classification performance of the model. AUROC, determined by calculating the area under the ROC curve, depicts the relationship between the true positive rate and the false positive rate across different thresholds, revealing the classifier’s performance under various thresholds.
3 Results
Firstly, we trained models using 59 features and 32 features respectively in the NHANES dataset and evaluated the models on a unified hold-out test set.
As shown in Table 4, all five models achieved satisfactory results, with nearly all evaluation metrics exceeding 0.8. Among all five models, the MLP demonstrated the best performance, achieving top-tier results in every performance evaluation metric. The LR shows the best in precision. The nonlinear ensemble models like XGBoost and Catboost, although they had high Recall values, performed worse than MLP and LR.
Additionally, when comparing the results of models with 59 features against those with 32 features in NHANES only, it is evident that, although there was a decline in performance across all models, the magnitude of this decline wasn’t significant. This implies that after eliminating body measurements like height, weight, and additional blood test features, the models were not significantly affected.
By evaluate the performance of the models trained on NHANES when applied to different populations of local dataset (with 32 features), as shown in Table 5, it indicated that the MLP still outperformed the others. Overall, compared to their counterpart on the NHANES dataset, all the models witnessed a substantial decline in performance. For instance, the AUROC score of the best-performing model, MLP, dropped from 0.89 to 0.71.
4 Discussion
In our experiments, we explored a variety of machine learning models for MetS classification based on demographic and blood test indicators. Through multiple model and dataset configurations, we discerned that the MLP model showed superior performance in this context. It is noted that the inclusion or exclusion of variables like height and weight, which are conventionally strongly associated with MetS, did not have a significant impact on model performance. Most crucially, our experiment exhibited considerable variability in model performance when applied across different populations. This underscores a potential distribution shift across various regions and demographic characteristics. Hence, transferring a model to be applied to a different population distribution necessitates a judicious approach. Enhancing the model’s generalization capability through continual learning strategies remains a future objective of this study.
References
Grundy, S.M., et al.: Diagnosis and management of the metabolic syndrome: an American heart association/national heart, lung, and blood institute scientific statement. Circulation 112, 2735–2752 (2005)
Mottillo, S., et al.: The metabolic syndrome and cardiovascular risk a systematic review and meta-analysis. J. Am. Coll. Cardiol. 56, 1113–1132 (2010)
Zhang, L., Guo, Z., Wu, M., Hu, X., Xu, Y., Zhou, Z.: Interaction of smoking and metabolic syndrome on cardiovascular risk in a Chinese cohort. Int. J. Cardiol. 167, 250–253 (2013)
Foster, K.R., Koprowski, R., Skufca, J.D.: Machine learning, medical diagnosis, and biomedical engineering research - commentary. Biomed. Eng. Online 13, 94 (2014). https://doi.org/10.1186/1475-925X-13-94
Kononenko, I.: Machine learning for medical diagnosis: history, state of the art and perspective. Artif. Intell. Med. 23(1), 89–109 (2001)
Zipf, G., Chiappa, M., Porter, K.S., Ostchega, Y., Lewis, B.G., Dostal, J.: National health and nutrition examination survey: plan and operations, 1999–2010. Vital Health Stat 1(56), 1–37 (2013)
National Health and Nutrition Examination Survey data. Hyattsville (MD): US Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Health Statistics (2016). https://www.cdc.gov/Nchs/Nhanes/survey_methods.htm
Alberti, K.G., Zimmet, P., Shaw, J.: IDF epidemiology task force consensus group. The metabolic syndrome-a new worldwide definition. Lancet 366(9491), 1059–1062 (2005). https://doi.org/10.1016/S0140-6736(05)67402-8
Zhu, F., et al.: Elevated blood mercury level has a non-linear association with infertility in US women: data from the NHANES 2013–2016. Reprod. Toxicol. 91, 53–58 (2020)
Kohavi, R.: A study of cross-validation and bootstrap for accuracy estimation and model selection. IJCAI. 14(2), 1137–1145 (1995)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Liu, C., Liu, J., Liu, Z., Yang, Y. (2024). Machine Learning-Based Metabolic Syndrome Identification. In: Qi, J., Yang, P. (eds) Internet of Things of Big Data for Healthcare. IoTBDH 2023. Communications in Computer and Information Science, vol 2019. Springer, Cham. https://doi.org/10.1007/978-3-031-52216-1_8
Download citation
DOI: https://doi.org/10.1007/978-3-031-52216-1_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-52215-4
Online ISBN: 978-3-031-52216-1
eBook Packages: Computer ScienceComputer Science (R0)