Abstract
The daily self-management of type 1 diabetes (T1D) has benefitted from the advancements in real-time continuous glucose monitoring and hybrid closed-loop insulin delivery. These technologies comprise, in parallel, significant sources of data providing insight into daily glucose control and insulin treatment. The concurrent real-time continuous monitoring of vital signs, 24/7, complements the exploitable information for one individual. In this study, we investigate whether respiratory, hemodynamic, and body temperature vital signs correlate linearly with the subcutaneous glucose concentration in T1D, and improve its short-term, up to 60-min ahead, prediction as compared to a univariate model. To verify our research hypothesis, (i) we approximate the prediction of glucose concentration via a long short-term memory (LSTM) function of the recent 30-min history of glucose and those vital signs with a Pearson’s r > 0.5, and (ii) contrast its performance with that of the univariate model. LSTM has been trained and tested individually, using a dataset with 22 T1D people monitored for 2 or 4 weeks. Our analysis identified that: (i) subcutaneous glucose concentration is linearly correlated principally with heart rate and systolic blood pressure, and (ii) the value of the vital signs lies in the improvement of the predictions in hypoglycaemia as the prediction horizon (PH) increases, where we observed a substantial reduction of erroneous predictions from 19% to 7% for a PH of 60 min.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Type 1 diabetes (T1D), as a chronic autoimmune disorder of the glucose-insulin metabolism, is characterised by an abnormal blood glucose concentration regulation leading to hyperglycaemia (defined as glucose concentration values above or equal to 180 mg/dL) [1]. The advancements in continuous glucose sensing technologies as well as insulin analogues and continuous subcutaneous insulin infusion technologies provide people living with diabetes (PLWD) and healthcare professionals with a continuous healthcare model where insulin treatment and other modifiable factors affecting diabetes management (e.g., diet, physical activity) are adjusted efficiently according to objective measures of the glucose control [2, 3]. Hyperglycaemia is an independent risk factor of long-term micro- and macro-vascular complications, which, in turn, account for increased morbidity and mortality rates in diabetes. Nonetheless, hypoglycaemia (defined as glucose concentration values below or equal to 70 mg/dL) remains a most significant barrier for PLWD since its acute life-threatening symptoms have a direct effect on the quality of their life (QoL) [4, 5].
Whilst the problem of short-term prediction of subcutaneous glucose concentration in T1D has been extensively investigated and modelled via linear or non-linear, machine-learning (ML)-based functions, provided the wealth of data amassed by the continuous glucose monitoring (CGM) systems, the meta-analysis of a number of recent systematic reviews points out that it remains still a challenging problem which calls for a more comprehensive representation of the biology, behaviour and context of PLWD within the prediction function [6, 7]. The addition to the model’s input information on the insulin therapy, carbohydrates consumption, physical activity or physiological characteristics (e.g., heart rate, galvanic skin response, skin temperature) has been shown to improve the error of predictions as compared to the univariate prediction models [8, 9]. In addition, the combination of mechanistic models of the processes of intestinal absorption of carbohydrates and the absorption of subcutaneously injected insulin increases the granularity of the information fed into the prediction function [10, 11]. An interesting finding of head-to-head comparison studies is that non-linear univariate models compare with linear ones with respect to prediction horizons up to 60 min, which might be reflective of the fact that short-term glucose regulation features linear dynamics or can be attributed to the existence of unmodelled inputs [12,13,14]. On top of these, the clinical impact of prediction errors has been also considered as not only a performance metric but also an optimisation metric embedded into the training of the ML model [15].
In this study, we examine for the first time in the literature whether daily haemodynamic changes, as they are captured by: heart rate, heart rate variability, systolic blood pressure, diastolic blood pressure, stroke volume, systemic vascular resistance, cardiac output, cardiac index, pulse pressure, mean arterial pressure, may improve the short-term prediction of subcutaneous glucose concentration in T1D. The transient cardiac stress associated with hypoglycaemic events is well documented in the literature, while its predictive capacity has not been studied yet. To this end, we contrast the predictive capacity of a univariate long short-term memory (LSTM) neural network with that of a multivariate LSTM fed with the cardiac indices alongside CGM values. A dataset of 29 T1D patients is leveraged for this purpose generated by the GlucoseML-Phase I observational prospective study, which enables an unbiased evaluation of the prediction models.
2 Materials and Methods
2.1 Materials
The GlucoseML-Phase I prospective study has been designed as an observational study aiming at the collection of real-world data from T1D patients following an intensive insulin therapy scheme (i.e., multiple daily injections (MDI) of insulin or continuous subcutaneous insulin infusion (CSII)) (as it is shown in Table 1), which data will comprise the training/validation/test sets upon which short-term prediction models of subcutaneous glucose concentration time series will be development and internally evaluated. Participants use the GlucoMen Day CGM Menarini®Footnote 1 system and the Biobeat® wrist monitorFootnote 2, and, in parallel, they manually record the carbohydrate content of daily meals and administered insulin therapy using specially designed logs; the GlucoseML-Phase I study encompasses special training sessions on carbohydrates counting to alleviate the errors introduced in the collection of data. The target number of patients to be recruited and the target duration of the study has been set to 30 patients and 4 weeks, respectively. In total, 32 patients were recruited among whom 26 patients (82%) completed the 4-weeks monitoring period, 3 patients (9%) completed the 2-weeks monitoring period, while 3 patients (9%) drop out. Table 1 describes the overall characteristics of the GlucoseML-Phase I study cohort, while Table 2 presents the average glucose statistics observed at the end of the study according to the Ambulatory Glucose Profile report.
2.2 Methods
The prediction function of the subcutaneous glucose concentration time series has been approximated by an LSTM network, defined, fine-tuned and trained using the GlucoseML-Phase I dataset. The LSTM function maps the feature vector \(x\left(t\right)\):
to the observed subcutaneous glucose concentration value over the next t + PH minutes, \(y\left(t+PH\right)\), where \({\left\{{v}_{i}\right\}}_{i=1}^{14}\) denotes the set of input variables (Table 3), \({h}_{{v}_{i}}\) is the history window (expressed in min) specified for \({v}_{i}\), \({\varDelta t}_{{\mathrm{v}}_{\mathrm{i}}}\) is the sampling interval of \({v}_{i}\), and \(PH\) is the prediction horizon (expressed in min) [16]. In this study, we have considered two input cases: (i) Case 1 constitutes a univariate prediction problem relying on the assumption that the recent CGM profile suffices to predict its future short-term course, and (ii) Case 2 forms a multivariate prediction problem accounting additionally for the relationship between glycaemic excursions and changes in vital signs expressing the cardiovascular autonomic nervous system function. We have considered an equal history window \({h}_{{v}_{i}}=30\,{\rm min}\) for all input variables \({v}_{i}\), while the sampling interval of the subcutaneous glucose concentration time series is reduced to the one of physiological vital signs, i.e., 5 min, by applying the Dynamic Time Warping method (\({\varDelta t}_{{\mathrm{v}}_{\mathrm{i}}}=5\,{\rm min}\) for all input variables \({v}_{i}\)) [17].
The LSTM network, comprising two LSTM layers of 4 units and one dense output layer of one unit, has been trained and tested individually for each patient using the time series data segments defined by the first 70% of time points and the remaining 30% of the entire time series, respectively. All input variables have been scaled to \([0, 1]\) over the training set. In Case 2, the linear relationship between the subcutaneous glucose concentration and each of the vital signs time series is examined using the Pearson’s correlation coefficient, with only those vital signs featuring a > 0.5 correlation with the glucose concentration feeding the LSTM model. The hyperparameters relating to batch size, number of epochs, and the optimization algorithm itself are finetuned to minimise the 2-fold cross-validated Root Mean Squared Error (RMSE) computed over the training set. The grid of hyper-parameters search space is presented Table 4. The selected model was trained using Adam optimiser with a batch size equal to 16 and iterated over 100 epochs.
3 Results
The performance of the prediction models was assessed using: (i) two pure error metrics, i.e., the RMSE and the Mean Absolute Percentage Error (MAPE), and (ii) the Continuous Glucose Error Grid Analysis (CG-EGA) which evaluates, in parallel, the clinical impact of the errors depending on the glycaemic range the actual glucose concentration value lies in [18]. It should be noted that the results reported herein concern 22 out of 29 patients of the GlucoseML study; the vital signals of 7 patients were excluded due to a considerable percentage (>50%) of missing values.
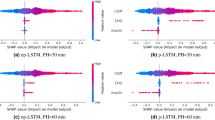
Via the Pearson’s correlation analysis, heart rate and blood pressure were selected for most patients, mean arterial pressure was retained as a feature for 2 patients, while stroke volume and skin temperature were retained as features only for one patient. Table 5 and Fig. 1 describe the distribution of test RMSEs and the test MAPEs associated with 15-min, 30-min and 60-min ahead predictions of subcutaneous glucose concentration when either Case 1 or Case 2 is applied. First, we observe that both input cases yield low errors in the case of a 15-min PH, retaining the average MAPE of 30-min and 60-min PHs below 5% and 8%, respectively. Second, we observe a modest but systematic improvement of the average prediction errors in Case 2, with their interquartile range being located lower for all prediction horizons except for the MAPE for a 60-min PH, which is also captured by the narrower standard deviation achieved in Case 2. The CG-EGA (Table 6, Fig. 2) confirms that the predictive capacity of the additional vital signs becomes evident in the hypoglycaemic range for all PHs, where Case 2 reduces apparently the Erroneous Predictions in this critical range. The contribution of Case 2 in the reduction of erroneous prediction in the hyperglycaemic or euglycemic ranges become more evident for 30-min and 60-min PHs.
4 Discussion and Conclusions
This study explores the predictive capacity of a set of cardiac indices monitored continuously 24/7 in the context of the GlucoseML study, with respect to the short-term prediction (setting the maximum prediction horizon to 60 min) of subcutaneous glucose concentration in T1D. The initial results achieved by an LSTM network, trained and tested over a dataset of 22 people living with T1D who are monitored for a period of 2 or 4 weeks, indicate that these variables can improve the clinical accuracy of predictions in the hypoglycaemic range in spite of the modest improvements in the overall RMSEs and MAPEs.
Clinical evidence supports that hypoglycaemia brings about temporal haemodynamic changes, stimulated by the autonomic nervous system, including: “an increase in heart rate and peripheral systolic blood pressure, a fall in central blood pressure, reduced peripheral arterial resistance (causing a widening of pulse pressure), and increased myocardial contractility, stroke volume, and cardiac output (7)” [19]. The results of our analysis coincide with the above statement in that: (i) Pearson’s correlation analysis identified a linear correlation between subcutaneous glucose time series and heart rate and systolic blood pressure for the majority of patients, and (ii) the introduction of such variables into the glucose predictive function improved the clinical accuracy of short-term glucose predictions, as it is captured by the CG-EGA analysis.
The selected LSTM model produces highly accurate predictions for both input cases, Case 1 and Case 2, and, although a head-to-head comparison of the different approaches cannot be directly applied since the underlying datasets and the associated clinical studies settings differs substantially, the results presented herein compare well with state-of-the-art performances attained for the same research problem. Nonetheless, we are currently refining the dimensionality reduction step, the prediction function and the training approach targeting the minimisation of the errors in the critical zones of hypoglycaemia and hyperglycaemia, fully exploiting not only linear but also nonlinear correlation between the glucose time series and the investigated herein cardiac indices time series, and, in parallel, investigating adaptive learning algorithms.
References
Frayn, K.N.: Metabolic Regulation: A Human Perspective. 3rd edn., pp. 306–308. Wiley-Blackwell, UK (2010)
Holt, R.I.G., et al.: The management of type 1 diabetes in adults. A consensus report by the American Diabetes Association (ADA) and the European association for the study of diabetes (EASD). Diab. Care 44(11), 2589–2625 (2021)
American Diabetes Association Professional Practice Committee: 6. Glycemic targets: standards of medical care in diabetes. Diab. Care 45(Supplement_1), S83–S96 (2022)
Amiel, S.A.: The consequences of hypoglycaemia. Diabetologia 64(5), 963–970 (2021). https://doi.org/10.1007/s00125-020-05366-3
Khunti, K., et al.: Hypoglycemia and risk of cardiovascular disease and all-cause mortality in insulin-treated people with type 1 and type 2 diabetes: a cohort study. Diab. Care 38(2), 316–322 (2014)
Tsichlaki, S., et al.: Type 1 diabetes hypoglycemia prediction algorithms: systematic review. JMIR Diab. 7(3), e34699 (2022)
Felizardo, V., et al.: Data-based algorithms and models using diabetics real data for blood glucose and hypoglycaemia prediction – a systematic literature review. Artif. Intell. Med. 118, 102–120 (2021)
Montaser, E., et al.: Seasonal local models for glucose prediction in type 1 diabetes. IEEE J. Biomed. Health Inform. 24(7), 2064–2072 (2020)
Rabby, M.F., et al.: Stacked LSTM based deep recurrent neural network with Kalman smoothing for blood glucose prediction. BMC Med. Inf. Decis. Making 21(1), 101 (2021)
Schiavon, M., et al.: Modeling subcutaneous absorption of long-acting insulin glargine in type 1 diabetes. IEEE Trans. Biomed. Eng. 67(2), 624–631 (2020)
Muñoz-Organero, M., et al.: Learning carbohydrate digestion and insulin absorption curves using blood glucose level prediction and deep learning models. Sens. (Basel) 21(14), 4926 (2021)
Xie, J.: Benchmarking machine learning algorithms on blood glucose prediction for type I diabetes in comparison with classical time-series models. IEEE Trans. Biomed. Eng. 67(11), 3101–3124 (2020)
Rodríguez-Rodríguez, I., et al.: On the possibility of predicting glycaemia ‘on the fly’ with constrained IoT devices in type 1 diabetes mellitus patients. Sens. (Basel) 19(20), 4538 (2019)
Vettoretti, M., et al.: Advanced diabetes management using artificial intelligence and continuous glucose monitoring sensors. Sensors 20(14), 3870 (2020)
Yotam, A., et al.: Clinically accurate prediction of glucose levels in patients with type 1 diabetes. Diab. Technol. Therap. 22, 562–569 (2020)
Brownlee, J.: Deep Learning for Time series Forecasting. Machine Learning Mastery (2018)
Shou, Y., et al.: Fst and exact warping of time series using adaptive segmental approximations. Mach. Learn. 58(2), 231–267 (2005)
Kovatchev, B.P., et al.: Evaluating the accuracy of continuous glucose-monitoring sensors: continuous glucose–error grid analysis illustrated by TheraSense freestyle navigator data. Diab. Care 27(8), 1922–1928 (2004)
Frier, B.M., et al.: Hypoglycemia and cardiovascular risks. Diab. Care 34(2), 132–137 (2011)
Acknowledgement
This research has been co‐financed by the European Regional Development Fund of the European Union and Greek national funds through the Operational Program Competitiveness, Entrepreneurship and Innovation, under the call RESEARCH – CREATE – INNOVATE (project code: T1EDK-03990).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 ICST Institute for Computer Sciences, Social Informatics and Telecommunications Engineering
About this paper
Cite this paper
Katsarou, D.N., Georga, E.I., Christou, M., Tigas, S., Papaloukas, C., Fotiadis, D.I. (2023). An Exploratory Study of the Value of Vital Signs on the Short-Term Prediction of Subcutaneous Glucose Concentration in Type 1 Diabetes – The GlucoseML Study. In: Tsanas, A., Triantafyllidis, A. (eds) Pervasive Computing Technologies for Healthcare. PH 2022. Lecture Notes of the Institute for Computer Sciences, Social Informatics and Telecommunications Engineering, vol 488. Springer, Cham. https://doi.org/10.1007/978-3-031-34586-9_30
Download citation
DOI: https://doi.org/10.1007/978-3-031-34586-9_30
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-34585-2
Online ISBN: 978-3-031-34586-9
eBook Packages: Computer ScienceComputer Science (R0)