Abstract
The COVID-19 pandemic led to an unprecedented volume of articles published in scientific journals with possible strategies and technologies to contain the disease. Academic papers summarize the main findings of scientific research, which are vital for decision-making, especially regarding health data. However, due to the technical language used in this type of manuscript, its understanding becomes complex for professionals who do not have a greater affinity with scientific research. Thus, building strategies that improve communication between health professionals and academics is essential. In this paper, we show a semi-automated approach to analyze the scientific literature through natural language processing using as a basis the results collected by the “Scientific Evidence Panel on Pharmacological Treatment and Vaccines – COVID-19” proposed by the Brazilian Ministry of Health. After manual curation, we obtained an accuracy of 0.64, precision of 0.74, recall of 0.70, and F1 score of 0.72 for the analysis of the using-context of technologies, such as treatments or medicines (i.e., we evaluated if the keyword was used in a positive or negative context). Our results demonstrate how machine learning and natural language processing techniques can greatly help understand data from the literature, taking into account the context. Additionally, we present a proposal for a scientific panel called SimplificaSUS, which includes evidence taken from scientific articles evaluated through machine learning and natural language processing methods.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
In 2020, the COVID-19 pandemic spread worldwide, causing irreparable human losses [1,2,3,4]. COVID-19 (CoronaVirus Disease 2019) is a respiratory disease caused by the SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus 2), a single-stranded sarbecovirus with a positive-sense RNA [5]. Thus, the scientific community responded with an unprecedented amount of studies seeking to understand the origins, mechanisms of action, and possible treatments for the disease [6,7,8].
Governments and health organizations aimed to provide the means to help the results of new studies reach physicians, health professionals, and society in general. For instance, the Brazilian Ministry of Health team proposed a panel to summarize the COVID-19 scientific literature available until that moment and show the results using user-friendly data visualizations [9]. The “Scientific Evidence Panel on Pharmacological Treatment and Vaccines - COVID-19” aimed to gather real-time information on technical-scientific publications from indexed and pre-printed journals that investigate the efficacy, safety, and effectiveness of drugs and biological products used for the treatment and prevention of COVID-19. Until the evaluated date (06/17/2022), the panel summarized information from 2,147 manually curated scientific articles. However, the large amount of data can be detrimental to interpreting the problem since the vast amount can lead to extremely different understandings of the information, making it difficult to transmit the knowledge.
For example, in one of the analyzes proposed by the panel, they raised more than 500 technologies used in the treatment mentioned in the articles. These technologies could be vaccines, therapies, drugs, among others. However, the proposed visualizations in their panel considered only the count of times a technology was mentioned. As the context was not considered, this could lead to misinterpretations. For example, the most cited technology in articles published up to that point was hydroxychloroquine, a drug used to treat malaria. Several studies pointed to the possibility of using hydroxychloroquine to combat COVID-19, in a strategy known as drug repositioning [10]. However, later studies and literature reviews showed that the efficiency of hydroxychloroquine in combating COVID-19 could not be proven [11,12,13,14,15].
In fact, the analysis of some of the articles cited in the panel already showed that both chloroquine and hydroxychloroquine did not have proven effectiveness [16]. However, considering only the number of times these technologies are cited in the literature may give a false impression that their use was effective. Additionally, a manual analysis of each technology and the context of mention in each article in real-time would require an unviable number of dedicated personnel.
We hypothesize that text mining and natural language processing (NLP) techniques could be used to identify the context in which technology is mentioned in the scientific literature. One such technique is sentiment analysis, often used to analyze whether users’ reviews about a certain product are positive or negative. In this context, this technique has the potential to promptly identify which papers cite each technology as effective or non-effective for a given disease, which could contribute to the faster adoption of more effective public policies by the health authorities.
Here we show the results of our NLP-based analyzes of the data presented in the “Scientific Evidence Panel on Pharmacological Treatment and Vaccines - COVID-19”. Our tool aims to collect, analyze, and present evidence taken from scientific articles evaluated through machine learning techniques and natural language processing. To evaluate our results, our team manually analyzed the citation context of each technology in the papers from the panel. Then, we performed a comparison with a panel produced by the Brazilian Ministry of Health.
2 Material and Methods
2.1 Data Collection
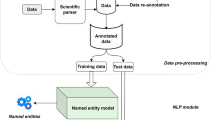
We collected metadata from 2,147 articles from the Scientific Evidence Panel on Pharmacological Treatment and Vaccines - CoViD-19, available at: https://infoms.saude.gov.br/extensions/evidencias_covid/evidencias_covid.html. Titles, abstracts, authors, DOI, journal name, and ISSN were obtained from PubMed API (https://www.ncbi.nlm.nih.gov/home/develop/api/) using in-house Python scripts. Qualis strata were collected from Sucupira (https://sucupira.capes.gov.br/sucupira/public/consultas/coleta/veiculoPublicacaoQualis/listaConsultaGeralPeriodicos.jsf) and journal impact factor values were obtained from SJC - Scimago Journal & Country Rank (https://www.scimagojr.com/journalrank.php). The list of technologies was also obtained from the Scientific Evidence Panel. Details were included in the Supplementary Material. We also used Drug Central’s database of FDA, EMA, PMDA Approved Drugs (https://drugcentral.org/download) to identify other technologies and treatments not considered in the initial database.
2.2 NLP Analyses
In natural language processing, sentiment analysis models usually perform the task of analyzing if a given text is referring to a certain product or technology in a positive or negative light. These models are commonly used in industries such as social media monitoring, customer service and market research, where many customer-generated texts (reviews) need to be analyzed to identify public mood and help inform strategic decision-making.
Considering the specific context of evaluating whether medications are referred to in the articles as effective, our first step was to use regular expressions search to identify which medications or therapies listed in the Drug Central’s FDA, EMA, and PMDA Approved Drugs database were mentioned in each paper. This approach is preferable to named entity recognition techniques since we are searching specifically for medications and therapies, not general entities.
Since not every paper is open-access, we only evaluated the titles and abstracts. After identifying the technologies cited in these sections, we separated the sentence in which they were cited as well as the next sentence and applied the Valence Aware Dictionary and sEntiment Reasoner (VADER) model to evaluate the sentiment of the two sentences [17]. If the technology was cited in more than one sentence, the procedure was repeated for each pair. Then, the average score was computed to indicate if the overall sentiment related to that technology was positive or negative.
Considering the aforementioned procedure, papers without abstracts or not in English were discarded before the processing. The overall results were then recorded in a CSV file, indicating which technologies were positively or negatively cited in each paper.
2.3 Manual Curation and Data Evaluation Metrics
Through a meticulous process of manual curation, each paper in the database received a label. Our team determined whether or not each study mentioned the highlighted treatment or technology as effective or not. Papers mentioning multiple technologies had multiple rows to allow for separate evaluation of each technology. On the other hand, papers that did not mention any technology or treatment present in Drug Central’s database were eliminated. This removal is in accordance with the behavior of the proposed method, which uses regular expression search to determine which therapies are mentioned by the paper and eliminates from analysis those that do not mention any valid treatment, as mentioned in the previous section.
To avoid biases in the analysis, the evaluators were not aware of the label assigned to each paper by the sentiment analysis model. The final result was a manually curated database that was used as the “gold standard” for assessing the accuracy, precision, recall, and F1-score of the sentiment analysis model by comparing manual and model classifications.
3 Results and Discussion
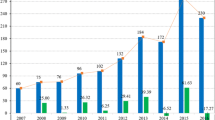
In this study, we evaluated technologies cited in articles, reviews, comments, and pre-prints published during the COVID-19 pandemic (Fig. 1A). We propose that using natural language processing approaches could benefit rapid analysis and help make more effective public policy decisions. Our main objective in this work is to allow a semi-automatic analysis of datasets obtained from the scientific literature to obtain an initial overview. This analysis should be semi-automatic as a completely automatic analysis could include biases that would impair our understanding. Furthermore, a fully manual analysis would be time-consuming.
Our initial analysis aimed to understand the importance of the means used to publish these manuscripts. For this, we use two different metrics: (1) the impact factor and (2) the Qualis stratum. The impact factor is a metric that considers the average citation of a journal in recent years. The Qualis stratum is a metric used by Brazilian agencies to assess the quality of journals in which Brazilian researchers have already published. Journals without an assigned impact factor or Qualis stratum tend to be less recognized by the scientific community. Therefore, this is an initial factor in the quality of the published manuscript.
From 2,147 manuscripts, we detected that almost half of them do not have an impact factor or Qualis stratum (Fig. 1B).
3.1 Technology Mentions
The “Scientific Evidence Panel on Pharmacological Treatment and Vaccines - COVID-19” provides a list of technologies cited in the manuscripts. For each manuscript, the authors manually selected technologies used to treat COVID-19. Then, they evaluated the most cited technologies but did not evaluate the context in which the keyword was mentioned.
In the original panel, a total of 564 technologies were detected. However, when analyzing the list of technologies, we found many repeated or overlapping terms (in Portuguese) such as “Colchicine”, “colchicine”, and “anti-inflammatory drugs”. In this initial form, the most cited technologies were: Hydroxychloroquine, tocilizumab, Lopinavir, Ritonavir, chloroquine, Azithromycin, remdesivir, corticosteroids, convalescent plasma, Angiotensin Converting Enzyme 2 Inhibitors (ACE 2), Angiotensin II Receptor Antagonists, prednisone, favipiravir, Umifenovir, heparin, Immunoglobulin, oseltamivir, Ivermectin, Ribavirin, Vitamin D, alpha interferon, Anakinra, cell therapy, interferon, darunavir, dexamethasone, BCG vaccine, vaccines, and so on.
Using the terms described in the Drug Central database, we identified 72 unique technologies mentioned in the titles and/or in the abstracts of the analyzed papers. Only 14 of the recognized therapies and medicines were cited in at least five different articles: hydroxychloroquine (90), tocilizumab (59), vaccines (35), azithromycin (28), chloroquine (25), remdesivir (22), anakinra (12), heparin (8), methylprednisolone (8), ribavirin (8), oxygen therapy (8), ivermectin (7), lopinavir (6) and famotidine (5). We evaluated the context using the sentiment analysis model and considered those whose context was classified as negative (negative sentiment score) as being referred to as non-efficient or prejudicial, while those described in a positive context (positive sentiment score) as efficient or beneficial for the COVID-19 treatment. The results were summarized as shown in Fig. 2.
3.2 Sentiment Analysis Model Performance Evaluation
The manually curated gold standard database consisted of 410 scientific articles that mentioned treatments and/or therapies for COVID-19. A total of 263 (~64%) of these papers highlighted the mentioned technologies as effective while 147 (~36%) of them cited the technologies as non-effective.
By comparing the results predicted by the NLP model with those obtained by manual curation, we obtained the results summarized in Table 1.
The comparison of the manual curation with the prediction indicates that the model’s predictions were 64.88% accurate. The precision and recall achieved were 73.71% and 70.34%, respectively. These results suggest that the model has reasonable accuracy and capacity to identify a particular context in which the technologies were cited in the literature.
Table 2 illustrates some examples of predictions and the manually curated attributions for each statement.
For instance, in line 1 of Table 2, we show the results for the context analysis for the technology “azithromycin” in the article “Systematic review on the therapeutic options for COVID-19: clinical evidence of drug efficacy and implications”, published by Abubakar et al. in 2020 in the Infection and Drug Resistance journal [18]. Our NLP analysis predicted that the word was used in a negative context (0). Indeed, a reviewer also manually attributed that this keyword was used in a negative context (0). We could verify this by analyzing the sentence in which it was cited: “[…] azithromycin produced no clinical evidence of efficacy […]”. Hence, we can conclude that the NLP analysis correctly predicted the context of mention.
In [19], the authors affirm that their study “did not support the use of hydroxychloroquine […]”. In this case, both our method and the reviewer attributed a negative context for the keyword “hydroxychloroquine”, predicting correctly once again. Also, in [20], our method again correctly predicted the using context for the “fingolimod” keyword.
In lines 4, 5, and 6 of Table 2, we can see the predictions for the using context for the words “vaccine”, “anakinra”, and “remdesivir”, in the papers [21, 22], and [23], respectively. In these cases, our method predicted a positive context, which was proved by manual curation.
The table also shows two failure samples, highlighted in lines 7 and 8. The analysis of these cases shows that the model sometimes fails to correctly classify if a treatment is effective or not in situations where it is described in sentences that also refer to the disease symptoms or external situations. In case 7, the prevalence of words and expressions usually seen in positive reviews (e.g., “hype”, “media announcements”, “more than 150”) might be the case for the incorrect classification as positive. Meanwhile, the description in case 8 has many words usually related to negative contexts but that is not related to the medication, such as “hyperinflammation”, “intervention”, “prevent”, “overcome” and “failure”, which may explain why the model attributed a negative context to the medication.
The examples above show that, despite achieving an acceptable performance in classifying which technologies and therapies are described mostly as effective or not effective in the literature, the model still fails in several common cases where the description of context and the disease symptoms are given close to the name of the medication. This suggests that a pre-trained model might not be the best option for the task selected, and building a more robust model trained on a larger database specifically designed for this use case might result in better performance for this task. We have future prospects to improve the model presented by testing it with other case studies.
SimplificaSUS Panel
As a secondary objective, we also present the SimplificaSUS panel. Our objective, in this case, is to provide a user-friendly tool that facilitates the understanding of scientific articles and that provides visualizations that complement the existing tools. However, this panel is only available in Portuguese. The tool is available at: http://simplificasus.com.
4 Conclusion
As the number of papers published in the literature continues to grow, machine learning techniques have become powerful tools to help us process and simplify knowledge. Here we investigated how sentiment analysis can be used to identify which technologies are presented as effective or not effective when used as a treatment option for a disease. In our case study, the pre-trained sentiment analysis model showed reasonable performance, indicating its potential as an early indication of the most promising therapies according to the literature. Thus, the use of machine learning and NLP can be helpful in summarizing information present in scientific articles and help guide such review efforts in the future. Finally, the manually curated database presented here can also serve as a basis for training more sophisticated models in the future, which may result in better tools for this task.
Data Availability
Supplementary material, data, and scripts are available at https://github.com/LBS-UFMG/SimplificaSUS.
References
Hu, B., Guo, H., Zhou, P., Shi, Z.-L.: Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 19(3), 141–154 (2021)
Pairo-Castineira, E., et al.: Genetic mechanisms of critical illness in COVID-19. Nature 591(7848) (2021). Art. no 7848. https://doi.org/10.1038/s41586-020-03065-y
Melms, J.C., et al.: A molecular single-cell lung atlas of lethal COVID-19. Nature 595(7865) (2021). Art. no 7865. https://doi.org/10.1038/s41586-021-03569-1
Pathak, G.A., et al.: A first update on mapping the human genetic architecture of COVID-19. Nature 608(7921) (2022). Art. no 7921. https://doi.org/10.1038/s41586-022-04826-7
Dos Santos, V.P., et al.: E-Volve: understanding the impact of mutations in SARS-CoV-2 variants spike protein on antibodies and ACE2 affinity through patterns of chemical interactions at protein interfaces. PeerJ 10, e13099 (2022). https://doi.org/10.7717/peerj.13099
Harper, L., et al.: The impact of COVID-19 on research. J. Pediatr. Urol. 16(5), 715–716 (2020). https://doi.org/10.1016/j.jpurol.2020.07.002
Glasziou, P.P., Sanders, S., Hoffmann, T.: Waste in covid-19 research. BMJ 369, m1847 (2020). https://doi.org/10.1136/bmj.m1847
Fraser, N., et al.: Preprinting the COVID-19 pandemic. bioRxiv, p. 2020.05.22.111294, 5 de fevereiro de 2021. https://doi.org/10.1101/2020.05.22.111294
Painel de Evidências Científicas sobre Tratamento Farmacológico e Vacinas - COVID-19. https://infoms.saude.gov.br/extensions/evidencias_covid/evidencias_covid.html. acesso em 20 de abril de 2023
Li, X., et al.: Is hydroxychloroquine beneficial for COVID-19 patients? Cell Death Dis. 11(7) (2020). Art. no 7. https://doi.org/10.1038/s41419-020-2721-8
Schwartz, I.S., Boulware, D.R., Lee, T.C.: Hydroxychloroquine for COVID19: the curtains close on a comedy of errors. Lancet Reg. Health – Am. 11 (2022). https://doi.org/10.1016/j.lana.2022.100268
Avezum, Á., et al.: Hydroxychloroquine versus placebo in the treatment of non-hospitalised patients with COVID-19 (COPE – Coalition V): a double-blind, multicentre, randomised, controlled trial. Lancet Reg. Health – Am. 11 (2022). https://doi.org/10.1016/j.lana.2022.100243
Maisonnasse, P., et al.: Hydroxychloroquine use against SARS-CoV-2 infection in non-human primates. Nature 585(7826) (2020). Art. no 7826. https://doi.org/10.1038/s41586-020-2558-4
Hoffmann, M., et al.: Chloroquine does not inhibit infection of human lung cells with SARS-CoV-2. Nature 585(7826) (2020). Art. no 7826. https://doi.org/10.1038/s41586-020-2575-3
Dhibar, D.P., et al.: The ‘myth of Hydroxychloroquine (HCQ) as post-exposure prophylaxis (PEP) for the prevention of COVID-19’ is far from reality. Sci. Rep. 13(1) (2023). Art. no 1. https://doi.org/10.1038/s41598-022-26053-w
Ghazy, R.M., et al.: A systematic review and meta-analysis on chloroquine and hydroxychloroquine as monotherapy or combined with azithromycin in COVID-19 treatment. Sci. Rep. 10(1) (2020). Art. no 1. https://doi.org/10.1038/s41598-020-77748-x
Hutto, C., Gilbert, E.: Vader: a parsimonious rule-based model for sentiment analysis of social media text. apresentado em Proceedings of the International AAAI Conference on Web and Social Media, pp. 216–225 (2014)
Abubakar, A.R., et al.: Systematic review on the therapeutic options for COVID-19: clinical evidence of drug efficacy and implications. Infect. Drug Resist., 4673–4695 (2020)
Rahmani, H., et al.: Comparing outcomes of hospitalized patients with moderate and severe COVID-19 following treatment with hydroxychloroquine plus atazanavir/ritonavir. DARU J. Pharm. Sci. 28(2), 625–634 (2020). https://doi.org/10.1007/s40199-020-00369-2
Gomez-Mayordomo, V., Montero-Escribano, P., Matías-Guiu, J.A., González-García, N., Porta-Etessam, J., Matías-Guiu, J.: Clinical exacerbation of SARS-CoV2 infection after fingolimod withdrawal. J. Med. Virol. 93(1), 546–549 (2021). https://doi.org/10.1002/jmv.26279
Kim, Y.C., Dema, B., Reyes-Sandoval, A.: COVID-19 vaccines: breaking record times to first-in-human trials. NPJ Vaccines 5(1), 34 (2020)
Karadeniz, H., Yamak, B.A., Özger, H.S., Sezenöz, B., Tufan, A., Emmi, G.: Anakinra for the treatment of COVID-19-associated pericarditis: a case report. Cardiovasc. Drugs Ther. 34(6), 883–885 (2020). https://doi.org/10.1007/s10557-020-07044-3
Davis, M.R., McCreary, E.K., Pogue, J.M.: That escalated quickly: Remdesivir’s place in therapy for COVID-19. Infect. Dis. Ther. 9(3), 525–536 (2020). https://doi.org/10.1007/s40121-020-00318-1
Roustit, M., Guilhaumou, R., Molimard, M., Drici, M.-D., Laporte, S., Montastruc, J.-L.: Chloroquine and hydroxychloroquine in the management of COVID-19: much kerfuffle but little evidence. Therapies 75(4), 363–370 (2020). https://doi.org/10.1016/j.therap.2020.05.010
La Rosée, F., et al.: The Janus kinase 1/2 inhibitor ruxolitinib in COVID-19 with severe systemic hyperinflammation. Leukemia 34(7) (2020). . Art. no 7. https://doi.org/10.1038/s41375-020-0891-0
Acknowledgments
The authors would like to thank Mariana Parise for her valuable contributions during the discussions on the elaboration of this manuscript. The authors also thank the funding agencies: CAPES, CNPq, and FAPEMIG. We also thank the Campus Party, Brazilian Ministry of Health, Fiocruz, and DECIT teams.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Carvalho, F. et al. (2023). Using Natural Language Processing for Context Identification in COVID-19 Literature. In: Reis, M.S., de Melo-Minardi, R.C. (eds) Advances in Bioinformatics and Computational Biology. BSB 2023. Lecture Notes in Computer Science(), vol 13954. Springer, Cham. https://doi.org/10.1007/978-3-031-42715-2_7
Download citation
DOI: https://doi.org/10.1007/978-3-031-42715-2_7
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-42714-5
Online ISBN: 978-3-031-42715-2
eBook Packages: Computer ScienceComputer Science (R0)