Abstract
The diagnosis of stomach cancer automatically in digital pathology images is a difficult problem. Gastric cancer (GC) detection and pathological study can be greatly aided by precise region-by-region segmentation. On a technical level, this issue is complicated by the fact that malignant zones might be any size or shape and have fuzzy boundaries. The research employs a deep learning-based approach and integrates many bespoke modules to cope with these issues. The channel refinement model is the attentional actor on the chin channel. While implementing the feature channel, the learnt channel weight can be used to eliminate unnecessary features. Calibration is essential to improve classification precision. The results of channel recalibration can be improved with the help of a re-calibration (MSCR) model. The top pooling layer of the network is where the multiscale attributes are sent. The outcomes of channel recalibration may be enhanced by using the channel weights found at various scales as input to the next channel recalibration perfect. Our unique gastric cancer segmentation dataset, carefully glossed down to the pixel level by medical authorities, is used for extensive experimental comparisons. The numerical comparisons with other approaches show that our strategy is superior.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- Deep learning
- Digital pathology image
- Gastric cancer detection
- Multiscale channel
- Pooling Layer
- Recalibration
1 Introduction
The Industrial Internet of Things (IIoT) is one of the most rapidly expanding net-works right now that can collect and exchange huge volumes of data utilizing sensors in a healthcare context [1]. Medical IoT, IoHT, or IoMT refers to the Internet of Things used in medicine and healthcare and is often referred to as a “expert application” [2]. The Internet of Medical Things (IoMT) is a framework for interconnecting digital resources in the healthcare industry. It is used to evaluate the physical properties of sensor nodes that collect data from the patient’s body using smart portable devices. Integrating AI approaches provides quick and flexible analysis and diagnoses of medical data, while IoMT paves the way for wireless and distant devices to connect securely over the Internet. Network topology, energy transfer, and processing power are just a few of the unknowns that IoT devices must deal with while sending data over the cloud [3].
The 5-year survival rate for people diagnosed with GC at an early stage can surpass 90%. However, the 5-year existence rate drops below 30% [4] because almost half of patients with GC have previously progressed to the advanced phase at the time of diagnosis. Pathologists typically perform this laborious and time-consuming procedure. Diagnostic accuracy is being significantly impacted on a global scale by a serious scarcity of pathologists and a large workload of diagnostics [5, 6]. Thus, a novel method must be developed to rapidly and precisely detect GC from diseased images. To stratify and select those who may benefit from adjuvant treatment, however, physical histological analysis of tumors specimens is currently not reliable enough [7]. The development of concise and reliable approaches to predict overall GC is, therefore, urgently needed in order to aid in the creation of tailored therapy plans and the maximization of their advantages.
Slowly but surely, oncologists have begun to take notice of deep learning. The discipline of cancer has benefited greatly from the advancements made by deep learning, which have been shown to be superior to those made by traditional machine learning methods. For many picture interpretation tasks, the (CNN) has proven to be the superior deep learning method. Several advancements have been achieved with the use of AI to detect tumors and forecast the prognosis of GC based on photographs of the disease in its various stages [8] collected from the IoT enabled microscopes and image sensors. To make the produced AI models more useful to clinical practice, we first identified a number of obstacles that needed to be solved. For example, a substantial sample size from many centres should be obtainable for training and verifying the proposed model to assure the robustness. The generated model needs to be practical for use in clinical practice while also being easy to use by those without an AI background or in regions with a less developed economy [9].
In this research, we built deep learning-based models using the refinement model as a starting point, and we dubbed them the multiscale channel squeeze-and-excitation model. The residual sections of the paper are structured as follows: The correlated texts are presented in Sect. 2. The suggested model and its experimental analysis are described briefly in Sects. 3 and 4 discussing the results. Finally, Sect. 5 presents conclusion and limitations.
2 Related Works
Whole-slide images (WSIs) of human sections are projected by Hu et al. [10] to highlight diagnostically relevant psychoneuroimmunology (PNI) regions. This framework is based on multi-task deep learning. The suggested system completes the task of recognizing PNI while also segmenting the gastric cancer region using a neural detection model and a PNI decision-making module.
Based on the attention mechanism, Guo et al. [11] proposed a compact micro fuzzy pathology detection algorithm; the YOLOv5 is enhanced under compact and micro fuzzy scenarios of cancer cell detection across the board in digital pathology; this algorithm is evaluated on the gastric cancer slice dataset. Test results show an F1 score of 0.616 and a mAP of 0.611 for the deep blur scenario, indicating that it can serve as a decision support for clinical judgement.
Ma et al. [12] offer a deep learning (DL)-based automatic system for diagnosing early gastric cancer (EGC). This work specifically constructs different DL architectures to enable the automatic interpretation of EGC pictures using a novel an-notated dataset obtained from a single-center. The experimental results on the submitted dataset revealed the potential uses of DL in helping of 0.64 for the included classification and segmentation assignments.
Based on ShuffleNetV2, Fu et al. [13] present a multi-dimensional convolutional lightweight network called MCLNet. The computational complexity, memory footprint, and GPU parallelism of the ShuffleNetV2 model are all quite modest. The problem is that ShuffleNetV2 only uses two-dimensional convolutions, so the amount of information recovered from the data is low. To compensate for the absence of 2-D in global feature extraction, employing 1-D increases information transmission between channels and enhances the information.
Using gastric cancer (GC) tissue slides, Lee et al. [14] attempted a fully automated classification of micro satellite instability (MSI) status. The (ROC) curve areas under the curves (AUCs) (TCGA) were 0.893 and 0.902, respectively. The 0.874 AUC achieved by the classifier on the external validation Asian FFPE cohort shows that the classifier trained with TCGA FFPE tissues is effective. It appears that DL has the potential to autonomously learn the most effective features for determining MSI status in GC tissue slides. This research proved that a DL-based MSI classifier could be useful for preliminary case screening.
For studies on gastric tumor image segmentation, Wang et al. [15] propose a stomach cancer lesion dataset. To get multi granular edge information and refinement, an encoder stage-specific boundary extraction refinement module is presented. The next step is to construct a selective fusion module that can be used to combine features from specific phases. The experimental results demonstrate that the projected method outdoes existing approaches on the CVC-Clinic DB and Kvasir-SEG datasets, with an accuracy of 92.3% and a similarity coefficient (DICE) of 86.9%.
3 Proposed System
3.1 Dataset
Our pathology pictures are taken from actual patient records. We have taken 500 pathological images at a resolution of 2048 by 2048 under an optical magnification of 20. Each picture was extracted from a larger slide of gastric tissue showing typical malignant spots in the stomach. Some samples from our dataset [16] are displayed in Fig. 1.
Model parameters were trained using the training set, and the testing set was used to validate the models’ predictions. There will be 350 training images and 150 test photographs used. Each image in the training set was cropped from its original 2048 by 2048 resolution to four 1024 by 1024 patches before being fed into the networks. By subtracting the dataset’s mean pixel value and dividing by the standard deviation, we have created uniform image patches.
The areas covered in yellow have cancer in the above Fig. 1. Size differences, hazy borders, and pliable characters are all made very obvious in these pictures. Five hundred abnormal photos with careful annotations make up our dataset.
3.2 Image Preprocessing
Excessive abnormalities from the staining method typically led to noise in the collected images. The following procedure was used during the preprocessing stage.
-
1.
Scaling down all deep learning models have a strict need that all input images have the same size computationally. Hence, the image was scaled down to a size of 224 × 224 pixels in order to shorten the processing time.
-
2.
Noises like additive, random, impulsive, and multiplicative noises removal are crucial. Gaussian noise, pepper noise, speckle noise, and Poisson noise all show up frequently in medical pictures. In this study, a median filter was employed to get rid of the salt and pepper noise over the entire slide image.
-
3.
Third, normalizing stains is a crucial step in the whole slide image (WSI) pre-processing phase of digital pathology. This research made advantage of the well-known Macenko stain normalization technique often applied on histopathology slides.
-
4.
Fourth, data augmentation is a technique for greatly expanding the types and quantities of data used to train models. To boost the amount of data without changing the appearance of the photographs, we rotated them by 90°, 180°, 270°, flipped them horizontally, and flipped them vertically. This resulted in a sixfold increase in the amount of information collected.
A stratified cross-validation technique was used to divide the raw data obtained in-to two equal parts: 80% for training, for testing. The sum of images in each class increases from 35 for Margin Negative to 49 for Margin Positive after 6 augmentation (with 90°, 180°, 270°, horizontal flip), omitting the testing data set, which must be the unique dataset.
Channel Recalibration Model:
To teach the network to pay attention only to relevant information, the channel recalibration model assigns a weight to each input channel. The input feature U is compressed in channel order by the channel recalibration model.
Once all of the channels in the input feature have been extruded, their respective weights can be calculated by activating the extrusion result using the subsequent formula.
where s is the feature channel’s weight; F ex (*,*) is the excitation function; z is the feature’s extruded result; (*) is the sigmoid function; (*) is the ReLU function [17], and (*) and (*) are the sigmoid and ReLU functions, respectively. The weights of the first fully associated layer (FC) are W1 and the second fully connected layer (FC) is W2.
Excitation’s first fully connected layer takes the sum of feature channels as a function of c and converts it to c/r, where r is the density ratio, with the result that only non-zero values are retained at output following the ReLU function. To maintain parity with the sum of channels in the input feature, the second completely connected layer brings back the number of feature channels to c. The sigmoid function is used to derive the final weight, with a range of 01.0 possible.
To summarise the above formula: It is the weight of the cth channel in the input feature, and x_c is the channel’s output characteristic following channel feature recalibration. The features of a given channel are multiplied by the channel weight, where F scale is a scaling function. By multiplying the eye of a given channel by the associated channel weight, Eq. (3) implements the recalibration of the feature channel, which in turn suppresses the characteristics that are irrelevant to the classification result, leading to higher precision in the classification.
Proposed Model Description:
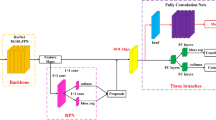
Channel recalibration models can be better-quality by using multiscale features instead of single-scale ones, and by combining the weights learnt at diverse scales to get the final feature channel weights. CNN with multiscale features are frequently used for target identification and recognition as well as picture semantic segmentation. Using feature information at various scales helps increase the precision of the final result. The input sent to the max pooling layer, where they are scaled differently using a pooling kernel size of 2. The feature channel weights across all scales are combined via maximum-value splicing.
Additive Fusion:
In order to do multiscale channel recalibration, the input features are increased by the obtained weights in the order of the respective channels, and the result is the channel weight produced by the additive fusion method using the following:
In the above formula \({\mathop U\limits^{\rm{\prime}}}_{2way\_add} \) is used to accomplish multiscale channel recalibration, the channel weight acquired by the preservative fusion technique 2 is the sum of the channel weights for the two feature scales, and this weight is then multiplied by the input topographies in the order of the respective channels. What follows is a procedure:
Maximum Fusion:
Maximum fusion, as contrast to additive fusion, chooses individual channels. The weight of the channel is equal to the sum of the highest values on each scale. The current procedure for recalibrating multiscale channels goes as follows:
Multiscale channel recalibration with extreme scale is represented as \({\mathop U\limits^{\rm{\prime}}}_{2way\_max}\) where max(,) is the maximum function. The channel weight is the sum of the channel weights under each scale, sorted in order of preference.
Splicing Fusion:
When fusion technique first maps the channel weights from each scale onto a final scale through a convolution layer, and then combines the results. If the batch picture size and the sum of channels in the input feature is C, then the channel weight size is NC11. Depending on which splicing coordinate axis is chosen, two distinct types of splicing fusion implementations can be identified.
(a) Splicing coordinates, or cat1, are located along the second co-ordinate axis (axis1). The current expression for the process of recalibration of multiscale channels is
where N is the number of input channels, C is the sum of output channels, F conv1 (*) is the mapping function for the convolutional layer conv1, the output of multiscale channel recalibration achieved by splicing and fusing cat1 at 2 scales.
(b) Splicing coordinates (cat2) are located along the third coordinate axis (axis3). The current expression for the process of recalibration of multiscale channels is
where \( {\mathop U\limits^{\rm{\prime}}}_{2way\_cat2}\) is the channel weights gotten in the two scales along the third co-ordinate axis, size N2C21, F conv2 (*) is convolution layer, where 21, and S (c cat2) is the consequence of the recalibration realised by splice and fusing cat2 at two scales. Input and output channel counts are both set to a value of C.
4 Results and Discussion
4.1 Implementation Details
TensorFlow is the foundation of our proposed model. All of the convolutions in a pretrained context have been batch normalised. Although it has been shown that a larger batch size is beneficial for segmentation, a fixed batch size of 8 is selected in order to seek for more effective designs. A regular (SGD) algorithm is used with a weight decay of 2e−4 for back propagation, picking a loss function based on the cross entropy of each pixel across all categories. The authors used the polynomial learning rate policy popularised by DeepLab. The training steps have been multiplied, and the learning rate has correspondingly adjusted.
where \(iter\) stands for a training step and \(maxiter\) for the total number of iterations used during training. Our starting power D is 0:9, and our learning rate is 1e−3. Two NVIDIA TITAN X graphics processing units are responsible for all the matrix calculations. One Intel Core I7-6900k octa-core processor running at 3.2 GHz has completed the remaining calculations. The computed memory size is 64G.
4.2 Validation Analysis of Proposed Model
In this section, the validation analysis are based on 70%–30% and 80%–20%, where Table 1 presents the analysis based on 70% of training data and 30% of testing data. The existing models focused on either their own dataset or simply classify the gastric cancer. Therefore, this research work considered the generic model and implemented with the dataset and results are averaged.
From the above table it is evitable that the proposed model showed an improvement over all other models in every metric.
All models perform well when the model’s training ratio is high which is observed from Table 2. Our approach clearly excels over the competing models in key respects. Our approach is more delicate with fine-grained characteristics and sensitive to regions of varying sizes. The models are graphically analysed in Figs. 2, 3, 4 and 5.
5 Conclusion and Future Work
For stomach cancer detection, the authors present a deep learning architecture that combines multi-scale modules with targeted convolutional operations. This study explores the classification task of stomach cancer pathology images and proposes a multiscale channel CNN with Res- network construction. The reliability of the feature channel weight learning process could be greatly improved through the fusing of feature weights learnt at various scales. By incorporating properties at several scales, network data can be made more useful. The obtained dataset has been subjected to rigorous comparative analyses, which prove that the proposed method is more precise and well-organized. Until then, further work is required on datasets and methods to advance the integration of deep learning and pathological diagnostics.
Although our approach has yielded promising results, it does have some restrictions which could be considered as future work. Diagnostic challenges will increase in clinical pathological picture analysis. There is still a great deal of in-depth work, from tests to clinical trials, that needs to be done.
References
Alansari, Z., Soomro, S., Belgaum, M.R., Shamshirband, S.: The rise of Internet of Things (IoT) in big healthcare data: review and open research issues. In: Saeed, K., Chaki, N., Pati, B., Bakshi, S., Mohapatra, D.P. (eds.) Progress in Advanced Computing and Intelligent Engineering. AISC, vol. 564, pp. 675–685. Springer, Singapore (2018). https://doi.org/10.1007/978-981-10-6875-1_66
Rayan, R.A., Tsagkaris, C., Papazoglou, A.S., Moysidis, D.V.: The internet of medical things for monitoring health. In: Internet of Things, pp. 213–228. CRC Press (2022)
Belgaum, M.R., Soomro, S., Alansari, Z., Musa, S., Alam, M., Su’ud, M.M.: Challenges: bridge between cloud and IoT. In: 2017 4th IEEE International Conference on Engineering Technologies and Applied Sciences (ICETAS), pp. 1–5. IEEE (2017)
Sung, H., et al.: Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J. Clin. 71(3), 209–249 (2021)
Morales, S., Engan, K., Naranjo, V.: Artificial intelligence in computational pathology–challenges and future directions. Digit. Sig. Process. 119, 103196 (2021)
Ramana, K., et al.: Early prediction of lung cancers using deep saliency capsule and pre-trained deep learning frameworks. Front. Oncol. 12 (2022)
Tie, J., et al.: Circulating tumor DNA analysis guiding adjuvant therapy in stage II colon cancer. New Engl. J. Med. 386(24), 2261–2272 (2022)
Vobugari, N., Raja, V., Sethi, U., Gandhi, K., Raja, K., Surani, S.R.: Advancements in oncology with artificial intelligence—A review article. Cancers 14(5), 1349 (2022)
Rajpurkar, P., Chen, E., Banerjee, O., Topol, E.J.: AI in health and medicine. Nat. Med. 28(1), 31–38 (2022)
Hu, Z., et al.: A multi-task deep learning framework for perineural invasion recognition in gastric cancer whole slide images. Biomed. Sig. Process. Control 79, 104261 (2023)
Guo, Q., et al.: Pathological detection of micro and fuzzy gastric cancer cells based on deep learning. Comput. Math. Methods Med. 2023 (2023)
Ma, L., Su, X., Ma, L., Gao, X., Sun, M.: Deep learning for classification and localization of early gastric cancer in endoscopic images. Biomed. Sig. Process. Control 79, 104200 (2023)
Fu, X., Liu, S., Li, C., Sun, J.: MCLNet: an multidimensional convolutional lightweight network for gastric histopathology image classification. Biomed. Sig. Process. Control 80, 104319 (2023)
Lee, S.H., Lee, Y., Jang, H.J.: Deep learning captures selective features for discrimination of microsatellite instability from pathologic tissue slides of gastric cancer. Int. J. Cancer 152(2), 298–307 (2023)
Wang, P., Li, Y., Sun, Y., He, D., Wang, Z.: Multi-scale boundary neural network for gastric tumor segmentation. Vis. Comput. 39(3), 915–926 (2023)
Sun, M., Zhang, G., Dang, H., Qi, X., Zhou, X., Chang, Q.: Accurate gastric cancer segmentation in digital pathology images using deformable convolution and multi-scale embedding networks. IEEE Access 7, 75530–75541 (2019)
Yan, J., Wang, B.: Two and multiple categorization of breast pathological images by transfer learning. In: 2021 6th International Conference on Intelligent Informatics and Biomedical Sciences (ICIIBMS), vol. 6, pp. 84–88. IEEE (2021)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Belgaum, M.R., Momina, S.M., Farhath, L.N., Nikhitha, K., Naga Jyothi, K. (2023). Development of IoT-Healthcare Model for Gastric Cancer from Pathological Images. In: Kadry, S., Prasath, R. (eds) Mining Intelligence and Knowledge Exploration. MIKE 2023. Lecture Notes in Computer Science(), vol 13924. Springer, Cham. https://doi.org/10.1007/978-3-031-44084-7_19
Download citation
DOI: https://doi.org/10.1007/978-3-031-44084-7_19
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-44083-0
Online ISBN: 978-3-031-44084-7
eBook Packages: Computer ScienceComputer Science (R0)