Abstract
When determining a lung nodule malignancy one must consider the spiculation represented by spike-like structures in the nodule’s boundary. In this paper, we develop a deep learning model based on a VGG16 architecture to locate the presence of spiculation in lung nodules from Computed Tomography images. In order to increase the expert’s confidence in the model output, we apply our novel Riemann-Stieltjes Integrated Gradient-weighted Class Activation Mapping attribution method to visualize areas of the image (spicules). Therefore, the attribution method is applied to the layer of the model that is responsible for the detection of the spiculation features. We show that the first layers of the network are specialized in detecting low-level features such as edges, the last convolutional layer detects the general area occupied by the nodule, and finally, we identify that spiculation structures are detected at an intermediate layer. We use three different metrics to support our findings.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Spiculation is one of the features used by medical experts to determine if a lung nodule is malignant [18, 30]. It is defined as the degree to which the nodule exhibits spicules, spike-like structures, along its border. This feature can be observed using imaging detection, which is easy to perform and causes less discomfort than alternative diagnosis methods such as a biopsy.
Automatic detection of features such as nodule spiculation by computer-aided detection (CAD) systems can help medical experts in the diagnosis process. For these systems to be adopted in clinical practice, their output has to be not only accurate, but also explainable in order to increase the trust between the human and the technology.
In this work, our main goal is to introduce a new approach to identify highly spiculated lung nodules using a convolutional network, and provide a visual explanation by highlighting the locations of the spicules. Furthermore, we want to determine what part of the network is responsible for the detection of spiculation. Convolutional neural networks (CNN) perform a bottom-up process of feature detection, beginning with low level features (such as edge detection) at the layers that are closer to the input, to high level features (such as the general location of an object) at layers closer to the output [12]. This is so because the kernels processing the outputs of each layer cover only a small area of the layer, and they can pay attention only to that small area. As the information flows from input to output it gets integrated into complex combinations of lower level features that can be interpreted as higher level features. In the particular problem studied here, i.e., detection of spiculation in lung nodules, we are interested in locating the defining elements of spiculation (the spicules) in the input image, and also what part of the network (which layer) plays the main role in detecting this particular feature. This is important because common attribution methods used to explain classification of images (like the ones discussed in the next section) are applied to a pre-selected layer of the network, so we need to determine which layer provides the strongest response to the presence of the feature that we are trying to detect. Our hypothesis is that spicule detection happens at some intermediate layer of the network, not necessarily the last one. In the process of testing this hypothesis we make the following contributions:
-
We use transfer learning on a network pretrained on a large dataset of images (ImageNet) to be used on CT scans of lung nodules to detect high/marked spiculation.
-
We apply a novel attribution method to locate the spicules in nodules classified with high/marked spiculation.
-
We identify the layer of the network that captures the “spiculation” feature.
The approach and methods used here are easy to generalize to other problems and network architectures, hence they can be seen as examples of a general approach by which not only the network output is explained, but also the hidden parts of the network are made more transparent by revealing their precise role in the process of feature detection.
2 Previous Work
Spiculation has been used for lung cancer screening [1, 17, 19, 20], and its detection plays a role in computer-aided diagnosis (CAD) [11]. New tools for cancer diagnosis have been made available with the development of deep learning, reaching an unprecedented level of accuracy which is even higher than that of a general statistical expert [8]. Following this success, the need of developing tools to explain the predictions of artificial systems used in CAD quickly arose. They take the form of attribution methods that quantify the impact that various elements of a system have in providing a prediction.
In the field of attribution methods for networks processing images there are a variety of approaches. One frequently used is Gradient-weighted Class Activation Mapping (Grad-CAM) [25], which produces heatmaps by highlighting areas of the image that contribute most to the network output. It uses the gradients of a target class output with respect to the activations of a selected layer. The method is easy to implement, and it has been used in computer-aided detection [15], but it does not work well when outputs are close to saturation levels because gradients tend to vanish. There are derivatives of Grad-CAM, such as Grad-CAM++ [3], that use more complex ways to combine gradients to obtain heatmaps, but they are still potentially affected by problems when those gradients become zero or near zero.
An attribution method that not only is immune to the problem of vanishing gradients, but can also be applied to any model regardless of its internal structure, is Integrated Gradients (IG) [27]. This method deals with the given model as a (differentiable) multivariate function for which we do not need to know how exactly its outputs are obtained from its inputs. IG is a technique to attribute the predictions of the model to each of its input variables by integrating the gradients of the output with respect to each input along a path in the input space interpolating from a baseline input to the desire input. One problem with this approach is that it ignores the internal structure of an explainable system and makes no attempt to understand the roles of its internal parts. While ignoring the internal structure of the model makes the attribution method more general, it deprives it from potentially useful information that could be used to explain the outputs of the model.
The attribution method used here, our novel Riemann-Stieltjes Integrated Gradient-weighted Class Activation Mapping (RSI-Grad-CAM), has the advantage of being practically immune to the vanishing gradients problem, and having the capability to use information from inner layers of the model [13].
3 Methodology
To detect spiculated nodules and localize the boundaries that present characteristics specific to spiculation, we employ the following steps:
-
1.
Train a neural network to classify lung nodules by spiculation level.
-
2.
Use an attribution method capable to locate the elements (spicules) that contribute to make the nodule having high spiculation.
-
3.
Provide objective quantitative metrics showing that the attribution method was in fact able to locate the spicules.
Additionally, we are interested in determining what part of the network is responsible for the detection of spiculation versus other characteristics of the nodule, such as the location of its contour (edge detection) and the general area occupied by the nodule. This is important because it tells us where to look in the network for the necessary information concerning the detection of the feature of interest (spiculation).
In the next subsections we introduce the dataset, deep learning model, and attribution method used. Then, we explain the metrics used to determine the impact of each layer of the network in the detection of spiculation.
3.1 Dataset
This work uses images taken from the Lung Nodule Image Database Consortium collection (LIDC-IDRI), consisting of diagnostic and lung cancer screening thoracic computed tomography (CT) scans with marked-up annotated lesions by up to 4 radiologists [9]. The images have been selected from CT scans containing the maximum area section of 2687 distinct nodules from 1010 patients. Nodules of three millimeters or larger have been manually identified, delineated, and semantically characterized by up to four different radiologists across nine semantic characteristics, including spiculation and malignancy. Given that ratings provided by the radiologists do not always coincide, we took the mode of the ratings as reference truth. Ties were resolved by using the maximum mode.
The size of each CT scan is \(512\times 512\) pixels, but most nodules fit in a \(64\times 64\) pixel window. Pixel intensities represent radio densities measured in Hounsfield units (HU), and they can vary within a very large range depending on the area of the body. Following guidelines from [7] we used the \([-1200,800]\) HU window recommended for thoracic CT scans, and mapped it to the 0–255 pixel intensity range commonly used to represent images.
The radiologists rated the spiculation level of each nodule in an ordinal scale from 1 (low/no spiculation) through 5 (high/marked spiculation) [5]. The number of nodules in each level is shown in Table 1. spiculation labels changed according to [5].
We aim to detect spike-like structures, similar to the binary split in [19], we combine the nodules in two classes to denote low-level spiculation (Class 1 - level 1) and high-level spiculation (Class 2 - levels 4 and 5). After eliminating nodules with sizes in pixels less than \(6\times 6\) (too small to significantly encode content information) and larger than \(64\times 64\) (there was only one nodule with that size in our dataset) we obtained a total of 1714 nodules in Class 1 and 234 nodules in Class 2. Each class is divided in training and testing sets in a proportion of 80/20. The final number of samples in each class is shown in Table 2. Images of \(64\times 64\) pixels with the nodule in the center are produced by clipping the original images. Figure 1 shows a few examples of images of nodules after clipping.
To achieve class balancing in the training set we add images obtained by random rotations and flips from the images of nodules with high spiculation (Class 2). In order to avoid losing the corners of the images during the rotations we work with clippings of size \(128\times 128\) pixels, and reclip to the final size \(64\times 64\) after rotation. We do not perform class balancing in the testing set.
3.2 Classifier Network
We built a classifier using transfer learning on a VGG16 network pretrained on ImageNet [2, 23, 26]. We used the base section of the VGG16 excluding its top fully connected layers, and added a global average pooling layer at the end, plus a fully connected layer with 512 outputs and ReLU activation function, followed by a fully connected layer with 1 output and sigmoid activation function. An n-class classifier network typically has n output units, but for a binary classifier one output unit suffices. The outputs are numerical, with target 0 representing Class 1, and target 1 representing Class 2.
We performed transfer learning in two steps:
-
1.
Model training: We froze all its layers except our two last fully connected layers and the last convolutional layer of each of its five blocks. The reason to retrain deep hidden layers is to help the network learn low level features of images that belong to a domain different from the original ImageNet on which it was pretrained.
-
2.
Model parameter tuning: We kept training the network with only the last (fully connected) layers unfrozen.
The loss function used in both trainings was the Mean Squared Error.
3.3 Attribution Method
We apply our novel attribution method, Riemann-Stieltjes Integrated Gradient-weighted Class Activation Mapping (RSI-Grad-CAM). This method can be applied to any convolutional network, and works as follows. First we must pick a convolutional layer A, which is composed of a number of feature maps, also called channels, \(A^1, A^2, \dots , A^{N}\) (where N is the number of feature maps in the picked layer), all of them with the same dimensions. If \(A^k\) is the k-th feature map of the picked layer, and \(A_{ij}^k\) is the activation of the unit in the position (i, j) of the k-th feature map, then, a localization map or “heatmap” is obtained by combining the feature maps of the chosen layer using weights \(w_k^c\) that capture the contribution of the k-th feature map to the output \(y^c\) of the network corresponding to class c. There are various ways to compute the weights \(w_k^c\). For example Grad-CAM, introduced in [25], uses the gradient of the selected output \(y^c\) with respect to the activations \(A_{ij}^k\) averaged over each feature map, as shown in Eq. (1). Here Z is the size (number of units) of the feature map.
Given that Grad-CAM is vulnerable to the vanishing gradients problem that occurs when the gradients are zero or near-zero [6], we propose to use our novel RSI-Grad-CAM method which handles the vanishing gradient problem. RSI-Grad-CAM computes the weights \(w_k^c\) using integrated gradients in the following way. First we need to pick a baseline input \(I_0\) (when working with images \(I_0\) is typically a black image). Then, given an input I, we consider the path given in parametric form \(I(\alpha ) = I_0 + \alpha (I - I_0)\), where \(\alpha \) varies between 0 and 1, so that \(I(0) =I_0\) (baseline) and \(I(1) = I\) (the given input). When feeding the network with input \(I(\alpha )\), the output corresponding to class c will be \(y^c(\alpha )\), and the activations of the feature map k of layer A will be \(A_{ij}^k(\alpha )\). Then, we compute the weights by averaging the integral of gradients over the feature map, as shown in Eq. (2).
The integral occurring in Eq. (2) is the Riemann-Stieltjes integral of function \(\partial y^c(\alpha )/\partial A_{ij}^k\) with respect to function \(A_{ij}^k(\alpha )\) (see [16]). For computational purposes this integral can be approximated with a Riemann-Stieltjes sum:
where \(\varDelta A(\alpha _{\ell }) = A_{ij}^k(\alpha _{\ell }) - A_{ij}^k(\alpha _{\ell -1})\), \(\alpha _{\ell } = \ell /m\), and m is the number of interpolation steps.
The next step consists in combining the feature maps \(A^k\) with the weights computed above, as shown in Eq. (4). Note that the combination is also followed by a Rectified Linear function \(\text {ReLU}(x) = \text {max}(x,0)\), because we are interested only in the features that have a positive influence on the class of interest.
After the heatmap has been produced, it can be normalized and upsampled via bilinear interpolation to the size of the original image, and overlapped with it to highlight the areas of the input image that contribute to the network output corresponding to the chosen class.
We also generated heatmaps using other attribution methods, namely Grad-CAM [25], Grad-CAM++ [3], Integrated Gradients [27], and Integrated Grad-CAM [24], but our RSI-Grad-CAM produced the neatest heatmaps, as shown in Fig. 3.
3.4 Metrics
In order to test the quality of our attribution method, we need to ensure that the metrics are not affected by limitations in the classification power of the neural network. Consequently, during the evaluation we use only sample images that have been correctly classified by the network (Table 4).
Since the spicules occur at the boundary of the nodule, we measure the localization power of our attribution method by determining to what extent the heatmaps tend to concentrate on the contour of the nodule (annotated by one of the radiologists) compared to other areas of the image. To that end, we compute the average intensity value of the heatmap along the contour (Avg(contour)), and compare it to the distribution of intensities of the heatmap on the whole image. Using the mean \(\mu \) and standard deviation \(\sigma \) of the intensities of the heatmap on the image, we assign a z-score to the average value of the heatmap along the contour using the formula
This provides a standardized value that we can compare across different images. We expect our attribution method will produce a larger \(z_{Avg(contour)}\) for high spiculation nodules compared to low spiculation nodules. We also study how the difference between high and low spiculation \(z_{Avg(contour)}\) varies depending on which layer we pick to apply our attribution method.
The first approach consisted in comparing boxplots of \(z_{Avg(contour)}\) for low and high spiculation (see Fig. 4). That provided a first (graphical) evidence of the power of our attribution method to recognize spiculation as a feature that depends on characteristics of the contour of a nodule. An alternative approach consists in comparing the cumulative distribution functions (cdfs) of \(z_{Avg(contour)}\) for high and low spiculation nodules (see Fig. 5).
Next, we measured the difference between the distributions of \(z_{Avg(contour)}\) for low and high spiculation nodules using three different approaches, two of them measuring distances between distributions, and the third one based on an hypothesis testing in order to determine which distribution tends to have larger values.
In the first approach we used the Energy distance between two real-valued random variables with cumulative distributions functions (cdfs) F and G respectively, given by the following formula [22, 28]:
Recall that the cdf F of a random variable X is defined \(F(x) = P(X\le x) = \) probability that the random variable X is less than or equal x.
Our second approach uses the 1-dimensional Wasserstein distance [10, 29], which for (1-dimensional) probability distributions with cdfs F, G respectively is given by the following formula [21]:
There are other equivalent expressions for Energy and 1-dimensional Wasserstein distances, here we use the ones based on cdfs for simplicity (they are \(L^p\)-distances between cdfs). The distributions of \(z_{Avg(contour)}\) are discrete, and cdfs can be used in this distribution, therefore formulas (6) and (7) are still valid [4].
The Energy distance (D) and Wasserstein distance (W) tell us how different two distributions are, but they don’t tell which one tends to take larger values. In our tests we gave a sign to the metrics equal to that of the difference of the means of the distributions. More specifically, assume the mean of distributions with cdfs F and G are respectively \(\mu _{F}\) and \(\mu _{G}\). Then, we define the signed metrics as follows:
where \(\text {sign}(x)\) is the sign function, i.e., \(\text {sign}(x) = x/|x|\) if \(x\ne 0\), and \(\text {sign}(0) = 0\).
The third approach consists in using the one-sided Mann-Whitney U rank test between distributions [14]. For our purposes this test amounts to the following. Let \(C_1\) be the set of samples with low spiculation, and let \(C_2\) be set of samples with high spiculation. For each sample \(s_1\) with low spiculation let \(u_1\) be the z-score of the average of the heatmap over the contour of \(s_1\). Define analogously \(u_2\) for each sample \(s_2\) with high spiculation. Let \(U_1=\) number of sample pairs \((s_1,s_2)\) such that \(u_1 > u_2\), and \(U_2=\) number of sample pairs \((s_1,s_2)\) such that \(u_1 < u_2\) (if there are ties \(u_1=u_2\) then \(U_1\) and \(U_2\) are increased by half the number of ties each). Then, we compute \((U_2-U_1)/(U_1+U_2)\), which ranges from \(-1\) to 1. If the result is positive that will indicate that \(u_2\) tends to take larger values than \(u_1\), while a negative value will indicate that \(u_1\) tends to take larger values than \(u_2\). The one-sided Mann-Whitney U rank test also assigns a p-value to the null hypothesis \(\text {H}_0 \equiv P(u_1 \ge u_2) \ge 1/2\) (where P means probability) versus the alternate hypothesis \(\text {H}_1 \equiv P(u_1 < u_2) > 1/2\).
4 Results
We used the VGG16 classifier network described in Sect. 3.2 trained using the following parameters (for training and fine tuning): learning rate \(= 0.00001\), batch size \(= 32\), number of epochs \(= 10\). The performance on the testing set is shown in the classification report, Table 3. The accuracy obtained was 91%.
Next, we provide evaluation results of our attribution method. In order to focus the evaluation on the attribution method rather than the network performance, we used only images of nodules that were correctly classified. The last column of Table 4 indicates the number of correctly classified nodules from each class.
We computed heatmaps using RSI-Grad-CAM at the final convolutional layer of each of the five blocks of the VGG16 network, using a black image as baseline. We used index 0 through 4 to refer to those layers, where 0 represents the last layer of the first block and 4 refers to the last layer of the last block, as shown in Table 5.
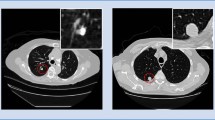
Figure 2 shows heatmaps produced by our RSI-Grad-CAM method at the final layer of the final convolutions block for a highly spiculated nodule. The contour of the nodule appears in red.
For comparison, Fig. 3 shows heatmaps produced by various attribution methods for a highly spiculated nodule at layer 3 (except for Integrated Gradients that applies to the layer input only).
We notice that in all the layers heatmaps tend to highlight more the contour of the highly spiculated nodule, but this is not yet evidence of detection of spicules in the contour. We expect the first layers of the network to be specialized in low level features such as edge detection, while the last layer may detect the general area occupied by the nodule. As stated in our hypothesis, we expect spicule detection to happen at some intermediate layer. The boxplots of \(z_{Avg(contour)}\) in (Fig. 4) provide evidence in favor of this hypothesis.
The comparison of the cdfs of the z-scores of average values of heatmaps on contours (Fig. 5) shows that the values tend to be larger for high spiculation nodules with respect to low spiculation nodules at layers 2 and 3, with the maximum difference at layer 3.
The Energy and Wasserstein distances between the distributions of z-scores of average values of heatmaps on contours for low and high spiculation nodules at each layer are shown in Table 6. In order to include information about what distribution takes higher values we multiplied the computed distance by the difference of means of the distributions to obtain signed metrics. The numbers still reach their maximum at layer 3, followed by layer 2.
The results for the 1-sided Mann-Whitney U rank test are shown in Table 7. The barplot shows that the maximum of \((U_2-U_1)/(U_1+U_2)\) happens at layer 3, which is consistent with the results obtained using Energy and Wasserstein metrics. The p-values obtained strongly favor the hypothesis that the z-scores of average values of heatmaps on contours are larger for high spiculation nodules at layers 2 and 3, with the maximum at layer 3.
5 Conclusions
We have used a modified VGG16 network retrained with a transfer learning technique to classify low and high spiculation nodules from images from the LIDC-IDRI database. Then, we applied the RSI-Grad-CAM attribution method to locate the elements of the images that contribute to the spiculation, i.e., spicules located at the boundary of the nodules. Furthermore, we were interested in determining what part of the network detects the spiculation feature. Common attribution methods are applied to a pre-selected layer of the network, so we need to determine which layer provides the strongest response to the presence of the feature that we are aiming to detect. Also, it is important to highlight that some features may be hard to locate directly in the model input, so methods based on heatmaps highlighting areas of the input may be less useful than the ones able to identify what internal parts of the model perform the detection of a given feature.
The metrics used to compare the distributions of average values of heatmaps on contours corresponding to low and high spiculation nodules were Energy distance, Wasserstein distance, and the 1-sided Mann-Whitney U rank test. All three test favor the hypothesis that the spiculation feature is detected at the last layer of the fourth convolutional block of the network (layer index 3, an intermediate hidden layer rather than the last one). In practice this means that, for our network, an explanation for the detection of spiculation can be provided by the heatmap produced at the last layer of the fourth convolutional block, since that heatmap tends to highlight the spicules occurring at the contour of the nodule.
The work performed here has been restricted to one network architecture (VGG16) performing binary classification, one image domain (images of lung nodules from the LICD-IDRI dataset), and one semantic feature (spiculation). Further work can be made to adapt the methods used here to other network architectures (e.g. ResNet, Siamese networks, etc.), multiclass classification (e.g. by adding the middle spiculation levels), data domains (e.g. natural language), and features (e.g. sample similarity).
References
Andrejeva, L., Geisel, J.L., Harigopa, M.: Spiculated masses, breast imaging. Oxford Medicine Online (2018). https://doi.org/10.1093/med/9780190270261.003.0025
Bengio, Y.: Deep learning of representations for unsupervised and transfer learning. In: Proceedings of the 2011 International Conference on Unsupervised and Transfer Learning Workshop, vol. 27, pp. 17–37 (2011)
Chattopadhyay, A., Sarkar, A., Howlader, P., Balasubramanian, V.N.: Grad-CAM++: generalized gradient-based visual explanations for deep convolutional networks. In: 2018 IEEE Winter Conference on Applications of Computer Vision (WACV) (2018). https://doi.org/10.1109/wacv.2018.00097
Devore, J.L., Berk, K.N.: Discrete random variables and probability distributions. In: Devore, J.L., Berk, K.N. (eds.) Modern Mathematical Statistics with Applications. STS, pp. 96–157. Springer, New York (2012). https://doi.org/10.1007/978-1-4614-0391-3_3
Hancock, M.C., Magnan, J.F.: Lung nodule malignancy classification using only radiologist-quantified image features as inputs to statistical learning algorithms: probing the lung image database consortium dataset with two statistical learning methods. J. Med. Imaging 3(4), 044504 (2016)
Hochreite, S.: The vanishing gradient problem during learning recurrent neural nets and problem solutions. Int. J. Uncertain. Fuzziness Knowl.-Based Syst. 06(02), 107–116 (1998)
Hofer, M.: CT Teaching Manual, A Systematic Approach to CT Reading. Thieme Publishing Group (2007)
Huanga, S., Yang, J., Fong, S., Zhao, Q.: Artificial intelligence in cancer diagnosis and prognosis: opportunities and challenges. Cancer Lett. 471(28), 61–71 (2020)
Armato III, S.G., et al.: Lung image database consortium: developing a resource for the medical imaging research community. Radiology 232(3), 739–748 (2004). https://doi.org/10.1148/radiol.2323032035
Kantorovich, L.V.: Mathematical methods of organizing and planning production. Manage. Sci. 6(4), 366–422 (1939)
Lao, Z., Zheng, X.: Multiscale quantification of tissue spiculation and distortion for detection of architectural distortion and spiculated mass in mammography. In: M.D., R.M.S., van Ginneken, B. (eds.) Medical Imaging 2011: Computer-Aided Diagnosis, vol. 7963, pp. 468–475. International Society for Optics and Photonics, SPIE (2011). https://doi.org/10.1117/12.877330
LeCun, Y., Bengio, Y., Hinton, G.: Deep learning. Nature 521, 436–444 (2015)
Lucas, M., Lerma, M., Furst, J., Raicu, D.: Visual explanations from deep networks via Riemann-Stieltjes integrated gradient-based localization (2022). https://arxiv.org/abs/2205.10900
Mann, H., Whitney, D.: On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 18(1), 50–60 (1947)
Mendez, D.M.M., Bermúdez, A., Tyrrell, P.N.: Visualization of layers within a convolutional neural network using gradient activation maps. J. Undergraduate Life Sci. 14(1), 6 (2020)
Protter, M.H., Morrey, C.B.: The Riemann-Stieltjes integral and functions of bounded variation. In: Protter, M.H., Morrey, C.B. (eds.) A First Course in Real Analysis. Undergraduate Texts in Mathematics. Springer, New York (1991). https://doi.org/10.1007/978-1-4419-8744-0_12
Nadeem, W.C.S., Alam, S.R., Deasy, J.O., Tannenbaum, A., Lu, W.: Reproducible and interpretable spiculation quantification for lung cancer screening. Comput. Methods Programs Biomed. (2020). https://doi.org/10.1016/j.cmpb.2020.105839
Paci, E., et al.: Ma01.09 mortality, survival and incidence rates in the italung randomised lung cancer screening trial (Italy). J. Thorac. Oncol. 12(1), S346–S347 (2017)
Qiu, B., Furst, J., Rasin, A., Tchoua, R., Raicu, D.: Learning latent spiculated features for lung nodule characterization. In: Annual International Conference of the IEEE Engineering in Medicine and Biology Society, pp. 1254–1257 (2020). https://doi.org/10.1109/EMBC44109.2020.9175720
Qiu, S., Sun, J., Zhou, T., Gao, G., He, Z., Liang, T.: Spiculation sign recognition in a pulmonary nodule based on spiking neural p systems. BioMed Res. Int. 2020 (2020). https://doi.org/10.1155/2020/6619076
Ramdas, A., Garcia, N., Cuturi, M.: On Wasserstein two sample testing and related families of nonparametric tests. Entropy 19(2), 47 (2017). https://doi.org/10.3390/e19020047
Rizzo, M.L., Székely, G.J.: Energy distance. Wiley Interdiscip. Rev. Comput. Stat. 8(1), 27–38 (2015)
Russakovsky, O., et al.: ImageNet large scale visual recognition challenge. Int. J. Comput. Vision 115(3), 211–252 (2015). https://doi.org/10.1007/s11263-015-0816-y
Sattarzadeh, S., Sudhakar, M., Plataniotis, K.N., Jang, J., Jeong, Y., Kim, H.: Integrated Grad-CAM: sensitivity-aware visual explanation of deep convolutional networks via integrated gradient-based scoring (2021). https://arxiv.org/abs/2102.07805
Selvaraju, R.R., Cogswell, M., Das, A., Vedantam, R., Parikh, D., Batra, D.: Grad-CAM: visual explanations from deep networks via gradient-based localization. Int. J. Comput. Vision 128(2), 336–359 (2019). https://doi.org/10.1007/s11263-019-01228-7
Simonyan, K., Zisserman, A.: Very deep convolutional networks for large-scale image recognition (2015). https://arxiv.org/abs/1409.1556
Sundararajan, M., Taly, A., Yan, Q.: Axiomatic attribution for deep networks. In: Precup, D., Teh, Y.W. (eds.) Proceedings of the 34th International Conference on Machine Learning. Proceedings of Machine Learning Research, vol. 70, pp. 3319–3328. PMLR (2017). https://proceedings.mlr.press/v70/sundararajan17a.html
Szekely, G.J.: E-statistics: the energy of statistical samples. Technical report, Bowling Green State University, Department of Mathematics and Statistics (2002)
Waserstein, L.N.: Markov processes over denumerable products of spaces, describing large systems of automata. Problemy Peredac̆i Informacii 5(3), 6–72 (1969)
Winkels, M., Cohena, T.S.: Pulmonary nodule detection in CT scans with equivariant CNNs. Med. Image Anal. 55, 15–26 (2019)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Lucas, M., Lerma, M., Furst, J., Raicu, D. (2023). Explainable Model for Localization of Spiculation in Lung Nodules. In: Karlinsky, L., Michaeli, T., Nishino, K. (eds) Computer Vision – ECCV 2022 Workshops. ECCV 2022. Lecture Notes in Computer Science, vol 13807. Springer, Cham. https://doi.org/10.1007/978-3-031-25082-8_30
Download citation
DOI: https://doi.org/10.1007/978-3-031-25082-8_30
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-25081-1
Online ISBN: 978-3-031-25082-8
eBook Packages: Computer ScienceComputer Science (R0)