Abstract
A notable challenge in modern biomedicine is the development of innovative therapies that mitigate resistance to conventional antibiotics. Antimicrobial peptides (AMPs), represent a promising source of new antibiotics due to their broad-spectrum activity and low incidence of resistance development, even against new infectious diseases. In nature, anuran amphibians (such as frogs and toads) are the largest source of recorded AMPs. However, salamanders and newts (such as the Mexican axolotl) have received less attention despite the fact that some promising bioactive peptides have been detected in their skin secretions. The axolotl is a critically endangered caudate amphibian endemic from Xochimilco’s Lake in Mexico, which is considered a biological model par excellence for genomic studies related to salamanders. In the present study, the wide availability of genomic resources of the axolotl was used to investigate the presence of possible new AMPs with biomedical potential, whose findings could be also extensive to other salamander species unexplored. In the axolotl, seventeen different coding transcripts for presumptive antimicrobial peptides, such as Leap2, Cathelicidins, β-Defensin, Hepcidins, Transferrins, and Cystatin-C were identified, whose transcriptional expression is mainly concentrated in the liver and spleen. This could potentially foster the development of new pharmacological treatments in the future.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Antimicrobial peptides (AMPs) are organic polymers of amino acids that are produced endogenously by numerous multicellular metazoans in order to protect the host from pathogenic microorganisms [1]. Therefore, AMPs are also known as defense peptides due to their involvement in innate immunity [1]. Among the various characteristics described for AMPs, there is a relatively short length (\(\sim\)12 to 100 residues), a net positive charge ranging from +2 to +9, as well as a certain amphipathicity that enables them to dissolve in aqueous environments [1]. By exhibiting antimicrobial and immunomodulatory activities, AMPs are humoral factors able to activate immune response mechanisms such as phagocytosis, prostaglandin release, neutralization of lipopolysaccharide effects, immune cells recruitment at inflammatory sites, wound healing, and promotion of angiogenesis, between others [1]. The largest source of AMPs found in literature records are anuran amphibians, which account for \(\sim\)30% (mainly frogs followed by toads), being some typical classes described of amphibian AMPs the β-Defensins, Cathelicidins, Hepcidins, Cystatins, Bombinins, Magainins, Dermaseptins, Esculentins, among many others [1, 2]. In fact, some classes of AMPs are characterized by containing disulfide bridges in their structure, being commonly expressed in epithelial cells and leukocytes. In addition, many AMPs also give to a prepropetide consisting of a secretion signal peptide, a linker proregion or protease inhibitor domain, as well as a highly variable mature peptide segment that is responsible for the antimicrobial biological function [2]. Particularly, most of scientific attention for the identification and characterization of novel bioactive peptides have been directed towards the anuran families of Pipidae and Bombinatoridae, given their global distribution and degree of diversification [3]. Meanwhile, genera from the orders Caudata and Apoda, which include tailed and limbless amphibians respectively, have been largely overlooked in this regard [3].
The axolotl (Ambystoma mexicanum) is a representative specie of salamander (caudate amphibian) that has garnered significant attention as an emergent biological model organism [4]. Native from Mexico, the axolotl exhibits interesting characteristics, such as its regenerative ability, neoteny, scar-free healing, and robust immune system, which has led to the availability of extensive genomic resources, make it an invaluable biological model [4]. These features impact multiple disciplines, including regenerative biomedicine, developmental biology, wound healing, immunology, biomedical engineering, and genomic sciences [4]. Indeed, the axolotl genome has been sequenced, being one of the few complete sequenced genomes of salamanders to date, providing comprehensive resources to study the genes and regulatory elements involved in regeneration, and immunity related to AMPs [5]. While several countries regulate and protect certain amphibian species, such as the axolotl (by NOM-059-SEMARNAT-2010 in Mexico), it is now possible to obtain antimicrobial peptides (AMPs) through genomic information implementing heterologous expression or similar biotechnological systems [6]. This allows sustainability and bioavailability to be guaranteed without endangering amphibian biodiversity, which is already threatened. Thus, the rich genomic dataset offers an unprecedented opportunity to discover new representative antimicrobial peptides in salamanders and potentially translate them into therapeutic strategies for human health to future.
Given the evident increase in resistance to conventional drugs by pathogens, the scientific community has been forced to develop and improve new anti-infective agents. Since AMPs are one of the best alternatives to address this problem, the search and characterization of new AMPs has become imperative, with amphibians being a promising source of these peptides, as previously mentioned [7]. The role of antimicrobial peptides in the fight against antibiotic-resistant bacterial infections seems set to be a turning point in the coming decades [7]. Therefore, the present study aims to identify new antimicrobial peptides present in the Mexican axolotl, through in silico screenings with bioinformatics tools and taking advantage of available genomic resources, which could represent a great alternative with biomedical potential in the pharmacological field, currently threatened due to increasing resistance to conventional drugs available.
2 Materials and Methods
2.1 Screening of Coding Transcripts for Bioactive Peptides in the Axolotl Transcriptome
We used online data from the UniProt repository to create a non-redundant reference database of bioactive antimicrobial peptides from amphibians. This database helped us to detect AMP-encoding axolotl transcripts via BLAST v2.12 with the options “task blastp-short -evalue 1e-3 -max_target_seqs 1” (bitscore \(\sim\)28). The axolotl transcriptomes used in this study were obtained from the NCBI GenBank database with the following TSA (transcriptome shotgun assembly) accessions: GFBM010000000, and GFZP01000000. All axolotl transcripts were merged and redundancy reduced with CD-HIT-EST v4.8.1 under the parameters “-c 0.95 -g 1 -b 50 -n 10 -A 0.95” before being employed for AMPs screening. We then used the EMBOSS v6.6 package to identify and translate all possible six-frame open reading frames (ORFs) in the axolotl transcripts, by running getorf with the parameters “-table 1 -minsize 30 -find 3”. After, we applied transeq to convert the ORFs into protein sequences for BLAST searches, first discarding any sequences with non-standard undefined amino acids.
2.2 Annotation and Domain Architecture Analysis of Candidate Sequences with Potential Antimicrobial Activity
The annotation of the selected axolotl transcripts was done with Trinotate v4.0. The annotated amphibian protein sequences were classified according to their gene family with InterProScan v5, SMART v9.0, and ScanProsite v20.0 to determine the composition and structural organization of the characteristic protein domains. In addition, Cyscon was used for the inference of disulfide bridges and PeptideCutter for cleavage sites. The results of domain architecture identification for the determined AMP classes were plotted with IBS v2.0. The prediction of antimicrobial properties was done with AMPlify v1.1 (which is an attentive deep learning model), and AmpGram v1.1 (based on n-gram encoding and random forests), using the default parameters.
2.3 Transcript Expression Analysis with Axolotl Transcriptomic Data
Raw mRNA-seq data publicly available at NCBI was acquired to study the gene expression patterns of previously identified axolotl AMPs in different organs and excised segments of structures. The corresponding accessions of the SRA studies used were SRP093628 and SRP065567. Quality profiling was done with FastQC v0.12.1. For adapter trimming and read filtering, the BBTools v37.62 BBDuck was used with “ktrim=r k=23 mink=11 hdist=1 tpe=t tbo=t qtrim=rl trimq=20 minlen=15 forcetrimleft=14”, and BBMap with “maxindel=1 minid=0.95”. RiboDetector v0.2.7 was employed to discard rRNA reads from data set. Then, sequencing reads were mapped to the axolotl transcriptome with Bowtie2 v2.5 using the parameters “--sensitive --dpad 0 --gbar 99999999 --mp 1,1 --score-min L,0,-0.1 --no-mixed --no-discordant -k 200 -I 1 -X 1000”. RSEM v1.3.1 was after assessed for estimating gene expression levels by expected counts with EM algorithm to mapped reads. Subsequently, abundance analysis was performed with EdgeR obtaining the counts per million (CPM) for axolotl AMP transcripts, and calculating the Fold Change based on housekeeping transcript abundance of amx-odc-1. Results were graphed in a bar plot and a heatmap with the libraries gplots v3.1.3 and RColorBrewer v1.1 of R v4.1.2.
3 Results and Discussion
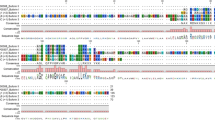
In the Mexican axolotl transcriptome, seventeen transcripts were identified encoding for at least 3 different classes of Transferrins (one Melanotransferrin, one Saxiphilin and one Serotransferrin), four Hepcidins, two Leap2 (Liver-Enriched Antimicrobial Peptide 2), six Cathelicidins, one Cystatin-C, and one β-Defensin. Through a domain architecture analysis (see Fig. 1), it was possible to verify the presence of characteristic functional peptide domains, as well as the presence of signal peptides. In this sense, it was observed that the vast majority of the axolotl bioactive peptides analyzed have a clear signal peptide of the Sec/SPI type, a standard secretory signal through the Sec pathway found in the endoplasmic reticulum and which directs the insertion of membrane proteins; destined in turn to the vesicle sorting pathway and which are cleaved by a signal peptidase I [8].
Domains organization schemes for sequences translated of axolotl transcripts identified encoding antimicrobial peptides; where domains are distinguished with different colors and names; predicted secretory signal in orange; red diamonds represent cleavage sites; yellow spheres correspond to binding sites. (A) Transferrins. (B) Liver-Enriched Antimicrobial Peptide 2. (C) β-Defensin. (D) Cystatin-C. (E) Cathelicidins. (F) Hepcidins.
Furthermore, the size range of the predicted axolotl antimicrobial proteins was between approximately 5 and 20 kDa, with exception of Transferrin proteins of around \(\sim\)80 kDa, all as expected [7]. Among the most outstanding findings, the presence of multiple axolotl paralogs for the Leap2 and Hepcidin classes can be mentioned. These are proteins intimately related to iron homeostasis (together with Transferrins), which display a classical tripartite organization of a signal peptide, with a linker variable proregion, and the functional peptide cleaved by a Furin during prepropeptide maturation [9, 10]. However, unlike what happened in mammals, there are at least two different paralogs for each class of Leap2 and Hepcidin (see Fig. 1, B and F panels), more similar to what is observed in fish and reptile species [9, 10]. In the case of Transferrins, these axolotl proteins showed two transferrin domains that are known to form two homologous lobes called N-lobe and C-lobe (see Fig. 1, panel A), the latter domain being the most important due to its high affinity for Fe3+ binding, as can be seen in Serotransferrin [11]. On the other hand, the axolotl β-Defensin and Cystatin-C identified (see Fig. 1, C and D panels) had three and two predicted disulfide bridges respectively, as reported in other studies [12, 13]. For axolotl Cathelicidins (see Fig. 1, panel E), these proteins presented a classical architecture with a secretory signal, followed by a Cathelin-like domain related to Cathepsin-L-like inhibition, commonly cleaved by Elastase or Proteinase-3 proteases to give rise a mature peptide highly variable in length [14].
When carrying out the prediction of antimicrobial activity with AMPlify and AmpGram [15, 16] for the annotated protein sequences of the axolotl, a high probability score was denoted for the mature forms of Hepcidins, as well as for some Cathelicidins and Leap2 (see Table 1). In other studies, some in vitro antibacterial activity has also been reported for similar amphibian Hepcidins, Cathelicidins, and Leap2, which is also consistent with our predictions shown [17].
Transcript expression profiling of axolotl sequences identified as presumptive AMPs. (A) Relative abundance in counts per million (CPM) for transcripts expressed in different organs, including biological structure segments. (B) Fold Change normalized with housekeeping amx-odc-1 abundance of axolotl transcripts and evidencing their similarities to each other. Fold changes (above 2) that were significant were highlighted in bold.
The axolotl organs and structure portions that revealed a high abundance of transcribed AMPs were the liver and spleen, followed by the tail (see Fig. 2, A and B panels). It has been documented in previous works that the vast majority of the regulatory factors of iron homeostasis in mammals are mainly produced by the liver, as was also evidenced by the abundance of messenger RNAs coding for axolotl leap-2, and several of hepcidins and transferrins [9,10,11]. Likewise, an organ that drew particular attention is the spleen. This organ, being histologically made up of red and white pulp, the latter abundant in leukocytes, was expected to show a high abundance of transcripts for several cathelicidins in the axolotl (see Fig. 2, panel A), since cathelicidin transcripts are usually expressed in epithelium and white cells of the immune system [14]. This transcriptional expression profiling, referring to the two previously contrasted conditions, allows us to note that the same trend is evolutionarily conserved even up to amphibians. Particularly, a significant change in abundance of the transcripts encoding hepcidin-b and leap-2ab is observed (see Fig. 2, panel B). More additional analyzes would be needed to unravel functional differences to the canonical ones described for the different paralogs identified in the present study.
4 Conclusion
Through a conservative comparative approach, using only public genomic resources, the identification of seventeen potential antimicrobial bioactive peptides in the axolotl was achieved. Furthermore, the structural organization of domains for the vast majority of the proteins encoded in the identified axolotl transcripts was observed to be highly conserved with respect to their counterparts reported in other species. Although predictions of antimicrobial activity are promising for many of the axolotl peptide sequences found, further exploratory analyzes are required to confirm this finding. Finally, the abundance of the axolotl transcripts coding AMPs analyzed in the transcriptional expression profiling, allowed the liver and spleen to be highlighted as important biological sources, information that could contribute to the isolation of such sequences or their products for future assays.
References
Jenssen, H., Hamill, P., Hancock, R.E.: Peptide antimicrobial agents. Clin. Microbiol. Rev. 19(3), 491−511 (2006)
Olascoaga Del Angel, K., et al.: Antimicrobial peptides, a promising alternative for the treatment of infectious diseases. Gac. Med. Mex. 154, 8 (2018)
Raaymakers, C., et al.: A new family of diverse skin peptides from the microhylid frog genus phrynomantis. Molecules 25(4), 912 (2020)
Bölük, A., Yavuz, M., Demircan, T.: Axolotl: a resourceful vertebrate model for regeneration and beyond. Dev. Dyn. 251(12), 1914–1933 (2022)
Varela-Rodríguez, H., et al.: Functional characterization of the lin28/let-7 circuit during forelimb regeneration in ambystoma mexicanum and its influence on metabolic reprogramming. Front. Cell Dev. Biol. 8, 562940 (2020)
Deng, T., et al.: The heterologous expression strategies of antimicrobial peptides in microbial systems. Protein Expr. Purif. 140, 52–59 (2017)
Xiao, Y., Liu, C., Lai, R.: Antimicrobial peptides from amphibians. Biomol. Concepts 2(1-2), 27–38 (2011)
Natale, P., Brüser, T., Driessen, A.J.: Sec and tat-mediated protein secretion across the bacterial cytoplasmic membrane distinct translocases and mechanisms. Biochem. Biophys. Acta. 1778(9), 1735–1756 (2008)
Kim, C.H., Kim, E.J., Nam, Y.K.: Subfunctionalization and evolution of liver ex-pressed antimicrobial peptide 2 (LEAP2) isoform genes in Siberian sturgeon (Acipenser baerii), a primitive chondrostean fish species. Fish Shellfish Immunol. 93, 161–173 (2019)
Barroso, C., Carvalho, P., Nunes, M., Gonçalves, J.F.M., Rodrigues, P.N.S., Neves, J.V.: The Era of antimicrobial peptides: use of hepcidins to prevent or treat bacterial infections and iron disorders. Front. Immunol. 12, 754437 (2021)
Mohd-Padil, H., Mohd-Adnan, A., Gabaldón, T.: Phylogenetic analyses uncover a novel clade of transferrin in nonmammalian vertebrates. Mol. Biol. Evol. 30(4), 894–905 (2013)
Jurczak, P., Groves, P., Szymanska, A., Rodziewicz-Motowidlo, S.: Human cystatin C monomer, dimer, oligomer, and amyloid structures are related to health and disease. FEBS Lett. 590(23), 4192–4201 (2016)
Correa, P.G., Oguiura, N.: Phylogenetic analysis of β-defensin like genes of bothrops crotalus lachesis snakes. Toxicon 69, 65–74 (2013)
Kościuczuk, E.M., et al.: Cathelicidins: family of antimicro bial peptides a review. Mol. Biol. Rep. 39(12), 10957–10970 (2012)
Li, C., Warren, R.L., Birol, I.: Models and data of amplify: a deep learning tool for antimicrobial peptide prediction. BMC. Res. Notes 16(1), 11 (2023)
Burdukiewicz, M., et al.: Proteomic screening for prediction and design of antimicrobial peptides with ampgram. Int. J. Mol. Sci. 21(12), 4310 (2020)
Vasconcelos, I.A., et al.: Salamanders and caecilians, neglected from the chemical point of view. Toxin Rev. 41(4), 1304–1332 (2022)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Cera-Domínguez, E., Arenas-Ballesteros, G., Varela-Rodríguez, L., Camarillo-Cisneros, J., Guzman-Pando, A., Varela-Rodríguez, H. (2024). Transcriptional Expression of Bioactive Antimicrobial Peptides with Biomedical Potential in Diverse Organs of the Mexican Axolotl. In: Flores Cuautle, J.d.J.A., et al. XLVI Mexican Conference on Biomedical Engineering. CNIB 2023. IFMBE Proceedings, vol 96. Springer, Cham. https://doi.org/10.1007/978-3-031-46933-6_15
Download citation
DOI: https://doi.org/10.1007/978-3-031-46933-6_15
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-46932-9
Online ISBN: 978-3-031-46933-6
eBook Packages: EngineeringEngineering (R0)