Abstract
Manual segmentation is still the gold standard in hippocampal segmentation due to its precision. However, it requires a considerable amount of time. Convolutional neural networks offer a less resource-intensive alternative. In this study, we propose a parallel convolutional neural network architecture for segmenting the hippocampus in patients with epilepsy and healthy patients based on magnetic resonance images. Our network design resembles a wavelet filter bank but utilizes adaptive filters generated by convolutional layers. By employing a limited number of convolutional layers, our approach achieves improved computational efficiency compared to existing network models in the literature. The performance evaluation was conducted using the Jaccard index, Dice coefficient, sensitivity, and precision, and compared to the widely used U-Net network segmentation. The results, based on similarity metrics, indicate that the parallel network demonstrates higher predictive similarity to the test dataset, achieving a precision score of 0.80 outperforming the U-Net network.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Around 50 million people in the world experience epilepsy, making it one of the most common neurological disorders, and organizations such as the World Health Organization recognize it as a major health problem. One of the most common form of human epilepsy is temporal lobe epilepsy. Nowadays, manual segmentation software allows professionals to perform automatic hippocampus segmentation in the temporal lobe in magnetic resonance imaging for epilepsy detection. These methods are costly and time-consuming, so neural network-based solutions have been used instead. The U-Net architecture is often used as the base network in several models for image segmentation tasks in the biomedical field [1,2,3].
In 2021, in [2] a network employing the U-Net architecture was introduced. In their work several alterations were implemented to the original U-Net structure: residual connections were integrated between the convolutional block’s input and output, to each convolutional layer, batch normalization was used, and instead of utilizing a single input, 2D adjacent patches were employed. The devised approach underwent training and validation using the HCUni-camp database as outlined in their study. Likewise, the method presented by [3] in 2019 utilized a 3D U-Net with a fusion of the anatomical planes’ outputs, which incorporates different segmentation and error correction steps employing replacing and refining networks. Finally, the Quick Nat software from [4] uses various 2D U-Net approximation for each image orientation and masks are manually refined. Free Surfer software is used for auxiliary data augmentation.
Prior research has showcased the utilization of intricate and resource-intensive networks for segmentation tasks, such as U-Net. These networks often pose challenges in terms of implementation, particularly when integrating algorithms into embedded systems or when dealing with volumetric data that necessitates a specialized server for processing. The complexity and computational demands of these networks impede their practicality in real-world scenarios, where efficiency and resource utilization are crucial. Additionally, the need for specialized hardware or dedicated servers for processing volumetric data further adds to the complexity and cost of implementing such segmentation algorithms. Addressing these limitations, our study aims to explore alternative approaches that prioritize simplicity and computational efficiency without compromising segmentation accuracy [5–7].
Hereby, we introduce a 2D Parallel convolutional neural network (CNN) for automating hippocampal segmentation for MRI images of the temporal lobe. Our proposed architecture exhibits several advantages over existing models, such as the well-known U-Net. Notably, our network comprises fewer components, resulting in improved computational efficiency. Moreover, it surpasses the time-consuming manual segmentation process in terms of speed. One key feature of our architecture is the utilization of multiple channels to process the input image. Each channel applies filters of varying sizes, allowing us to capture diverse levels of detail comparable to a wavelet filter bank. However, unlike fixed wavelet filters, our adaptive filters are dynamically determined through the convolutional layers. This approach enables our network to adapt and extract relevant features from the input image, optimizing the accuracy of the segmentation process. By incorporating these adaptive filters within our architecture, we enhance the network’s ability to capture intricate hippocampal structures and boundaries more effectively. By proposing this 2D Parallel CNN, we aim to provide a more computationally efficient and accurate solution for hippocampal segmentation in MRI images.
2 Methods
In this section, we provide a comprehensive description of the Parallel and U-Net architectures, along with the preprocessing steps, training process, and evaluation techniques utilized in our study.
2.1 Database
Several databases were searched on the Kaggle site. The Hippocampus Segmentation in MRI Image database was selected, which contains 50 MRI images in ANALYZE format with T1W weighting, from 40 epileptic and 10 non-epileptic patients. A magnetic field of 1.5 T was applied to 20 epileptic and 10 non-epileptic subjects, with the remaining subjects a 3 T magnetic field was used: in both cases with impaired recovered gradient echo sequence (SPGR).
In Fig. 1 is depicted the anatomical planes in a sample from the set of images from the database. Note that the following is included for segmentation, subiculum, head, body, and tail of the hippocampus. In contrast, the alveus and fimbria were not included: neither the amygdala nor the temporal horn of the lateral ventricle [8].
2.2 Preprocessing
Originally the database contained images size 516. However, the images were modified with a resize function to 128 × 128, to speed up the training and evaluation of the networks. Only the coronal slices were chosen, in a range of 40 to 70 slices, where the hippocampal segmentation is best appreciated. Figure 2 shows both images already modified. Additionally, the label’s set images were binarized with a threshold of 0.5. Finally, data partitioning was performed. The train_test_split function was used on a total of 755 images, to obtain a test set of 90% and a training set of 10%.
2.3 Architecture for Segmentation
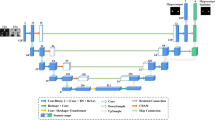
To accomplish the hippocampus segmentation, we proposed a modified and adapted deep learning network architecture based on the framework presented in reference, [9, 10]. Figure 3 shows a schematic of the network. Our architecture consists of three channels, each incorporating a total of nine convolutional layers. The purpose of utilizing multiple channels is to process the input image using filters of varying sizes, enabling us to capture different levels of detail akin to a wavelet filter bank. However, unlike fixed wavelet filters, our adaptive filters are obtained through the convolutional layers.
Specifically, the three channels operate at different resolutions to extract distinct representations from the input image. The first channel operates at a coarse resolution using a 9 × 9 filter, enabling the acquisition of broader contextual information. The second channel operates at a medium resolution using a 4 × 3 filter size, facilitating the extraction of intermediate-level features. Lastly, the third channel employs a fine resolution with a 2 × 2 filter size, allowing for the capture of fine-grained details.
The information from each channel is then concatenated and fed into a series of convolutional networks. By incorporating multiple channels, we aim to obtain diverse representations of the input signal, which in turn aids the final layers of the network in accurately segmenting the different regions of the hippocampus.
The sigmoid activation function is employed in the network. The rationale behind this is worth noting. It facilitates the tendency of the output image to become binary, which aligns perfectly with the requisite for our specific two-class segmentation.
To compare the proposed network, we used a U-Net, with the following attributes. For the contracting path, a Conv2D layer of 32 kernels of size 3 × 3 with ReLU activation was used, followed by a MaxPooling layer of size 2 × 2. Another three layers were added for the downstream phase, with 64, 128, and 256 filters of the same size as the input layer, with their respective reduction layers. The intermediate layer also consisted of Conv2D using 512 kernels of size 3 × 3 with ReLU activation. For the expansion pathway, three layers with 64, 128, and 256 filters of the same size as the input layer were used, with their respective up sampling layer, which was performed with the Conv2DTranspose function, with 25 filters of size 2 × 2. Finally, the output layer consisted of a convolution with a filter of size 1 × 1 with sigmoid activation.
Both networks employed as loss function the binary crossentropy, and as optimizer the Adam algorithm, and accuracy metric. In the training phase of the U-Net network, it underwent 70 epochs, a batch size of 50 and with a 0.001 learning rate. On the other hand, the second network underwent a longer training period of 400 epochs, a batch size of 40 and with a 0.0001 learning rate. The choice of binary crossentropy loss function is because the models were trained to perform binary classification tasks in this case the pixels were classified into two classes: hippocampus or not. The Adam optimizer, known for its efficiency and ability to handle large-scale optimization problems, was employed to update the weights of the network in the training.
Dice coefficient, Jaccard index (IoU), sensitivity and precision were obtained to assess the performance of both networks.
3 Results and Discussion
This section presents the results from the networks with 697 samples from the training set, of which 20% was used as the validation set. The U-Net was trained with 70 epochs and the parallel network with 400 epochs.
3.1 Test Set Result
We compare the coronal slices image, and the results of the hippocampal segmentation predictions of the images with higher accuracy, with the masks of the U-Net test set in Fig. 4. Both show the segmentation of the left and right lobes of the hippocampus. However, in the test set the edges are well defined while in the predictions it is seen to take neighboring pixels belonging to the amygdala as part of the hippocampus.
Analogous, Fig. 5 put side by side the coronal slices image, its corresponding segmentation from the test set and the prediction made by the parallel network. The segmentation of both hippocampal lobes can be distinguished in yellow, where the contours are more similar to the test set compared to the first network, however, it can be seen that the network segmented pixels corresponding to the amygdala.
3.2 Evaluation Metrics
Table 1 reports the average values of the evaluation metrics obtained by the U-Net and parallel network. In contrast, Table 2 shows the average values obtained by using two automatic segmentation algorithms to predict segmentation: Classifier Fusion software and Labelling (CFL) and Brain Parser using the same database The U-Net and parallel convolutional neural networks evaluation metrics have similar values for the IoU segmentation metrics and the Dice coefficient to those resulting from using the two segmentation algorithms CFL and Brain Parser. In the case of the parallel neural network, the average in the segmentation coefficients is higher, indicating a more accurate segmentation. In the case of the sensitivity metric, a value of 0.92 was reported when using Brain Parser, higher than that achieved in the two proposed neural networks. On the other hand, when comparing the precision of the two designed neural networks, it is observed that higher values were achieved than those obtained by the algorithms.
In addition, the work in [2] obtained a Dice of 0.71 using Quick Nat network [4], a Dice of 0.76 for Ext2D, and a Dice of 0.74 for Hippodeep [12]. However, these networks were using a different database they were not included in Table 2.
The value obtained in the metrics is relatively low due to the smaller area to be segmented in each image concerning the entire image; consequently, the loss function obtains a good value even when the hippocampal pixels have not been segmented since the rest of the pixels (background) are the majority. An essential factor for segmentation is image quality. In this case, the current resolution of the images, 0.78 × 0.78 × 2.00 mm3, does not allow adequately differentiating the edges so that the hippocampus blends with the amygdala. This causes a lower Dice coefficient due to false positive cases. In addition, the presence of a high number of anisotropic voxels makes segmentation difficult. Other factors are intrinsic or extrinsic lesions such as hippocampal atrophy, which affect the hippocampal texture area [10, 11].
To achieve the best contrast and higher resolution in structural MRI in patients with epilepsy is recommended an MRI basic protocol that includes 3D-T1 sequences, coronal T2 and FLAIR slices, and axial FLAIR and T2 slices.
4 Conclusion
The gold standard for hippocampal segmentation has long been manual segmentation, primarily due to its high precision. However, this method is time-consuming, requiring a significant investment of time and effort. To address this issue, CNNs have emerged as a promising alternative that significantly reduces the time required for segmentation. In this study, two specific network architectures were compared: 1) the U-Net architecture, and 2) a Parallel architecture. Both networks aimed to perform hippocampal segmentation using magnetic resonance imaging data obtained from both epileptic and healthy patients. To compare the performance of the two models, evaluation metrics such as the IoU and Dice coefficient were utilized.
The U-Net network yielded the following metrics: a mean IoU value of 0.48, an average Dice coefficient of 0.60, an average sensitivity of 0.61, and a precision of 0.68. On the other hand, the parallel network demonstrated superior performance, with a mean IoU value of 0.63, a mean Dice coefficient of 0.75, an average sensitivity of 0.75, and a precision of 0.8. These results clearly indicate that the parallel network outperforms the U-Net network in terms of producing more precise segmentations of the hippocampus.
However, despite these encouraging findings, there is still room for improvement to enhance the overall reliability of the developed networks. For instance, it is recommended to augment the training dataset by including a larger number of data samples specifically obtained from the hippocampus of epileptic patients. Additionally, adopting an imaging protocol that provides higher resolution images could potentially improve the accuracy of the segmentation. Moreover, incorporating sagittal slices into the training process could lead to more precise delineation of the hippocampal boundaries, particularly in capturing the leading edges.
By addressing these suggested improvements, the performance and reliability of the networks could be significantly enhanced, making them even more valuable tools in the field of hippocampal segmentation. Continued research and development in this area are vital to refine these neural network models further and unlock their full potential in medical image analysis applications.
References
WHO Homepage. https://www.who.int/news-room/fact-sheets/detail/epilepsy. Accessed 11 June 2023
Carmo, D., Silva, B., Yasuda, C., Rittner, L., Lotufo, R.: Alzheimer’s disease neuroimaging initiative: Hippocampus segmentation on epilepsy and Alzheimer’s disease studies with multiple convolutional neural networks. Heliyon 7(2), e06226 (2021). https://doi.org/10.1016/j.heliyon.2021.e06226.
Ataloglou, D., Dimou, A., Zarpalas, D., Daras, P.: Fast and precise hippocampus segmentation through deep convolutional neural network ensembles and transfer learning. Neuro informatics 17(4), 1–20 (2019)
Abhijit G, R. et al.: QuickNAT: a fully convolutional network for quick and accurate segmentation of neuroanatomy. NeuroImage 186, 713–727 (2019)
Kumar, A., Komaragiri, R., Kumar, M.: Time–frequency localization using three-tap biorthogonal wavelet filter bank for electrocardiogram compressions. Biomed. Eng. Lett. 9(3), 407–411 (2019)
Saad, O.M., Shalaby, A., Sayed, M.S.: Automatic discrimination of earthquakes and quarry blasts using wavelet filter bank and support vector machine. J. Seismol. 23(2), 357–371 (2018)
Samantaray Aswini, K., Rahulkar Amol, D.: New design of adaptive Gabor wavelet filter bank for medical image retrieval. IET Image Proc. 14(4), 679–687 (2020)
Jafari-Khouzani, K., Elisevich, K., Patel, S., Soltanian-Zadeh, H.: Database of magnetic resonance images of nonepileptic subjects and temporal lobe epilepsy patients for validation of hippocampalsegmentationtechniques. Neuroinformatics 9, 335–346 (2011)
Cordero Lares, A.K.: Diseño de una arquitectura de red para la segmentación de regiones tumorales en pulmón por medio de tomografía computarizada, aplicando redes convolucionales. Dissertation, University of Juarez, pp. 19–20 (2019)
Medina Bonilla, E.: Diseño de un sistema termográfico para el análisis de tablillas electrónicas fabricadas en la industria automotriz. Dissertation, University of Juarez (2019)
Alvarez-Linera, J.: Structural magnetic resonance imaging in epilepsy. Radiologia 54(1), 9–20 (2012). https://doi.org/10.1016/j.rx.2011.07.007
Thyreau, B., Sato, K., Fukuda, H., Taki, Y.: Segmentation of the hippocampus by transferring algorithmic knowledge for large cohort processing. Med. Image Anal. 43, 214–228 (2018)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Huizar, A.A.G., Muñoz, J.M.M. (2024). Design of a Convolutional Neural Network for Hippocampal Segmentation in Epileptics and Healthy Patients. In: Flores Cuautle, J.d.J.A., et al. XLVI Mexican Conference on Biomedical Engineering. CNIB 2023. IFMBE Proceedings, vol 96. Springer, Cham. https://doi.org/10.1007/978-3-031-46933-6_13
Download citation
DOI: https://doi.org/10.1007/978-3-031-46933-6_13
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-46932-9
Online ISBN: 978-3-031-46933-6
eBook Packages: EngineeringEngineering (R0)