Abstract
Human papillomavirus (HPV) accounts for 60% of head and neck (H &N) cancer cases. Assessing the tumor extension (tumor grading) and determining whether the tumor is caused by HPV infection (HPV status) is essential to select the appropriate treatment. Therefore, developing non-invasive, transparent (trustworthy), and reliable methods is imperative to tailor the treatment to patients based on their status. Some studies have tried to use radiomics features extracted from positron emission tomography (PET) and computed tomography (CT) images to predict HPV status. However, to the best of our knowledge, no research has been conducted to explain (e.g., via rule sets) the internal decision process executed on deep learning (DL) predictors applied to HPV status prediction and tumor grading tasks. This study employs a decompositional rule extractor (namely DEXiRE) to extract explanations in the form of rule sets from DL predictors applied to H &N cancer diagnosis. The extracted rules can facilitate researchers’ and clinicians’ understanding of the model’s decisions (making them more transparent) and can serve as a base to produce semantic and more human-understandable explanations.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- Local explainability
- Global explainability
- Feature ranking
- rule extraction
- HPV status explanation
- TNM explanation
1 Introduction
Despite recent advances in head-and-neck (H &N) cancer diagnosis and staging, understanding the relationship between human papillomavirus (HPV) status and such cancers is still challenging. An early diagnosis of HPV could dramatically improve the patient’s prognosis and enable targeted therapies for this group, enhancing their life quality and treatment effectiveness [15]. Moreover, consolidating the diagnosis of cancer staging made by doctors for cancer growth and spread could help better understand how to treat the specific patient, adapting the therapy to the severity of the disease and HPV status.
The term “head and neck cancer” describes a wide range of cancers that develop from the anatomical areas of the upper aerodigestive tract [35]. Considering the totality of H &N malignancies, this type of cancer is the \(7^{th}\) leading cancer by incidence [7]. The typical patients affected by H &N cancers are older adults who have used tobacco and alcohol extensively. Recently, with the ongoing progressive decrease in the use of these substances, the insurgences of such cancers in older adults is slowly declining [13]. However, occurrences of HPV-associated oropharyngeal cancer are increasing among younger individuals (e.g., in North America and Northern Europe) [17].
Diagnosis of HPV-positive oropharyngeal cancers in the United States grew from 16.3% to more than 71.7% in less than 20 years [8]. Fortunately, patients with HPV-positive oropharyngeal cancer have a more favorable prognosis than HPV-negative ones since the former are generally healthier, with fewer coexisting conditions, and typically have better responses to chemotherapy and radiotherapy. Therefore, promptly detecting HPV-related tumors is crucial to improve their prognosis and tailor the treatments [30]. The techniques used in clinical practice to test the presence of HPV have obtained promising results. Still, they are affected by drawbacks, including a high risk of contamination, time consumption, high costs, invasiveness, and possibly inaccurate results [5].
The TNM staging technique is used to address the anatomic tumor extent using the “tumor” (T), “lymph node” (N), and “metastasis” (M) attributes, where “T” denotes the size of the original tumor, “N” the extent of the affected regional lymph nodes, and “M” the absence or presence of distant metastasis [42]. Accurate tumor staging is essential for treatment selection, outcome prediction, research design, and cancer control activities [24]. TNM staging is determined employing diagnostic imaging, laboratory tests, physical exams, and biopsies [25]. Radiomics relies on extracting quantitative metrics (radiomic features) within medical images that capture tissue and lesion characteristics such as heterogeneity and shape [33].
In recent decades, the employment of radiomics has shown great benefits in personalized medicine [16]. Radiomics is adopted as a substitute for invasive and unreliable methods, and they are applied in many contexts, including H &N cancer, with a particular interest in tumor diagnostic, prognostic, treatment planning, and outcome prediction. While the application of radiomics to predict TNM staging has never been addressed in the literature, the possibility of using them to predict the presence or absence of HPV has been recently explored. For example, recent research has shown that it is possible to predict HPV status by using deep learning (DL) techniques that exploit radiomics features [6].
Nevertheless, to the best of our knowledge, no study has yet focused on explaining internal DL predictors’ behavior through a rule extraction process, investigating and assessing the roles of the features and how they compose the rules leading the DL predictor decision. Therefore, there is a need for more investigations to fully understand the main tumor characteristics leading DL models to generate their prediction. To explain DL predictors’ behavior, this study investigates the use of a tool for decompositional rules generation in deep neural networks (namely DEXiRE [9]) to generate rules from Positron Emission Tomography (PET) and Computed Tomography (CT) images in the context of H &N cancers.
In the context of ML models, transparency can be defined as the degree of understanding of the models’ internal decision mechanisms, and overall behaviors can be simulated [18, 45, 52]. To increase transparency in ML models in general and in DL models in particular, we have employed decompositional rule extractors (a.k.a. DEXiRE) because this method can express neural activations in terms of logical rules that both human and artificial agents can understand, thus improving the understanding of the internal decision process executed by the model. The main contribution of this work is to apply the DEXiRE, explainable artificial intelligence (XAI) technique to explain through rule sets the internal decision process executed by DL predictors. Thus, clinicians and researchers can understand the predictors’ behavior to improve them in terms of performance and transparency.
In particular, we extracted radiomics features from PET-CT scans and trained several machine learning (ML) and deep learning (DL) predictors in classification tasks. In turn, we leverage DEXiRE to extract rule sets from a DL model trained on radiomics features extracted offline from PET-CT images. DEXiRE determines the most informative neurons in each layer that lead to the final classification (henceforth, which features and in which combination have contributed to the final decision). Finally, we have assessed and discussed the rule sets.
The rest of the paper is organized as follows: Sect. 2 presents the state of the art of H &N cancer diagnosis using radiomics features and DL models. Section 3 describes the proposed methodology. Section 4 presents and analyses the results. Section 5 discusses the overall study. Finally, Sect. 6 concludes the paper.
2 State of the Art
The metabolic response captured on PET images enables tumors’ localization and tissues’ characterization. PET images are frequently employed as a first-line imaging tool for studying H &N cancer [27]. Moreover, PET is widely used in the early diagnosis of neck metastases. Indeed, PET highlights the metabolic response of the tumors since their early stages — which cannot be seen with other imaging techniques [1]. Thus, PET and CT scans are often used for several applications in the context of H &N cancer.
The most relevant study concerning tumor segmentation includes Myronenko et al. [39], which in the HECKTOR challenge (\(3^{rd}\) edition), created an automatic pipeline for the segmentation of primary tumors and metastatic lymph nodes, obtaining the best result on the challenge with an average aggregate Dice Similarity Coefficient (\(DSC_{agg}\)) of 0.79. [2].
Rebaud et al. [46] predicted the risk of cancer recurrence’s degree using radiomics features and clinical information, obtaining an encouraging concordance index score of 0.68. Among the classification contributions, it is worth mentioning Pooja Gupta et al. [21], who developed a DL model to classify CT scans as tumoral (or not), reaching 98.8% accuracy. Martin Halicek et al. [22] developed a convolutional neural network classifier to classify excised, squamous-cell carcinoma, thyroid cancer, and standard H &N tissue samples using Hyperspectral imaging (with an 80% accuracy). Konstantinos P. Exarchos et al. [12] used features extracted from CT and MRI scans in a classification scheme to predict potential diseases’ reoccurrence, reaching 75.9% accuracy. Only recently, researchers have also focused on HPV status prediction. Ralph TH Leijenaar et al. [31] predict HPV status in oropharyngeal squamous cell carcinoma using radiomics extracted from computed tomography images (with an Area Under the Curve value of 0.78). Bagher-Ebadian et al. [6] construct a classifier for the prediction of HPV status using radiomics features extracted from contrast-enhanced CT images for patients with oropharyngeal cancers (The Generalized Linear Model shows an AUC of 0.878). Bolin Song et al. [51] develop and evaluate radiomics features within (intratumoral) and around the tumor (peritumoral) on CT scans to predict HPV status (obtaining an AUC of 0.70). Chong Hyun Suh et al. [53] investigated the ability of machine-learning classifiers on radiomics from pre-treatment multiparametric magnetic resonance imaging (MRI) to predict HPV status in patients with oropharyngeal squamous cell carcinoma (logistic regression, the random forest, XG boost classifier, mean AUC values of 0.77, 0.76, and 0.71, respectively). However, to the best of our knowledge, no studies have yet involved XAI techniques to unveil the underlying rules, mechanisms, and features leading the ML/DL predictors to their outcomes in such a context. Explaining why/how the models have been obtained is imperative, especially in the medical (diagnosis and decision support systems) domains. Having transparent (i.e., explainable models) models foster understandability, transparency, and trust.
Henceforth, contributions to the XAI field aim to explain the decision-making process carried out by AI algorithms to increase their transparency and trustworthiness [3, 11]. XAI is fundamental in safe-critical domains like medicine, where clinicians and patients require a thorough understanding of decision processes carried out by automatic systems to trust them [38].
AI algorithms, including decision trees, linear models, and rule-based systems, are explainable-by-design, meaning that predictions can be expressed as rules, thresholds, or linear combinations of the input features making the decision process transparent and interpretable [36]. However, algorithms like DL models and support vector machines with non-linear kernels are characterized by non-linear relationships between the input and the output, which improves performance and generalization — making the explanation process more challenging [19]. Therefore, a post-hoc approach is necessary to explain the decision-making process in complex and non-linear algorithms non-explainable-by-design (a.k.a. black-boxes). The post-hoc explanation approach is a third-party method that uses the model structure and input-output relationship to explain AI models [37, 50]. Post-hoc explanations can be classified into local and global. The former interprets one sample at a time — see methods based on sensibility analysis like Local Interpretable Model-agnostic Explanation (LIME) [34], local feature importance and utility CIU [14, 28], and methods based on local surrogate models [40]. While local explanations have the drawback of being valid only for one example or a small set of examples in the input space, global explanation methods aim to explain the overall predictor’s behavior covering as many samples as possible. Global explanations methods include global surrogate models [47, 57], global feature importance and attribution [20, 43], and rule extraction methods [4].
Rule extraction methods can follow three main approaches. First, decompositional methods look inside the predictors’ structure to induce rules; algorithms like FERNN [49], ECLAIRE [56], and DEXiRE [9] are examples of this approach. Second, pedagogical approaches extract rules based on the relationship between input features and predictions (e.g., TREPAN [10]). Finally, eclectic methods combine decompositional and pedagogical methods to produce explanations (e.g., Recursive Rule-extraction (RX) [23]).
The following section describes the methodology used to produce post-hoc explanations of DL models through binarizing neurons and rule induction methods in the medical domain.
3 Methodology
This section presents the approach undertaken to explain the decision-making process carried out by a DL predictor trained on radiomics features to predict the HPV status and the TNM staging through rules. In particular, we executed experiments in two classification tasks:
- T1::
-
HPV diagnosis targets the binary variable HPV_status, which describes if a given patient has an HPV tumor or not (HPV_status = 1/0).
- T2::
-
Cancer staging consists of assigning to a tumor a grade value (an integer number between 1 and 4) based on the TNM tumor grading system, which is composed of three measures: Tumor primary size and extent, Nearby lymph nodes infiltrated, and Metastasis [26, 48, 54]. Tumor grading can be modeled as a multiclass classification task for machine learning.
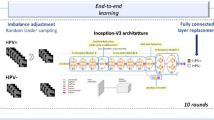
Figure 1 schematizes the experimental pipeline employed to extract the underlying rules leading the predictors trained in T1 and T2 to their outcomes. The experimental pipeline starts with the feature extraction process from PET-CT images. Then, these features have been preprocessed, and exploratory data analysis (EDA) is required to understand the data and the task and choose the appropriate predictors. In turn, predictors are trained and fined tuned using 5-fold cross-validation. Next, the rule set extraction takes place using the training set and pre-trained DL predictor. Finally, the rule set is evaluated using the test set and compared with the baseline predictors’ performance.
3.1 Experimental Pipeline
Below, a brief description of the data set used in this study.
Dataset Description. The dataset used in this study is the HEad and neCK TumOR (HECKTOR 2022) dataset. The dataset was introduced to compare and rank the top segmentation and outcome prediction algorithms using the same guidelines and a comparable, sufficiently large, high-quality set of medical data [2, 41]. The patients populating the data set have histologically proven oropharyngeal H &N cancer and have undergone/planned radiotherapy and/or chemotherapy and surgery treatments. The data originates from FluoroDeoxyGlucose (FDG) and low-dose non-contrast-enhanced CT images (acquired with combined PET-CT scanners) of the H &N region. The primary tumor (GTVp) and lymph nodes (GTVn) segmentations were also provided with the images. The records from this dataset have been collected in eight different centers and contain 524 training examples and 359 for testing. For this study, for task T1 (HPV status prediction), we have removed all those samples with unknown values or without HPV status values (target variable), obtaining a value of 527 patients in total (train + test sets). For Task T2, TNM staging, we have chosen only patients from the TNM 7th edition, obtaining a value of 640 patients in total (train + test sets).
Experimental Pipeline Description. The steps composing the experimental pipeline shown in Fig. 1 are characterized as follows:
-
S1 Feature extraction: The features have been extracted from 3D PET-CT volumes using the Python library Pyradiomics, a tool to calculate radiomics features from 2D and 3D medical images [55]. In this study, the features for T1 and T2 have been extracted from a bounded box surrounding the interest area in the image and not from the whole image, reducing the computational cost of the feature extraction process and producing meaningful features that allow to classify and grade the tumors.
-
S2: Preprocessing: The extracted features have been preprocessed to make them suitable for training ML/DL predictors.
The feature preprocessing is structured as follows:
-
S2.1 Remove non-informative features: Non-informative features like duplicated, empty and constant ones have been removed. Additionally, features like the position of the bonding box have been removed because they are not informative for tasks T1 and T2. Features nb_lymphonodes and nb_lessions have also been removed, given that such features are directly related to the target tumor grade. Inferring the target variable from them is trivial, and it yields to ignore other radiomics features.
-
S2.2 Encode categorical features: Categorical features like gender have been encoded into numerical values using one hot encoding procedure.
-
S2.3 Transform complex numbers to real: Some features calculated with Pyradiomics include complex numbers in which the imaginary part is always zero. To provide a uniform and suitable number format for ML predictors, features with complex numbers have been transformed to float point data types by taking only their real parts.
-
S2.4 Removing NaN columns: Not-a-Number (NaN) columns are numeric columns with missing values and cannot be employed in an ML predictor. Although some missing values can be imputed, in this case, we have decided not to do so to avoid introducing bias and additional uncertainty in the predictors.
-
S2.5 Feature Normalization: Once NaN columns have been removed, the features have been normalized using the standard scale method, which scales all the features to the same range and thus avoids biases from scale differences between features.
-
-
S3: Exploratory Data Analysis (EDA):
The objective of the exploratory data analysis is to identify the most relevant characteristics and the structure of the data set to select the most appropriate predictors for each task. In EDA, a correlation between the features and the target variable has been calculated to identify possible linear relationships between the input features and the target variable. Additionally, correlation analysis has been performed between the features to identify correlation and collinearity. Finally, a distribution analysis has been applied to the target variables, showing a considerable imbalance between the classes.
-
S4: Data split: To maintain reproducible experimentation and fair comparison between the different predictors, the data set was split into 80% for training and 20% for testing using the same random seed and stratified sampling. Additionally, to this data partition, we also employ the HECKTOR challenge partition, where the test set is composed of centers MDA, USZ, and CHB.
-
S5: Feature selection and model tuning:
Due to the high number of features after the preprocessing step, \(\sim 2427\) for task T1 and \(\sim 2035\) for task T2, a feature selection process has been applied to reduce the complexity, avoid the curse of dimensionality [29] and focus on those features strongly related to the target variable. The feature selection process has been executed over the train set.
The feature selection (FS) process encompasses the following steps:
-
FS1 For each input feature, calculate the univariate correlation coefficient with the target variable.
-
FS2 Rank features based on the absolute value of the correlation coefficients calculated in step FS1.
-
FS3 Choose the highest top 20 features based on the rank.
-
FS4 Filter the train and test parts with selected features.
Once the features were selected, we trained four models (M1-M4) in the same setup, as described below.
-
M1 Support Vector Machine (SVM) with a linear kernel and a C parameter of C=10, and a class_weight parameter set in weighted.
-
M2 Decision Tree (DT) with a maximum depth of 50 levels and with impurity Ginny metric as bifurcation measure.
-
M3 Random forest (RF) model with 100 estimators.
-
M4 DL predictor is a Feed-forward neural network in which hyperparameters have been selected employing 5-fold cross-validation and three candidate architectures, with hidden layers ranging from 2 to 6. The number of neurons in each hidden layer varies from \(2^2\) to \(2^8\) on a logarithmic scale.
These models have been chosen due to their similar performance and interpretability.
-
-
S6: Rule extraction process: To extract logic rules from a pre-trained DL predictor, we have employed the algorithm DEXiRE [9]. Such an algorithm extracts boolean rules from a DL predictor by binarizing network activations and then inducing logical rules using the binary activations and the rule inductor algorithms. Finally, the inducted rules are recombined (from local to global) and expressed in terms of input features (see Fig. 2 – DEXiRE pipeline).
-
S7: Metrics evaluation:
To measure the predictors’ performance, we have employed the following performance metrics (PM):
-
PM1 Cohen-Kappa (CK-score): Cohen-kappa score measures the agreement level between the conclusions of two experts. Its value oscillates between -0,20 to 1.0. Negative or low CK-score values indicate not or slight agreement, whereas high values indicate total or strong agreement [32].
-
PM2 F1-score: F1-score belongs to a family of metrics (F-measure) or (F-score). Its value oscillates between 0.0 to 1.0. Higher F1-score values indicate high performance. The F1-score calculates the harmonic mean between the precision and recall measures, combining specificity and sensibility measures. For the case of the multiclass F1-score reported, we employed the weighted mean to consider the imbalance of the dataset.
Additionally, to measure the ability of the rule set to explain the original model, we have used the following measures:
-
PM3 Fidelity: Fidelity measures how similar are the predictions from the rule set to the prediction of the original DL predictor.
-
PM4 Rule length: Rule length measures the number of atomic unique Boolean terms in the rule set.
All the models have been trained and evaluated on the same train and test partitions to make analysis and comparison of results possible. In addition, DNNs have been tested and compared against a set of baseline models with similar capabilities, i.e., Support Vector Machine (SVM), Decision Tree, and Random Forest.
-
DEXiRE pipeline to extract rules from DL predictor [9].
4 Results and Analysis
It is worth recalling that the overall objective of this study is to explain a DL predictor’s behavior (HPV status prediction or tumor staging in H &N cancer) through rule sets extracted employing the decompositional rule extraction algorithm DEXiRE. Table 1 presents the rule sets’ average performance for task T1 (HPV diagnosis) and T2 (Cancer staging), employing the metrics described in step S7 (see Sect. 3).
Datasets for tasks T1 and T2 are highly unbalanced, which can affect the predictors’ and rule sets’ performance. We have executed three experiments with different balancing and partitions for each task to test the possible effect of high dataset imbalance on the rule generation process and the rule set’s performance.
Table 1 summarizes DEXiRE’s rule sets for task T1 (HPV diagnosis) and T2 (Tumor staging) in three different dataset balance configurations. First, the imbalance dataset, in which partitions have been randomly selected while maintaining the proportions of the target variables. The dataset has been balanced with an oversampling technique (SMOTE) in the second configuration. In the third configuration, the dataset follows HECKTOR’s challenge partitions, which are focused on medical centers’ generalization. The rest of the Table 1 is organized as follows, the third column summarizes the average and standard deviation of the rule length (number of features involved in the rule), with values ranging from 7.6 to 14.8 terms for task T1 and 11.2 to 13.6 terms for task T2. The fourth column presents the fidelity measure, which describes the similarity degree between the rule sets’ predictions and those from the original model. The highest fidelity value for T1 is \(\approx \) 80%, while for task T2 is \(\approx \) 73%. The fifth column shows the obtained F1-score, with values above \(\approx \) 80% for task T1 and around \(\approx \) 70% for task T2. Finally, the last column apprises the Cohen-kappa score \(\approx \) 12% for T1 and above 20% for T2.
Appendix A shows the rule set recording the highest performance for each experiment for both T1 and T2.
4.1 Task T1 HPV Diagnosis
Task T1 performs a binary classification employing the radiomics features to predict whether a given patient is HPV positive or not. Results obtained for different datasets’ configuration are described in the following subsections.
Experiment with Imbalanced Dataset. The dataset has not been modified in this setting, retaining its natural imbalance of 90% positive and 10% negative samples. Table 2 shows the results obtained by the baseline models, the DL predictor, and the DEXiRE’s rule set concerning performance metrics PM1 to PM4 (Sect. 3 – step S7). The first column shows the F1-score is reported with all the values over approximately 80%, and the DL predictor obtained the best score (91%). The second column shows the Cohen-Kappa score (CK-score), whose values range from 12% to 28%. Once again, the DL predictor obtains the maximum score. The rule length and fidelity metrics concern only the rule set. The average rule length for this experiment is 8.8 boolean terms, and the fidelity is 76%.
Experiment with Balance Dataset. To test the effect of an artificial balancing dataset technique, we have executed an experiment with a balanced training set employing the oversampling method SMOTE, which allows the drawing of new samples from the minority class based on the neighbors. Table 3 shows the obtained results. In particular, the first column shows the F1-score (all the values are over approximately 80%, and the DL predictor got the best score of 91%). The second column shows the CK-scores, ranging from 13% to 63%. Again, the SVM has obtained the maximum score with \(\approx \) 63%. The average rule length for this experiment is 14.8 boolean terms, and the fidelity is \(\approx \) 80%.
Experiment with the HECKTOR Partition. An essential task within PET-CT medical image analysis is the ability to generalize the results of the prediction models to different medical centers with equipment from different manufacturers and slightly different protocols. This challenge still demands further research and more flexible and robust techniques. To test the rule sets’ generalization ability to various centers, we have used the partitions employed in the HECKTOR 2022 challenge, which provides a reproducible inter-center generalization scenario. Table 4 summarizes the results, recording the highest F1-score of 0,9724, obtained by the random forest predictor, followed by the SVM with an F1-score of 0,9432, the decision tree with a value of 0,9329, the DL predictor with a value of 0,9025, and the rule set with a value of 0,8840. Concerning the CK-score, the highest result is obtained by the SVM predictor with a value of 0.2732, followed by the decision tree with a value of 0.2004, the random forest with a value of 0.1447, the neural network with a value of 0.1259, and the rule set with a value of 0.0241. The F1-score has shown variations of up to 11%. Similarly, the CK-score shows variations of up to 11%.
4.2 Task T2 Cancer Staging
Task T2 performs a multiclass classification to stage and grade tumor. The cancer stage scale is progressive, ranging from a minimum value of 1 to a maximum of 4. In this dataset, the imbalance between the target classes is enormous, but the case of class 1, which has a total of 4 samples over 640, is particularly noteworthy. With such a few samples, applying any effective technique to balance the dataset without introducing bias and errors is very difficult. For this reason, we have decided to remove class 1 from the dataset and perform the subsequent experiments with three categories that, although still imbalanced, provide enough information to apply balance techniques and train the predictors effectively.
Experiments with Imbalanced Dataset. Table 5 presents the results obtained by the baseline models, the DL predictor, and the rule set concerning performance metrics PM1 to PM4 (Sect. 3 step S7) with the imbalanced dataset. The first column shows the F1-score with all the values over approximately 70%, and the DL predictor obtained the best score (76%). The second column reports the CK-score, whose values range from 8% to 27%. Here again, the DL predictor obtains the maximum score. The average rule length for this experiment is 11.2 boolean terms, and the fidelity is 73%.
Experiments with Balanced Dataset. Table 6 presents the results obtained by the baseline models, the DL predictor, and the rule set. The first column shows the F1-score with all the values over \(\approx \) 70%, and the random forest predictor obtained 84% as the best score. The second column reports the CK-score, whose values range from 12% to 35%. Again, the random forest has obtained the maximum score with \(\approx \) 34%. The average rule length for this experiment is 13.6 boolean terms, and the fidelity is \(\approx \) 72%.
Experiments with HECKTOR Partition. Table 7 summarizes the results obtained using the imbalanced dataset with the HECKTOR partition. The highest reported F1-score is 84%, obtained by the random forest predictor, followed by the DL predictor with an F1-score of 80%, the SVM with 78%, the rule set with 77%, and the decision tree with 74%. The random forest predictor obtained the highest CK-score with a value of 0.3469, followed by the rule set with 0.2359, the decision tree with 0.2186, the neural network with a value of 0.2085, and the SVM with 0.1274. The F1-score shows variations up to 10%. Similarly, the CK-score shows variations up to 50%.
5 Discussion
This section elaborates on the results and performance obtained during the explanation of DL predictors (i.e., HPV diagnosis and tumor staging).
5.1 On Rules and Metrics for Task T1
Looking at the results obtained in the three experimental setups for task T1, the balanced dataset generated the rule set with the overall best performance, yet having the highest number of terms. Thus, the obtained results suggest a correlation between rule sets’ length and performance. However, more extended rule sets are challenging to be understood, reducing their quality as explainers. Balancing the rule set’s predictive ability and complexity (number of terms) is necessary yet not immediate.
5.2 On Rules and Metrics for Task T2
Looking at the results obtained in the three experimental setups, the average rule set length in this task is higher w.r.t. T1, reflecting the increased complexity of this task. Moreover, the overall average rule sets’ performance of fidelity and F1-score are sensibly below T1’s results. However, T2’s CK-score is higher than T1’s. This difference can be attributed to the different rule induction methods employed by DEXiRE for the binary and multiclass cases. While the former uses one-rule learning, the latter uses decision trees, which produce more robust and flexible rules.
5.3 Good Explanations, but What About the Predictors?
The results allow inferring that rule sets are good explainers, since they mimic the behavior of the original DL predictor on the training set with high quality. However, the results obtained in the test set and the CK-score for most experiments show results below other predictors. Indicating a limit to the generalization ability of the rule sets concerning more robust structures such as DL models and kernel methods.
5.4 Beyond Metrics
Using more than one metric to evaluate the rule sets generation is a good practice. However, it is possible to observe discrepancies between the consistently high F1-score and the consistently low CK-score. Such discrepancy is due to the data set imbalance that affects only the F1-score.
Indeed, Table 2 (imbalanced dataset) shows the rule set might not be the best, but from the F1-score perspective, it competes with the other predictors — although the CK-score is relatively poor. The performance differences between the rule set’s efficacy measured with the F1-score and the one measured with the CK-score can be explained because the F1-score is the harmonic average of precision and recall. Therefore, in an imbalanced dataset, high precision and recall values on the majority class could produce a high F1-score, even if the class imbalance biases the metric. However, this is not the case for the CK-score. Indeed, it is based on the agreement between two experts, discounting the random influence. Proof of this explanation can be found in Table 3, where the results reported are obtained after balancing the dataset. In this table, the rule set’s F1-score performance and CK-score are similar to the ones obtained by other models, including decision trees, SVM, and random forest.
5.5 The Influence of Data Partition on Rule Sets
In task T1, the rule set performance metrics are similar to those obtained by decision trees, SVM, and random forest — except for the HECKTOR partition. Thus, we can infer that the partitions’ selection affects the models’ performance and the rule extraction process. Moreover, the generalization ability of rule sets is limited to the samples in the training set.
The selection of data splits in tasks T1 and T2 can influence the performance evaluation because of the disparity between sample distribution in different centers. Despite the generalization ability of ML models and the regularization terms, overfitting for a particular center is an issue to be considered. In particular, the rule sets are less flexible, and they tend to overfit the training set to approximate the behavior of the original model on the train set as much as possible.
5.6 Imbalanced Datasets in Medical Domain and Bias Predictors
Imbalanced datasets are common in the medical domain. Such a condition is exacerbated in clinical studies, mainly because of the study of rare diseases or because screening trials focus on ill individuals. Indeed, this is the case for the datasets employed in task T1 HPV diagnosis and T2 cancer staging. Figures 3 and 4 show the sample counting for each target class.
A significant imbalance in the dataset can cause poor performance on the predictors, overfitting, and biases. This is because many optimization algorithms in ML/DL predictors privilege majority class and global accuracy over minority classes. As mentioned above, even the rule sets are affected by this phenomenon. During rule extraction (step S6), some rule sets have been generated biased to predict only the majority class. Over the years, different solutions have been proposed to solve the imbalance in medical datasets. Rahman and Davis [44] proposed to balance medical datasets employing SMOTE. Although this approach works well, it can only be applied in some cases. For example, in task T2, this approach could not be employed in class 1 because there are not enough samples (4) to perform the interpolation.
Rule sets provide domain-contextualized explanations, a logical language that human and artificial agents can understand. Logical language constitutes an advantage in safe-critical domains like medical diagnoses and prognoses because clinicians can validate this knowledge based on their expertise and through a rigorous reasoning process and extract conclusions that can be employed to support their daily work. DL predictors are not extensively used in clinical diagnosis due to the need for more transparency and domain-contextualized explanation of their internal behaviors that enable clinicians to trust their predictions. However, with the introduction of logic and semantic explanations, trust in DL predictors is expected to increase, and they could become part of daily clinical workflows, improving efficiency and effectiveness and helping clinicians and patients to understand their diagnoses.
5.7 Decompositional Rule Extraction Advantages
Decompositional rule extraction methods have several advantages over other post-hoc XAI methods. In particular, the DEXiRE rule extraction algorithm has the following advantages.
-
Logical rule sets can be understood by human and artificial agents. Moreover, they simplify the knowledge exchange between agents in heterogeneous environments.
-
Rule sets, as symbolic objects, can be easily shaped into other symbolic objects like arguments or natural language explanations.
-
Decompositional rule extraction algorithms generate rules by inspecting every neuron activation and better reflecting the internal behavior of the DL predictor.
-
Rule sets can be formally verified to assess their correctness.
-
Besides extracted rule sets, DEXiRE can provide intermediate rule sets that describe the logical behavior in hidden layers, enabling model refinement and better understanding.
-
Alongside the rule sets, DEXiRE can provide activation paths that describe the most frequent neural activation patterns to a given input, identifying the neurons that contribute more to the predictors’ final decision.
-
Rule sets can also be employed to perform inference and reasoning.
5.8 Limitations and Shortcomings
Despite the significant advancement in XAI in recent years, several challenges still need to be solved to apply XAI methods in safe-critical domains like medical diagnosis. The following briefly describes some limitations and shortcomings when using the DEXiRE algorithm in the medical domain.
-
It is not possible to extract rule sets from every DL predictor. This is due to the non-linearity and complexity of the DL predictors’ decision functions, which boolean rules cannot accurately approximate in all cases.
-
DEXiRE algorithm is a very flexible algorithm able to extract rule sets from a wide range of DL architectures. However, currently, DEXiRE can only be applied to classification tasks. More research is required to extend DEXiRE to other machine learning tasks like regression or reinforcement learning.
-
Rule sets depend on the models’ architecture and data set partitions, making them less flexible in responding to never-before-seen cases or outliers. For this reason, we propose to use rule sets to understand and validate DL models rather than to perform large-scale inference processes.
To overcome these limitations, we have proposed several research paths, described at the end of the Conclusions and Future Work section.
6 Conclusions and Future Work
This study can conclude that the DEXiRE method enables the extraction of rule sets from DL predictors, aiming to make data-driven classifiers more transparent and facilitating the understanding of the motivations behind models’ predictions to researchers and clinicians. In particular, it extracted rules from DL predictors trained on HPV diagnosis (T1) and TNM staging (T2) for H &N cancer, employing the decompositional rule extraction tool, namely DEXiRE. For both analyzed tasks T1 and T2 (HPV status and TNM staging), we conducted three experiments with imbalanced (original), balanced (SMOTE), and HECKTOR (inter-center) data partitioning. Finally, the rule sets and their performance metrics have been compared with baseline predictors to test their generalization, prediction, and explaining abilities. Elaborating on the obtained results and analysis, we can summarize the following:
-
Concerning the F1-score metric, the extracted rule sets have shown similar performance among the predictors (i.e., SVM, decision tree, and random forest) and slightly lower performance of those obtained from the DL predictors.
-
Concerning the CK-score, the extracted rule sets performance has shown better results in the multiclass task (T2) than in the binary classification (T1). This is because DEXiRE uses explainable layers (ExpL) and one-rule learning as rule induction methods for binary classification and decision trees for the multiclass case, inducing more robust rule sets.
-
The rule sets are less flexible than ML/DL predictors. Therefore, they have a limited generalization capability and are more useful for providing post-hoc explanations than for making large-scale inferences.
-
The decompositional rule extraction algorithm DEXiRE is affected by data partitions and dataset imbalance — since they impact the entropy, frequency of neuron activation, and terms’ thresholds.
-
Longer rule sets have shown better predictive performance and fidelity. However, it is harder to comprehend longer rule sets. A balance between performance and explainability is necessary for an optimal rule set.
Finally, we envision the following future works:
(i) To conduct further experiments focused on the inter-center rule set generalization, extracting rule sets based on the data from certain medical centers and applying them to other medical centers, (ii) Tumor staging task can also be analyzed using regression models. Thus we intend to extend DEXiRE enabling the explanation of regression DL predictors, (iii) To reduce the effect of unbalanced datasets in DEXiRE, we have proposed to extend DEXiRE to include sample and class weight to deal with imbalanced datasets, and (iv) To make DEXiRE more flexible and robust, we intend to extend it using fuzzy logic, which would allow a better approximation of the DL predictors’ decision function.
References
Özel et al., H.: Use of pet in head and neck cancers (2015). https://doi.org/10.5152/tao.2015.863
Andrearczyk, V., Oreiller, V., Boughdad, S., Rest, C.C.L., Elhalawani, H., Jreige, M., Prior, J.O., Vallières, M., Visvikis, D., Hatt, M., et al.: Overview of the hecktor challenge at miccai 2021: automatic head and neck tumor segmentation and outcome prediction in pet/ct images. In: Head and Neck Tumor Segmentation and Outcome Prediction: Second Challenge, HECKTOR 2021, Held in Conjunction with MICCAI 2021, Strasbourg, France, September 27, 2021, Proceedings, pp. 1–37. Springer (2022)
Arrieta, A.B., et al.: Explainable artificial intelligence (xai): concepts, taxonomies, opportunities and challenges toward responsible ai. Inform. Fusion 58, 82–115 (2020)
Augasta, M.G., Kathirvalavakumar, T.: Rule extraction from neural networks-a comparative study. In: International Conference on Pattern Recognition, Informatics and Medical Engineering (PRIME-2012), pp. 404–408. IEEE (2012)
Augustin, J.G., et al.: Hpv detection in head and neck squamous cell carcinomas: What is the issue? 10 (2020). https://doi.org/10.3389/fonc.2020.01751
Bagher-Ebadian, H., et al.: Application of radiomics for the prediction of hpv status for patients with head and neck cancers. Med. Phys. 47(2), 563–575 (2020). https://doi.org/10.1002/mp.13977
Bray, F., et al.: Global cancer statistics 2018 (2018). https://doi.org/10.3322/caac.21492
Chaturvedi, A.K., et al.: Human papillomavirus and rising oropharyngeal cancer incidence in the united states. J. Clin. Oncol. 29(32), 4294–4301 (2011). https://doi.org/10.1200/JCO.2011.36.4596
Contreras, V., et al.: A dexire for extracting propositional rules from neural networks via binarization. Electronics 11(24) (2022). https://doi.org/10.3390/electronics11244171, https://www.mdpi.com/2079-9292/11/24/4171
Craven, M.W., Shavlik, J.W.: Understanding time-series networks: a case study in rule extraction. Int. J. Neural Syst. 8(04), 373–384 (1997)
Das, A., Rad, P.: Opportunities and challenges in explainable artificial intelligence (xai): A survey. arXiv preprint arXiv:2006.11371 (2020)
Exarchos, K.P., Goletsis, Y., Fotiadis, D.I.: Multiparametric decision support system for the prediction of oral cancer reoccurrence. IEEE Trans. Inf Technol. Biomed. 16(6), 1127–1134 (2012). https://doi.org/10.1109/TITB.2011.2165076
Fitzmaurice, C., et al.: The global burden of cancer 2013. JAMA Oncol. 1(4), 505–527 (2015)
Främling12, K.: Contextual importance and utility in r: the ‘ciu’package (2021)
Galati, L., et al.: Hpv and head and neck cancers: Towards early diagnosis and prevention. Tumour Virus Research p. 200245 (2022)
Gillies, R.J., Schabath, M.B.: Radiomics improves cancer screening and early detection. Cancer Epidemiol., Biomarkers Prevent. 29(12), 2556–2567 (2020). https://doi.org/10.1158/1055-9965.EPI-20-0075
Gillison, M.L., Chaturvedi, A.K., Anderson, W.F., Fakhry, C.: Epidemiology of human papillomavirus-positive head and neck squamous cell carcinoma. J. Clin. Oncol. 33(29), 3235–3242 (2015). https://doi.org/10.1200/JCO.2015.61.6995
Graziani, M., et al.: A global taxonomy of interpretable AI: unifying the terminology for the technical and social sciences. Artif. Intell. Rev. 56(4), 3473–3504 (2023)
Guidotti, R., Monreale, A., Ruggieri, S., Turini, F., Giannotti, F., Pedreschi, D.: A survey of methods for explaining black box models. ACM Comput. Surv. (CSUR) 51(5), 1–42 (2018)
Gupta, P., et al.: Explain your move: Understanding agent actions using specific and relevant feature attribution. In: International Conference on Learning Representations (ICLR) (2020)
Gupta, P., Kaur Malhi, A.: Using deep learning to enhance head and neck cancer diagnosis and classification, pp. 1–6 (2018). https://doi.org/10.1109/ICSCAN.2018.8541142
Halicek, M., et al.: Deep convolutional neural networks for classifying head and neck cancer using hyperspectral imaging. J. Biomed. Opt. 22(6), 060503 (2017). https://doi.org/10.1117/1.JBO.22.6.060503
Hayashi, Y., Yukita, S.: Rule extraction using recursive-rule extraction algorithm with j48graft combined with sampling selection techniques for the diagnosis of type 2 diabetes mellitus in the pima indian dataset. Inform. Med. Unlocked 2, 92–104 (2016)
Huang, S.H., O’Sullivan, B.: Overview of the 8th edition tnm classification for head and neck cancer. Current Treatment Options in Oncology (2017). https://doi.org/10.1007/s11864-017-0484-y
Institute, N.C.: Cancer staging (2022). https://www.cancer.gov/about-cancer/diagnosis-staging/staging
Institute, N.C.: Cancer staging (2022). https://www.cancer.gov/about-cancer/diagnosis-staging/staging
Junn, J.C., Soderlund, K.A., Glastonbury, C.M.: Imaging of head and neck cancer with ct, mri, and us. Seminars in Nuclear Medicine 51(1), 3–12 (2021). https://doi.org/10.1053/j.semnuclmed.2020.07.005, https://www.sciencedirect.com/science/article/pii/S0001299820300763 imaging Options for Head and Neck Cancer
Knapič, S., Malhi, A., Saluja, R., Främling, K.: Explainable artificial intelligence for human decision support system in the medical domain. Mach. Learn. Knowl. Extract. 3(3), 740–770 (2021)
Köppen, M.: The curse of dimensionality. In: 5th Online World Conference on Soft Computing in Industrial Applications (WSC5). vol. 1, pp. 4–8 (2000)
Lechner, M., Liu, J., Masterson, L., et al.: Hpv-associated oropharyngeal cancer: epidemiology, molecular biology and clinical management. Nat. Rev. Clin. Oncol. 19(3), 306-327 (2022). https://doi.org/10.1038/s41571-022-00603-7
Leijenaar, R.T., et al.: Development and validation of a radiomic signature to predict HPV (p16) status from standard CT imaging: a multicenter study. Br. J. Radiol. 91(1086), 20170498 (2018). https://doi.org/10.1259/bjr.20170498
McHugh, M.L.: Interrater reliability: the kappa statistic. Biochemia medica 22(3), 276–282 (2012)
Mayerhoefer, M.E. et al.: Introduction to radiomics. J. Nuclear Med. 61(4), 488–495 (2020)
Mishra, S., Sturm, B.L., Dixon, S.: Local interpretable model-agnostic explanations for music content analysis. In: ISMIR. vol. 53, pp. 537–543 (2017)
Mody, M., Rocco, J.W., Yom, S.S., Haddad, R.I., Saba, N.F.: Head and neck cancer: high-end technology is no guarantee of high-quality care (2022). https://doi.org/10.1016/S0140-6736(22)00426-3
Molnar, C.: Interpretable machine learning. Lulu. com (2020)
Moradi, M., Samwald, M.: Post-hoc explanation of black-box classifiers using confident itemsets. Expert Syst. Appl. 165, 113941 (2021)
Muddamsetty, S.M., Jahromi, M.N., Moeslund, T.B.: Expert level evaluations for explainable ai (xai) methods in the medical domain. In: Pattern Recognition. ICPR International Workshops and Challenges: Virtual Event, January 10–15, 2021, Proceedings, Part III. pp. 35–46. Springer (2021)
Myronenko, A., Siddiquee, M.M.R., Yang, D., He, Y., Xu, D.: Automated head and neck tumor segmentation from 3D pet/CT (2022). arXiv:2209.10809
Nóbrega, C., Marinho, L.: Towards explaining recommendations through local surrogate models. In: Proceedings of the 34th ACM/SIGAPP Symposium on Applied Computing, pp. 1671–1678 (2019)
Oreiller, V., et al.: Head and neck tumor segmentation in pet/CT: the hecktor challenge. Med. Image Anal. 77, 102336 (2022)
of Otolaryngology. Head, A.A., Foundation, N.S.: Tnm staging of head and neck cancer and neck dissection classification (2014)
Puri, N., et al.: Explain your move: Understanding agent actions using specific and relevant feature attribution. arXiv preprint arXiv:1912.12191 (2019)
Rahman, M.M., Davis, D.N.: Addressing the class imbalance problem in medical datasets. Int. J. Mach. Learn. Comput. 3(2), 224 (2013)
Raji, I.D., Yang, J.: About ml: Annotation and benchmarking on understanding and transparency of machine learning lifecycles. arXiv preprint arXiv:1912.06166 (2019)
Rebaud, L., Escobar, T., Khalid, F., Girum, K.B., Buvat, I.: Simplicity is all you need: Out-of-the-box nnunet followed by binary-weighted radiomic model for segmentation and outcome prediction in head and neck pet/CT (09 2022). https://doi.org/10.13140/RG.2.2.30709.04328/1
Sabbatini, F., Ciatto, G., Calegari, R., Omicini, A.: On the design of psyke: A platform for symbolic knowledge extraction. In: WOA, pp. 29–48 (2021)
van der Schroeff, M.P., de Jong, R.J.B.: Staging and prognosis in head and neck cancer. Oral Oncol. 45(4–5), 356–360 (2009)
Setiono, R., Leow, W.K.: Fernn: an algorithm for fast extraction of rules from neural networks. Appl. Intell. 12(1–2), 15–25 (2000)
Slack, D., Hilgard, A., Singh, S., Lakkaraju, H.: Reliable post hoc explanations: Modeling uncertainty in explainability. Adv. Neural. Inf. Process. Syst. 34, 9391–9404 (2021)
Song, B., et al.: Radiomic features associated with hpv status on pretreatment computed tomography in oropharyngeal squamous cell carcinoma inform clinical prognosis. Front. Oncol. 11, 744250 (2021). https://doi.org/10.3389/fonc.2021.744250
Strobel, M.: Aspects of transparency in machine learning. In: Proceedings of the 18th International Conference on Autonomous Agents and MultiAgent Systems, pp. 2449–2451 (2019)
Suh, C., Lee, K., Choi, Y.: Oropharyngeal squamous cell carcinoma: radiomic machine-learning classifiers from multiparametric MR images for determination of HPV infection status (2020). https://doi.org/10.1038/s41598-020-74479-x
Takes, R.P., et al.: Future of the TNM classification and staging system in head and neck cancer. Head & neck 32(12), 1693–1711 (2010)
Van Griethuysen, J.J., et al.: Computational radiomics system to decode the radiographic phenotype. Can. Res. 77(21), e104–e107 (2017)
Zarlenga, M.E., Shams, Z., Jamnik, M.: Efficient decompositional rule extraction for deep neural networks. arXiv preprint arXiv:2111.12628 (2021)
Zhu, X., Wang, D., Pedrycz, W., Li, Z.: Fuzzy rule-based local surrogate models for black-box model explanation. IEEE Trans. Fuzzy Syst. (2022)
Acknowledgments
This work is supported by the Chist-Era grant CHIST-ERA19-XAI-005, and by (i) the Swiss National Science Foundation (G.A. 20CH21_195530), (ii) the Italian Ministry for Universities and Research, (iii) the Luxembourg National Research Fund (G.A. INTER/CHIST/19/14589586), (iv) the Scientific, and Research Council of Turkey (TÜBİTAK, G.A. 120N680).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Appendix Rule Sets
Appendix Rule Sets
In this appendix, examples of the rule sets extracted from DL predictors, in each experiment are presented.
1.1 Rule Sets for Task T1
Table 8 presents the best rule set extracted from DL predictor trained with imbalanced (original) dataset.
Table 9 presents the best rule set extracted from DL predictor trained with SMOTE balanced dataset using.
Table 10 presents the best rule set extracted from DL predictor trained with the HECKTOR partition.
1.2 Rule Sets for Task T2
Table 11 presents the best rule set extracted from DL predictor trained with the imbalanced dataset.
Table 12 presents the best rule set extracted from DL predictor trained with the balanced dataset.
Table 13 presents the best rule set extracted from DL predictor trained with the balanced dataset.
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Contreras, V., Bagante, A., Marini, N., Schumacher, M., Andrearczyk, V., Calvaresi, D. (2023). Explanation Generation via Decompositional Rules Extraction for Head and Neck Cancer Classification. In: Calvaresi, D., et al. Explainable and Transparent AI and Multi-Agent Systems. EXTRAAMAS 2023. Lecture Notes in Computer Science(), vol 14127. Springer, Cham. https://doi.org/10.1007/978-3-031-40878-6_11
Download citation
DOI: https://doi.org/10.1007/978-3-031-40878-6_11
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-40877-9
Online ISBN: 978-3-031-40878-6
eBook Packages: Computer ScienceComputer Science (R0)