Abstract
Medical teams can use an electrocardiogram (ECG) as a quick test to examine the electrical activity and rhythm of the heart to look for irregularities that may be indicative of diseases. This work aims to summarize the outcomes of several artificial intelligence techniques developed to identify ECG data by gender automatically. The analysis and processing of ECG data were collected from 219 individuals (112 males, 106 females, and one other) aged between 12 and 92 years in different geographical regions, located mainly in the municipalities of the center of Portugal. These data allowed to discretize gender by the analysis of ECG data during the experiment performed and were acquired with the BITalino (r)evolution device, connected to a personal computer, using the OpenSignals (r)evolution software. The dataset describes the acquisition conditions, the individual’s characteristics, and the sensors used as the data acquired from the ECG sensor.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
The early detection of cardiovascular issues is essential for reducing the fatality rate associated with cardiovascular diseases [3, 18], which are among the major causes of death worldwide [6, 27]. For this reason, we believe that experts should adopt automatic electrocardiogram (ECG) analysis to aid in diagnosing cardiovascular diseases [11, 19, 20].
The research on the gender identification is a new subject that is currently starting with more research studies, but the number of studies available in the literature is very small [2, 24]. However, if the studies were performed, we can classify the results according with the gender.
We investigated the most recent machine learning techniques for categorizing and analysis of ECG signals and found that few studies have been unsuccessful in achieving high precision rates [2, 24]. Due to this, we decided to do our research using a dataset we had built and some of the methods we had discovered during our search.
We implemented eight different methods to analyze the results: Nearest Neighbors, Linear SVM, RBF SVM, Decision Tree, Random Forest, Neural Networks, AdaBoost, and Naive Bayes. Based on the machine learning methods, the data was classified by Gender.
2 Methods
2.1 Study Design and Participants
Using a BITalino (r)evolution device [10], and the OpenSignals (r)evolution software, [7], 219 individuals from Portugal’s continental region provided the ECG recordings used in this study. All volunteers in this study disclosed at least one previously diagnosed health condition, such as allergies, hypertension, cholesterol, diabetes, arrhythmia, asthma, unspecified heart problems, and unspecified brain problems; the most frequently disclosed conditions are hypertension and diabetes. The patient spends 30 s sitting down and 30 s standing during the ECG recordings, which at least takes about 60 s. This procedure tries to emphasize the differences between individuals (gender) and allow a minimum effort to avoid a significant change in the overall ECG result. Ethics Committee from Universidade da Beira Interior approved the study with the number CE-UBI-Pj-2021-41.
2.2 Feature Extraction
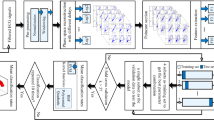
The NeuroKit Python module [29] was used to automatically extract important features from the ECG recordings for this study (Fig. 1), including P, Q, R, S, and T peaks, as well as the onsets and offsets of P, T, and R waves. Based on the features that were automatically retrieved and used in this investigation, the following features were manually calculated:
-
RR interval → \({{Peak R}_{N}-Peak {R}_{N-1}}\)
-
PP interval → \(Peak {{P}_{N}-Peak {P}_{N-1}}\)
-
P duration → \(Offset {P-Onset P}\)
-
PR interval → \(Onset {R-Onset P}\)
-
PR segment → \(Onset {R-Offset P}\)
-
QRS duration → \(Offset {R-Onset R}\)
-
ST segment → \(Onset {T-Offset R}\)
-
ST-T segment → \(Offset {T-Offset R}\)
-
QT duration → \(Offset {T-Onset R}\)
-
TP interval → \(Onset {P-Offset T}\)
-
R amplitude → \({{Peak R}_{N}-Peak {S}_{N}}\)
-
T amplitude → \({Peak T}_{N}-Peak {S}_{N}\)
-
P amplitude → \({Peak P}_{N}-Peak {Q}_{N}\)
2.3 Description of the Method
We have implemented eight machine learning methods to analyze the results in the dataset that can predict some results.
Nearest Neighbors.
K-Nearest Neighbors (K-NN) is a non-parametric technique that uses data from many classes to predict how the new sample point will be classified [26]. This algorithm does not use the training data points to draw any conclusions.
Linear SVM.
The Support Vector Machine attempts to generate the best line or decision boundary to divide n-dimensional space into classes, so new data points are assigned to the correct category when added [5]. Support vectors are the extreme points/vectors that help this algorithm generate the best decision boundary and give the method its name [5]. The term linear SVM refers to linearly separable data, which can be divided into two classes using a single straight line [25].
RBF SVM.
The Radial Basis Function (RBF) is the default kernel function in many kernelized learning algorithms [21]. It’s very similar to the K-Nearest Neighborhood Algorithm, but instead of storing the entire dataset during training, the RBF SVM only needs to store the support vectors [12]. Linear SVM differs from RBF SVM in that the latter is not a parametric model like linear SVM, is more complex depending on the size of the database, and is more expensive to train [16].
Decision Tree.
It is a rule-based supervised machine learning classifier that generates questions based on dataset properties and may categorize new entries depending on the answers [4]. It is a tree-based method because every question it creates has a binary response, which divides the database into halves [9]. A tree-like graph may be seen as the result of these divisions [9].
Random Forest.
It is a machine learning algorithm widely applied to classification and regression issues [1]. It works by building decision trees on various samples throughout the training phase [1]. The outcome is determined based on the class with the most tree selections.
Neural Networks.
It is commonly a multilayer perceptron with three layers: an input, a hidden layer, and an output layer [13, 15]. The last two layers are made up of nodes that function as neurons and make use of a nonlinear activation function. A multilayer perceptron can categorize data that is not linearly separable and uses backpropagation for training [22].
AdaBoost.
It is an ensemble learning method that combines the results of various classifier algorithms to increase their effectiveness and predictive ability [14]. The output of the AdaBoost meta-algorithm is the outcome of this weighted sum.
Naive Bayes.
Based on the Bayes theorem, it is a probabilistic machine learning method classifier [23, 28]. Its simplicity and lack of a complex iterative parameter calculation make it suitable for diagnosing cardiac patients in medical science [8]. This algorithm performs well and is popular because it frequently outperforms the most advanced classification techniques.
3 Results
3.1 Data Acquisition
For this study, we collected data from 219 volunteers (112 men, 106 women, and one other) aged between 12 and 92 years old. All participants provided informed consent to the experiments, allowing us to share the results anonymously. The agreement also provided informed consent to the participants regarding the risks and purpose of the study. Ethics Committee from Universidade da Beira Interior approved the study with the number CE-UBI-Pj-2021-041. The dataset used in this research is publicly available at [17].
Data were acquired with the BITalino (r)evolution device with a 1 kHz sample frequency, connected to a personal computer, using OpenSignals (r)evolution software. Each volunteer’s data was stored in two files: one JSON file referring to the characteristic data of the volunteer plus their lifestyle and a text file with the test data recorded over time. These files were stored in an individual folder per volunteer.
The volunteer needed to stand for 30 s and then sit in a chair for 30 s while the data was collected.
The dataset is available in a Mendeley Data repository, which contains two files for each individual, with 219 folders. Each folder has a JSON file containing a description of the data acquisition conditions, the individual’s characteristics, and the sensors used, and a TXT text file including the data acquired from the ECG sensor.
The Fig. 2 flowchart illustrates the data processing till the classification results.
3.2 Results by Gender
We started with the extraction of the different analyzed variables related to the ECG data, such as RR interval, PP interval, P duration, PR interval, PR segment, QRS duration, ST segment, ST-T segment, QT duration, TP interval, R amplitude, T amplitude, and P amplitude. Table 1 presents the average of the features extracted. Before the analysis by gender, we found that the data of the individuals with the IDs 20, 25, 31, 33, 35, 38, 39, 54, 153, 195, and 202 were invalid, so it was necessary to exclude these 11 individuals from the analysis.
In Table 2, we compare the results of each classifier utilized during this study. The Decision Tree algorithm achieved the highest accuracy at 62.90%. Linear SVM, Adaboost, and Naive Bayes are right next, with an accuracy of 61.29%
Nearest Neighbors method correctly identified 32 male and female out of 62 volunteers in this study. It achieved an accuracy of 51.61%, a precision of 51.51%, a recall of 54.84%, and an F1-score of 53.14%. More details of this method are in the confusion matrix presented in Table 3.
As seen in Table 3, the Linear SVM classifier correctly identified 38 males and females out of 62, making it one of the methods that identified the highest numbers. This algorithm reached an accuracy of 61.29%, a precision of 51.72%, a recall of 78.13%, and an F1-score of 62.24%.
From the ECG recordings in our dataset, the RBF SVM method was capable, as seen in Table 3, of accurately predicting 28 males and females. It achieved the lowest accuracy out of any method at 48.38%, a precision of 38.46%, a recall of 16.67%, and an F1-score of 23.26%.
The Decision Tree algorithm could identify the correct gender in 4 instances. As seen in Table 3, it correctly classified 19 of the recordings as belonging to males and 20 of them as belonging to a female. Overall, this method was the one that could have the highest percentages. It achieved an accuracy of 62.90%, a precision of 61.29%, a recall of 63.33%, and an F1-score of 62.29%.
The Random Forest classifier correctly predicted 35 results. As seen in Table 3, it identified 15 males and 20 females. This method reached an accuracy of 56.45%, a precision of 57.69%, a recall of 48.39%, and an F1-score of 52.63%.
As seen in Table 3, the Neural Network classifier utilized in this study correctly identified 14 males and 18 females, counting 32. This method reached an accuracy of 51.61%, a precision of 45.16%, a recall of 51.85%, and an F1-score of 48.27%.
As seen in Table 3, the AdaBoost algorithm correctly classified 38 of the 62 volunteers. It achieved an accuracy of 61.29%, a precision of 46.67%, a recall of 63.63%, and an F1-score of 53.85%.
The Decision Tree algorithm presented in Table 3 could identify the gender achieving 38 males and females out of 62 volunteers. Overall, it achieved an accuracy of 61.29%, a precision of 46.67%, a recall of 63.63%, and an F1-score of 53.85%.
4 Discussions and Conclusions
We tested 8 methods on a dataset of ECG recordings to see how well they could categorize the data. We could use the confusion matrix produced by the application to our dataset to calculate each of these eight methods’ accuracy, precision, recall, and F1-score. The Decision tree attained the best accuracy at 62.90%, followed by the Linear SVM, AdaBoost, and Naive Bayes approach at 61.29% of accuracy.
With these outcomes, we concluded that Decision Trees was the technique that performed the best overall, which accurately identified 39 of the 62 results. Our initial expectations met the outcomes of this investigation, as the best-performing approach could accurately classify more than 50% of males and females in these ECG recordings.
This study used a small database, which could influence the results presented in these studies. We expect to get more data to consolidate the results in the future.
References
Alazzam, H., Alsmady, A., Shorman, A.A.: Supervised detection of IoT botnet attacks. In: Proceedings of the Second International Conference on Data Science, E-Learning and Information Systems, pp. 1–6 (2019)
AlDuwaile, D.A., Islam, M.S.: Using convolutional neural network and a single heartbeat for ECG biometric recognition. Entropy 23, 733 (2021)
Ali, F., et al.: A smart healthcare monitoring system for heart disease prediction based on ensemble deep learning and feature fusion. Inf. Fus. 63, 208–222 (2020)
Almuhaideb, S., Menai, M.E.B.: Impact of preprocessing on medical data classification. Front. Comput. Sci. 10(6), 1082–1102 (2016). https://doi.org/10.1007/s11704-016-5203-5
Amarappa, S., Sathyanarayana, S.V.: Data classification using support vector machine (SVM), a simplified approach. Int. J. Electron. Comput. Sci. Eng. 3, 435–445 (2014)
Balakumar, P., Maung-U, K., Jagadeesh, G.: Prevalence and prevention of cardiovascular disease and diabetes mellitus. Pharmacol. Res. 113, 600–609 (2016)
Batista, D., Plácido da Silva, H., Fred, A., Moreira, C., Reis, M., Ferreira, H.A.: Benchmarking of the BITalino biomedical toolkit against an established gold standard. Healthc. Technol. Lett. 6, 32–36 (2019)
Celin, S., Vasanth, K.: ECG signal classification using various machine learning techniques. J. Med. Syst. 42, 1–11 (2018)
Chio, C., Freeman, D.: Machine Learning and Security: Protecting Systems With data and Algorithms. O’Reilly Media, Inc. (2018)
Da Silva, H.P., Guerreiro, J., Lourenço, A., Fred, A.L., Martins, R.: BITalino: a novel hardware framework for physiological computing. In: PhyCS, pp. 246–253 (2014)
Escobar, L.J.V., Salinas, S.A.: e-Health prototype system for cardiac telemonitoring. In: 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, pp. 4399–4402. IEEE (2016)
García, V., Mollineda, R.A., Sánchez, J.S.: On the k-NN performance in a challenging scenario of imbalance and overlapping. Pattern Anal. Appl. 11, 269–280 (2008)
Gautam, M.K., Giri, V.K.: A neural network approach and wavelet analysis for ECG classification. In: 2016 IEEE International Conference on Engineering and Technology (ICETECH), Coimbatore, India, pp. 1136–1141. IEEE (2016)
Hastie, T., Rosset, S., Zhu, J., Zou, H.: Multi-class AdaBoost. Stat. Interface 2, 349–360 (2009). https://doi.org/10.4310/SII.2009.v2.n3.a8
Haykin, S.: Neural Networks: A Comprehensive Foundation, 1st edn. Prentice Hall PTR, Hoboken (1994)
Hsu, C.-W., Lin, C.-J.: A comparison of methods for multiclass support vector machines. IEEE Trans. Neural Netw. 13, 415–425 (2002)
Pires, I.M., Garcia, N.M., Pires, I., Pinto, R., Silva, P.: ECG data related to 30-s seated and 30-s standing for 5P-Medicine project. Mendeley Data (2022). https://data.mendeley.com/datasets/z4bbj9rcwd/1
Jindal, H., Agrawal, S., Khera, R., Jain, R., Nagrath, P.: Heart disease prediction using machine learning algorithms. In: IOP Conference Series: Materials Science and Engineering. IOP Publishing, p. 012072 (2021)
Kakria, P., Tripathi, N.K., Kitipawang, P.: A real-time health monitoring system for remote cardiac patients using smartphone and wearable sensors. Int. J. Telemed. Appl., 1–11 (2015). https://doi.org/10.1155/2015/373474
Kannathal, N., Acharya, U.R., Ng, E.Y.K., Krishnan, S.M., Min, L.C., Laxminarayan, S.: Cardiac health diagnosis using data fusion of cardiovascular and haemodynamic signals. Comput. Methods Programs Biomed. 82, 87–96 (2006). https://doi.org/10.1016/j.cmpb.2006.01.009
Maji, S., Berg, A.C., Malik, J.: Classification using intersection kernel support vector machines is efficient. In: 2008 IEEE Conference on Computer Vision and Pattern Recognition, pp. 1–8. IEEE (2008)
Pires, I.M., Garcia, N.M., Flórez-Revuelta, F.: Multi-sensor data fusion techniques for the identification of activities of daily living using mobile devices. In: Proceedings of the ECMLPKDD (2015)
Prescott, G.J., Garthwaite, P.H.: A simple Bayesian analysis of misclassified binary data with a validation substudy. Biometrics 58, 454–458 (2002)
Ramaraj, E.: A novel deep learning based gated recurrent unit with extreme learning machine for electrocardiogram (ECG) signal recognition. Biomed. Signal Process. Control 68, 102779 (2021)
Suthaharan, S.: Support vector machine. In: Suthaharan, S. (ed.) Machine Learning Models and Algorithms for Big Data Classification. Integrated Series in Information Systems, vol. 36, pp. 207–235. Springer, Boston (2016). https://doi.org/10.1007/978-1-4899-7641-3_9
Tran, T.M., Le, X.-M.T., Nguyen, H.T., Huynh, V.-N.: A novel non-parametric method for time series classification based on k-nearest neighbors and dynamic time warping barycenter averaging. Eng. Appl. Artif. Intell. 78, 173–185 (2019)
Vogel, B., et al.: The Lancet women and cardiovascular disease commission: reducing the global burden by 2030. Lancet 397, 2385–2438 (2021)
Webb, G.I., Boughton, J.R., Wang, Z.: Not so Naive Bayes: aggregating one-dependence estimators. Mach. Learn. 58, 5–24 (2005). https://doi.org/10.1007/s10994-005-4258-6
Neurophysiological Data Analysis with NeuroKit2 — NeuroKit2 0.2.1 documentation. https://neuropsychology.github.io/NeuroKit/. Accessed 10 July 2022
Acknowledgments
This work is funded by FCT/MEC through national funds and co-funded by FEDER – PT2020 partnership agreement under the project UIDB/50008/2020 (Este trabalho é financiado pela FCT/MEC através de fundos nacionais e cofinanciado pelo FEDER, no âmbito do Acordo de Parceria PT2020 no âmbito do projeto UIDB/50008/2020).
This article is based upon work from COST Action CA19136 - International Interdisciplinary Network on Smart Healthy Age-friendly Environments (NET4AGE-FRIENDLY), supported by COST (European Cooperation in Science and Technology). More information in www.cost.eu.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 ICST Institute for Computer Sciences, Social Informatics and Telecommunications Engineering
About this paper
Cite this paper
Bastos, E.S. et al. (2023). Preliminary Study on Gender Identification by Electrocardiography Data. In: Spinsante, S., Iadarola, G., Paglialonga, A., Tramarin, F. (eds) IoT Technologies for HealthCare. HealthyIoT 2022. Lecture Notes of the Institute for Computer Sciences, Social Informatics and Telecommunications Engineering, vol 456. Springer, Cham. https://doi.org/10.1007/978-3-031-28663-6_4
Download citation
DOI: https://doi.org/10.1007/978-3-031-28663-6_4
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-28662-9
Online ISBN: 978-3-031-28663-6
eBook Packages: Computer ScienceComputer Science (R0)