Abstract
Radiology plays an essential role in the identification of pathologies; however, image interpretation and the guarantee of accurate diagnoses continue to represent a challenge that involves expert radiologists. This study proposes a model to identify pneumonia in chest X-rays using the deep transfer learning technique, where five pre-trained network architectures and a classifier are tested. The images used in this work are categorized into bacterial pneumonia, viral pneumonia, and normal cases. The pre-trained models used include DenseNet201, MobileNet, VGG16, VGG19 and ResNet50. A support vector machine is used as a classifier. Results show that the ResNet50 model performs best in binary classification (pneumonia vs. non-pneumonia) with 98.1% accuracy and 98.7 F1-score. For multiclassification, VGG19 performs best with an accuracy of 84.7% and an average F1-score of 81.1%. The methodology employed proved to be competent and outstanding when compared to other studies in the state of the art.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
The probability of lung disease is notably elevated, particularly in lower-middle-income nations undergoing development, where countless individuals are exposed to poverty and air pollution. Based on estimations by the World Health Organization (WHO), over 4 million premature deaths happen every year due to household air pollution, which can lead to the emergence of health issues like asthma and pneumonia. Pneumonia is an infectious condition impacting either one or both lungs, resulting in the air sacs, known as alveoli, becoming filled with fluid or pus. Pneumonia can be caused by bacteria, viruses, or fungi. This illness poses a significant public health concern and stands as a leading contributor to illness and death in both Mexico and worldwide. It is estimated that pneumonia caused 921,000 deaths in children under 5 years of age in 2015 globally, which represents 15% of all deaths in children under 5 years of age worldwide, a considerable figure related to this disease [1].
To diagnose pneumonia, a doctor reviews your medical history, performs a physical exam, and orders diagnostic tests, such as a chest X-ray. Diagnosing pulmonary diseases like pneumonia from chest X-rays or computed tomography scans can be challenging, often needing experienced physicians or radiology specialists to interpret the signs due to similar-looking diseases in the images. This sometimes requires additional time or studies for an accurate diagnosis. Moreover, issues like low resolution or varying characteristics of the images can further complicate identification. Therefore, developing diagnostic systems to assist in decision-making for lung disease diagnosis is valuable [2].
In the last decade, deep learning methods, specifically convolutional neural networks (CNNs), have been chosen as they are able to automatically learn multiple invariant features in signals or images for a given task. Because of their feature extraction ability, CNNs have proven to perform well in many applications showing strong robustness against geometric distortions, skew, scale, etc. Furthermore, CNNs trained with large amounts of data (images) in a large scenario of a demanding task can be used to extract image features from another particular context and perform efficiently; this technique is known as deep transfer learning (DTL). The objective of this work is to develop an algorithm capable of identifying pneumonia in chest X-ray images based on the DTL technique, using pre-trained convolutional network architectures as feature extractors and a machine learning classification model. A recent review reported that, regarding the use of DTL in medical image classification tasks, the VGG16 and DenseNet networks have been used more frequently in lung X-ray studies [3]. In this way, the authors of this study decided to explore the performance that the different networks would have for classification and define which of them had the best performance. Thus, five networks were tested that present differences in terms of complexity, number of parameters, depth, size, etc. It is important to highlight that while there are numerous studies that utilize DTF, a majority of them are centered around fine-tuning pre-trained networks. Notably, our proposal surpasses the algorithms found in the current state of the art [4,5,6,7,8].
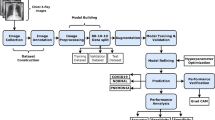
2 Methodology
This section outlines the steps taken to create and evaluate the proposed classification model. It starts by introducing the database employed, followed by clarifying the method for extracting features via pre-trained convolutional neural networks. The training of the classifier is then elaborated upon, and lastly, the conducted performance tests on the generated models are described.
The algorithms were developed using the Python programming language. The Keras library was used to handle the convolutional neural networks [9], and the Sklearn library was used to develop the support vector machine-based classifier model [10].
2.1 Database
This project utilized images sourced from the Kaggle website, a repository that offers a wide range of beneficial databases for data science projects. The specific database used, named “Chest X-ray Images”, comprises X-ray images of patients with bacterial pneumonia, viral pneumonia, and patients without the disease [11].
The database is organized into training and test sets, and contains images labeled as normal, bacterial pneumonia and viral pneumonia. The images were stored in grayscale 8-bit depth.jpeg format, and their sizes ranged from 494 × 151 pixels to 2024 × 2036 pixels (width × height). Table 1 below provides a summary of the number of images for each class in each set of the database.
2.2 Feature Extraction by Deep Transfer Learning
Feature extraction by DTL refers to a technique in deep learning where pre-trained neural network models are used as a starting point to extract relevant features from different datasets. Transfer learning leverages the knowledge and representations learned by a model on a large dataset to improve performance on a smaller or different dataset.
In DTL, the initial layers of a pre-trained model, typically trained on a large-scale dataset (such as ImageNet), are used as a feature extractor. These initial layers are responsible for learning low-level features such as edges, textures, and shapes, which are generally applicable across various visual tasks. By freezing the parameters of these layers and removing the final classification layers, the pre-trained model can be transformed into a feature extraction network [3]. When constructing a classifier model using the technique of deep transfer learning with feature extraction, it is necessary to employ a machine learning model that is trained taking the extracted features from the pre-trained networks as input and subsequently performs inference on the test observations. Commonly used machine learning models include support vector machines, random forests, k-nearest neighbors, among others. More details about this technique can be found at [3, 12].
In this study, the pre-trained networks VGG16, VGG19, ResNet50, DenseNet201, and MobileNet were utilized for feature extraction from images. These networks were pre-trained on the ImageNet dataset, a renowned dataset for object recognition [13].
The networks were fed resized database images of 224×224 pixels, with intensity values scaled between 0 and 1. Each network produced an output feature vector with sizes of 512 (VGG16 and VGG19), 1024 (DenseNet and MobileNet), and 2048 (ResNet50) for each image.
2.3 Classifier Training
A Support Vector Machine (SVM) with a Gaussian radial basis kernel function was chosen as the classification model. Five models were trained, each corresponding to a network used for feature extraction. A matrix of feature vectors from the training set images was used as input for SVM training. The regularization hyperparameter ‘C’ of the SVM loss function was determined through a grid search with possible values, utilizing 5-fold cross-validation.
2.4 Experimentation and Evaluation
Two experiments were conducted: the first focused on creating multiclassification models to distinguish normal, bacterial pneumonia, and viral pneumonia classes within the database. The second experiment involved binary classification models to identify normal images from those with pneumonia, without specifying the type of pneumonia.
For the second experiment, bacterial and viral pneumonia image files were pooled, and the task consisted of discriminating patients with pneumonia vs. normal. Since in this experiment, the number of total pneumonia images (3875) almost tripled the number of normal case images (1341) in the training set, it was decided to artificially create a larger number of normal case images with the intention of balancing the number of images from both classes. To achieve this, the data augmentation technique was applied. In this process, a subset of normal class images (N = 2540) underwent random rotations (within the range of 0 to ± 15°), zooming (within the range of 0 to ± 15%), horizontal and vertical shifts (within the range of 0 to ± 10%), and shearing (within the range of 0 to ± 15%). These augmented images were then incorporated into the training set alongside the pre-existing ones.
To assess the effectiveness of the constructed classifier models, evaluation metrics including classification accuracy (Acc), recall (Rec), precision (Pre), and F1-score (F1) were computed solely using images from the test set. Equations 1 to 4 were employed to calculate these metrics.
where tp and tn mean true positives and true negatives respectively, and fp and fn mean false positives and false negatives respectively. To calculate the metrics in the multilabel classification task, the strategy of one vs. all was used.
3 Results
This section presents and describes the results of this study. Various deep learning architectures were utilized for feature extraction and subsequent training and testing of classification models. The performance of these models in relation to the two proposed experiments is presented, employing the performance metrics discussed earlier.
Table 2 shows the performance obtained by the multiclassification models using the different pre-trained neural network architectures.
As can be seen in Table 2, the model trained with the features extracted using the VGG19 network presented the highest accuracy in the classification of the 3 classes of images. While this model demonstrates high sensitivity (recall) in detecting patterns of bacterial pneumonia and normal cases with 89.6% and 98.2% respectively, its sensitivity in identifying images of viral pneumonia is notably low at 54.5%. In fact, all models exhibited strong sensitivity for normal cases and bacterial pneumonia, yet their sensitivity for identifying viral pneumonia was consistently lower. Notably, the model utilizing ResNet50 exhibited the highest sensitivity for this class, albeit reaching only 57.4%. On the other hand, the DenseNet201 and MobileNet models demonstrated high precision in detecting viral pneumonia at 96.1% and 94.3% respectively. However, their low sensitivity indicates they may be misclassifying many viral pneumonia cases as normal or bacterial pneumonia. This is further supported by their low F1-score values.
Figure 1 shows the confusion matrix for evaluating the model using the VGG19 network. It reveals that the model committed 5 errors in identifying normal patient images, where one was wrongly labeled as viral pneumonia and 4 as bacterial pneumonia. For bacterial pneumonia identification, the model inaccurately classified 14 instances as normal and 44 as viral pneumonia. The main confusion within the model is evident between the two pneumonia categories.
A summary of the evaluation of the models in relation to the binary classification task (normal vs. pneumonia) is presented in Table 3.
As indicated in Table 3, the ResNet50 model exhibited the highest performance, boasting a precision of over 99.3% in identifying positive pneumonia cases (regardless of the pneumonia type) and an F1-score of 98.7%. Notably, all models demonstrated outstanding performance, achieving sensitivities exceeding 96% and F1 scores surpassing 96%.
Figure 2 shows the confusion matrix of the model test using the ResNet50 network. Notably, model misclassification was observed in only 20 out of a total of 1,034 images, resulting in a classification error rate of 1.9%. It is worth highlighting that the model also exhibits substantial specificity, achieving a remarkable value of 98.1%.
Prior to training the models, data augmentation was applied to balance the numbers of normal and pneumonia cases using the methodology described in Sect. 2.4. Initial results without data augmentation indicated that the models never exceeded 91% accuracy (with specificity values below 80%). Consequently, the decision was made to employ the data augmentation technique.
Table 4 presents a summary of the best results obtained in previous studies that utilize the same database as our work to perform the binary classification task (normal vs. pneumonia).
Table 4 displays that our model, based on ResNet50 + SVM, achieved the highest precision and F1-score (99.3% and 98.7%) among reviewed studies. It was second only to Hossain et al. in accuracy and sensitivity. Notably, our model’s advantage lies in simplicity, using features from one network, unlike Hossain’s model which combines outputs from five networks: ResNet18, Xception, InceptionV3, DenseNet121, and MobileNetV3.
4 Conclusions
Identifying lung diseases like pneumonia in X-ray images through automated computer vision algorithms is challenging due to the resemblance of patterns and characteristics to other diseases in these images.
This project developed a model to identify pneumonia in X-ray images, utilizing deep transfer learning for feature extraction and an SVM classifier. Two tasks were executed: a multi-classification task for identifying bacterial, viral pneumonia, and normal images, and a binary classification task for pneumonia versus normal cases. The VGG19 model showed the best performance in the three-class classification task with an average precision of 82.4%, sensitivity of 80.8%, and F1-score of 81.1%. Furthermore, the ResNet50-based model outperformed other models in both this work and state-of-the-art models for binary classification, achieving a precision of 99.3% and an F1 score of 98.7%.
The results of this project demonstrate the reliability of the use of models designed using the deep transfer learning technique for the task of detecting pneumonia in X-ray images. Bearing in mind that timely identification of the disease is essential for adequate treatment and thus achieve the recovery of the patient, the development of this type of models would open the possibility of clinical applications in which they can serve as a support tool in decision-making for the diagnosis of this type of pathology.
References
McAllister, D., et al.: Global, regional, and national estimates of pneumonia morbidity and mortality in children younger than 5 years between 2000 and 2015: a systematic analysis. Lancet Glob. Heal. 7(1), e47–e57 (2019)
Bharati, S., Podder, P., Mondal, M.: Hybrid deep learning for detecting lung diseases from X-ray images. Informatics Med. Unlocked 20, 100391 (2020)
Morid, M., Borjali, A., Del Fiol, G.: A scoping review of transfer learning research on medical image analysis using ImageNet. Comput. Biol. Med. 128, 104115 (2021)
Liang, G., Zheng, L.: A transfer learning method with deep residual network for pediatric pneumonia diagnosis. Comput. Methods Programs Biomed. 187, 104964 (2020)
Jain, R., Nagrath, P., Kataria, G., Kaushik, V., Hemanth, D.: Pneumonia detection in chest X-ray images using convolutional neural networks and transfer learning. Measurement 165, 108046 (2020)
Patel, M., Sojitra, A., Patel, Z., Bohara, M.: Pneumonia Detection Using Transfer Learning. Int. J. Eng. Res. Technol. 10(10), 252–261 (2021)
Hashmi, M., Katiyar, S., Keskar, A., Bokde, N., Geem, Z.: Efficient pneumonia detection in chest xray images using deep transfer learning. Diagnostics 10(6), 417 (2020)
Dey, N., Zhang, Y., Rajinikanth, V., Pugalenthi, R., Raja, N.: Customized VGG19 architecture for pneumonia detection in chest X-rays. Pattern Recognit. Lett. 143, 67–74 (2021)
Chollet F., et al.: Keras, 2015. https://keras.io, Accessed: 15 May 2023
Pedregosa, F., et al.: Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011)
Mooney, P.: Chest x-ray images (pneumonia), kaggle (2018). https://www.kaggle.com/datasets/paultimothymooney/chest-xray-pneumonia, Accessed 15 Mar 2023
Tan C., et al.: A survey on deep transfer learning, In: International Conference on Artificial Neural Networks, pp. 270–279 (2018)
Russakovsky, O., et al.: ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. 115(3), 211–252 (2015)
Sharma, S., Guleria, K.: A Deep Learning based model for the Detection of Pneumonia from Chest X-Ray Images using VGG-16 and Neural Networks. Procedia Comput. Sci. 218, 357–366 (2023)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Campos-Lopez, Z., Diaz-Roman, J., Mederos-Madrazo, B., Gordillo-Castillo, N., Cota-Ruiz, J., Mejia-Muñoz, J. (2024). Identification of Pneumonia with X-ray Images Using Deep Transfer Learning. In: Flores Cuautle, J.d.J.A., et al. XLVI Mexican Conference on Biomedical Engineering. CNIB 2023. IFMBE Proceedings, vol 96. Springer, Cham. https://doi.org/10.1007/978-3-031-46933-6_4
Download citation
DOI: https://doi.org/10.1007/978-3-031-46933-6_4
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-46932-9
Online ISBN: 978-3-031-46933-6
eBook Packages: EngineeringEngineering (R0)