Abstract
Diffusion MRI (dMRI) of the developing brain can provide valuable insights into the white matter development. However, slice thickness in fetal dMRI is typically high (i.e., 3–5 mm) to freeze the in-plane motion, which reduces the sensitivity of the dMRI signal to the underlying anatomy. In this study, we aim at overcoming this problem by using autoencoders to learn unsupervised efficient representations of brain slices in a latent space, using raw dMRI signals and their spherical harmonics (SH) representation. We first learn and quantitatively validate the autoencoders on the developing Human Connectome Project pre-term newborn data, and further test the method on fetal data. Our results show that the autoencoder in the signal domain better synthesized the raw signal. Interestingly, the fractional anisotropy and, to a lesser extent, the mean diffusivity, are best recovered in missing slices by using the autoencoder trained with SH coefficients. A comparison was performed with the same maps reconstructed using an autoencoder trained with raw signals, as well as conventional interpolation methods of raw signals and SH coefficients. From these results, we conclude that the recovery of missing/corrupted slices should be performed in the signal domain if the raw signal is aimed to be recovered, and in the SH domain if diffusion tensor properties (i.e., fractional anisotropy) are targeted. Notably, the trained autoencoders were able to generalize to fetal dMRI data acquired using a much smaller number of diffusion gradients and a lower b-value, where we qualitatively show the consistency of the estimated diffusion tensor maps.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Neonatal and fetal brain development involves complex cerebral growth and maturation both for gray and white matter [4, 10]. Diffusion MRI (dMRI) has been widely employed to study this developmental process in vivo, including neonates and fetuses [16, 18, 28]. As the diffusion weighted signal is sensitive to the displacement of water molecules, several models have been proposed for estimating the underlying anatomy such as diffusion tensor imaging (DTI) or spherical deconvolution methods [2, 6, 32]. The accuracy of these models is dependant on the angular and spatial resolution of the acquisitions that is typically limited for the neonate and fetal subjects [19, 22]. Stochastic motion and low signal-to-noise ratio (SNR) due to the small size of the developing brain often translate to degraded images with low spatial resolution. Additionally, slice thickness in fetal dMRI is typically high, varying between 3–5 mm, to freeze the in-plane motion, and hence reduces the sensitivity of the dMRI signal to the underlying anatomy. This highlights the need for methods to interpolate or synthesize new slices that were either (1) corrupted because of motion or (2) acquired using anisotropic voxel sizes. Interpolation is often performed either at scanner level or in post-processing [19], and has been demonstrated to be relevant for raw signal recovery and for subsequent analysis such as tractography [11]. Similarly, super-resolution (SR) methods that aim at increasing dMRI resolution can be applied at the acquisition-reconstruction level [27, 29] or at post-processing [5, 7, 12]. The latter used supervised learning methods, which require high resolution training data that is often unavailable for the developing brain. Additionally, these methods focus on enhancing the resolution homogeneously over all dimensions and were not assessed for anisotropic voxels, commonly acquired for fetuses and neonates [19, 22]. Additionally to the raw dMRI signal interpolation, other representations such as Spherical Harmonics (SH) could be of interest. SH are a combination of smooth orthogonal basis functions defined on the surface of a sphere able to represent spherical signals, such as the dMRI signal acquired using uniformly distributed gradient directions [13, 15]. Previous work used deep learning methods to map the SH coefficients from one shell to another [20, 24]. However, no prior work, to the best of our knowledge, relies on the SH decomposition to enhance the spatial image resolution.
In this study, we have used unsupervised learning to extend the application of autoencoders for through-plane super-resolution [21, 30] in the image domain to spherical harmonics domain where we synthesize SH coefficients of missing slices. As such, our network has access to both angular and spatial information. In contrast to training with non-DWI volumes [21], we have additionally trained a second network on spherical averaged dMRI images to complement and compare its performance in relation to the SH trained network. Moreover, we have compared both methods to conventional interpolation methods both using raw dMRI signals and their SH representation. The comparison was performed both on the raw dMRI signal; and on fractional anisotropy (FA) and mean diffusivity (MD) maps derived from the estimated diffusion tensors. Finally, we verified that the SH networks trained on pre-term data successfully generalized to fetal images, where we present the coherence of the synthesized slices.
2 Methodology
2.1 Materials
Neonatal Data - The developing Human Connectome Project (dHCP) dataFootnote 1 were acquired in a 3T Philips Achieva scanner in a multi-shell scheme (b \(\in \{0, 400,1000,2600\}\) s/mm\(^{2}\)). Details on acquisition parameters can be found in [17]. The data was denoised, motion and distortion corrected [3] and has a final resolution of \(1.17 \times 1.17 \times 1.5\) mm\(^3\) in a FOV of \(128 \times 128 \times 64\) mm\(^3\). In addition to \(b=0\) s/mm\(^{2}\) images (b0), we have selected the corresponding 88 volumes with \(b=1000\) s/mm\(^{2}\) (b1000) from all pre-term subjects (31) defined with less than 37 gestational weeks (GW) ([29.3, 37.0], mean = 35.5). In the anatomical dataset, brain tissue labels and masks [26] were provided.
Fetal Data - The fetal data were acquired with the approval of the ethics committee. Acquisitions were performed at 1.5T (GE Healthcare) with a single shot echo planar imaging sequence (TE = 63 ms, TR = 2200 ms) using \(b=700\) s/mm\(^{2}\) (b700) and 15 directions. The acquisition FOV was \(256\times 256\times 14-22\) mm\(^3\) for a resolution of \(1\times 1\times 4-5\) mm\(^3\). Three axial and one coronal acquisitions were performed for each subject. Four subjects were used in our study: two of 35 and 29 GW where three axial volumes were used, and two young subjects of 24 GW where one axial volume was used. We have only used axial acquisitions to avoid any confounding factor due to interpolation in the registration that would be needed between the orthogonal orientations. Volumes were corrected for noise [34], bias-field inhomogeneities [33] and distortions [1, 25] and did not require any motion correction.
2.2 Model
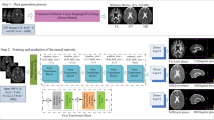
Network Architecture - Our network is composed of four blocks in the encoder and four blocks in the decoder, where each block consists of two layers of \(3\times 3\) convolutions, a batch normalization and an Exponential Linear Unit (ELU) activation function [9]. After each block of the encoder, a \(2\times 2\) average pooling operation was performed and the number of feature maps was doubled after each layer. Hence starting from 32 feature maps to 256 while three additional 3\(\,\times \,\)3 convolutions were added in the last block with 512, 256 and M feature maps respectively, \(M \in \{16,32,64,128\}\). The last M feature maps were considered as the latent space of our autoencoder. The decoder goes back to original input dimensions by means of either \(3\times 3\) transposed convolutions with strides of 2 or by \(2\times 2\) nearest neighbor interpolations (mutually exclusive), where the number of feature maps decreases by two after each layer from 512 to 32. A last 1\(\,\times \,\)1 convolution with sigmoid activation function was performed to generate the predicted image.
Training - Using the same architecture, we have trained three networks, with different inputs: b0 images (b0-net), average b1000 (Avg-b1000-net) (see Raw signal networks subsection) and a maximum SH order (\(L_{max}\)) of 4 (SH4-net) (see Spherical harmonics networks subsection). Input images were first normalized to the range [0, 1] by \(x = \frac{x - x_{min}}{x_{max} - x_{min}}\) where \(x_{min}\) and \(x_{max}\) are the minimum and maximum intensities respectively in a given slice. All networks were trained using an Nvidia GeForce RTX 3090 GPU in the TensorFlow framework (version 2.4.1) with Adam optimizer [23] for 200 epochs using mean squared error loss function, a batch size of 32 and a learning rate of \({5\times 10^{-5}}\). The validation was performed on 15% of the training data. The number of feature maps of the latent space was optimized using Keras-tuner [8] and the checkpoint with the minimal validation loss was finally selected for inference.
Raw Signal Networks - While b0-net was trained using b0 images, Avg-b1000-net was trained on average b1000 images, as training directly on individual b1000 images did not consistently converge [21]. We have thus trained Avg-b1000-net on average b1000 images with the aim of increasing the SNR and reducing variability. The average was computed over n randomly selected volumes, \(n\in \{3,6,15,30,40\}\). Empirically, higher n means a lower risk of network divergence, at the cost of increased smoothness/risk of losing image detail. Therefore n must be tuned. In the end, b0-net was used to infer b0 images whereas Avg-b1000-net was used to infer b1000 volumes.
Spherical Harmonics Network - We have fit SH representations by using \(L_{max}\) = 4 to the dMRI signal using Dipy [14] and fed the resulting 15 SH coefficients, slice by slice, to SH4-net. Let us note that we preliminary computed the mean squared error difference with respect to the ground truth data when estimating SH and projecting back to original grid from SH bases of \(L_{max} \in \{4, 6, 8\}\). As differences were relatively low between them (9.80, 8.64 and 9.95 for \(L_{max} \in \{4, 6,8\}\) respectively, scale \(\times 10^{-4}\)) and we aim at further testing on fetal data (where only 15 DWI are available) we selected to stick in what follows to \(L_{max}\) = 4.
Inference in Neonates - For all networks (b0-net, Avg-b1000-net and SH4-net), nested cross validation was performed where the 31 subjects were split into 8 folds. For each subject and each volume in the testing set, we removed N intermediate slices, \(N \in \{1,2\}\) that were considered as the ground truth we aim to predict. Using the two adjacent slices, we input each separately to the encoder part of the network to get the M latent feature maps. These feature maps were averaged using an equal weighting for \(N=1\) and a \(\{\frac{1}{3},\frac{2}{3}\}\), \(\{\frac{2}{3},\frac{1}{3}\}\) weighting for \(N=2\) (Fig. 1). The missing slices were then recovered by using the decoder part from the resulting latent feature maps. The output of the network was then mapped back to the range of input intensities. This was performed using histogram matching (using cumulative probability distributions) between the network output as a source image and the (weighted) average of the two adjacent input slices as a reference image. Finally, the histogram matched output of SH4-net was projected back to the original grid of 88 directions to recover the dMRI signal in the image domain.
Evaluation in Neonates - The inferred slices of Avg-b1000-net were compared to conventional interpolations, namely trilinear, tricubic and B-spline of \(5^{th}\) order [1, 31]. The comparison was performed separately for one and two missing slices (\(N \in \{1,2\}\)) using the mean squared error (MSE). As all interpolation baselines produce similar results with a slight overperformance for the linear method (for \(N=2\), MSE of 0.003164, 0.003204 and 003211 for linear, cubic and B-spline respectively), the former was chosen for further comparison with autoencoders. The two networks were additionally compared for FA and MD maps that were extracted from the diffusion tensors , as estimated in Dipy [14]. The DTI fit used the synthesized b0 by b0-net. The linear baseline was further compared with SH4-net and with the signal recovered from the same interpolation of the SH coefficients. The comparison was also extended for DTI maps (FA, MD). To compute them, DTI fit of SH4-net relied on the b0 as synthesized by b0-net, and the linear SH4 used corresponding linear interpolated b0. All comparisons were done using MSE for FA and MD maps in white matter, cortical gray matter, and corpus callosum. Moreover, we have fit SH representations of the ground truth signal by using \(L_{max} = 4\) which were compared after projecting back to the original grid of 88 gradient unit vectors to the original DWI signal, separately for (\(N \in \{1,2\}\)). This was considered as the lower bound error of SH4-net.
Application to Fetal DWI - After fitting the SH coefficients with \(L_{max}\)=4 to the fetal data. We have used SH4-net, i.e., trained on pre-term neonates to infer SH coefficients of middle (\(N \in \{1,2\}\)) slices of fetal subjects. The inference was performed in a similar manner as for neonates (Fig. 1). Cropping of fetal images to \(128\times 128\) voxels was necessary before feeding them to the encoder. Then, we generated the diffusion tensor based on this new DWI signal and b0 using b0-net, and visually assessed the consistency of the new slices in MD and FA maps for the four subjects. Only qualitative evaluation was performed for fetal enhancement because of the lack of ground truth.
3 Results
Based on the validation loss, the optimal number of feature maps in the latent space was found to be 32 for b0-net and Avg-b1000-net, and 64 for SH4-net. For Avg-b1000-net, averaging \(n=15\) DWI was also found to be optimal. Moreover, the transposed convolution in the decoder did not reduce the validation loss as compared to performing a nearest neighbor interpolation. Hence all networks used the latter in the decoder part to avoid unnecessary overparameterization of the network.
3.1 DWI Assessment
Autoencoder average b1000 trained network (Avg-b1000-net) produces superior results compared to linear interpolation (Fig. 2). The difference is higher for the case of two slices removed (\(N=2\)).
Mean squared error (MSE) on dMRI images of autoencoder enhanced using Avg-b1000-net slices (AE-1, AE-2 for \(N=1,2\) respectively) and for the baseline interpolation (linear on raw signal: Lin-1, Lin-2) and for SH4-net and SH linearly interpolated (Lin4-1, Lin4-2 for \(N=1,2\) respectively). The lower bounds for the SH errors (SH4GT) were also included as a reference. (Method-1, Method-2 for synthesizing/interpolating \(N=1\) and \(N=2\) slices, respectively)
Comparing raw and SH domain enhancement (Fig. 2), we first observe that independently of the method (autoencoder or linear), working directly on the raw signal outperforms working on SH and projecting back to signal. In fact, autoencoder Avg-b1000-net outperforms linear interpolation, and for \(N=1\) it is closely comparable to the SH encoding (SH4GT-1 in Fig. 2). While the SH autoencoder enhancement underperforms the classical SH linear interpolation for \(N=1\), SH4-net slightly outperforms linear-SH for \(N=2\). This gap between \(N=1\) and \(N=2\) for SH linear and autoencoder can be explained by the rich information that the autoencoder was exposed to in the training phase from similar images compared to the interpolation that has solely access to local information.
3.2 FA and MD in Newborns
Comparing DTI scalar maps (Fig. 3) for the same previous configurations (see Fig. 2), we notice that the autoencoder enhancement outperforms the linear interpolation in all brain regions (except MD for cortical gray matter when removing one slice, i.e. \(N=1\)) regardless of whether raw signal or SH was used. This outperformance is significant (paired Wilcoxon signed-rank test) for FA in all SH configurations, and for MD in one third of all configurations. The difference is typically more pronounced when we remove two slices (\(N=2\)). Let us note that, opposite of what we observed at the DWI signal level, SH4-net outperforms linearly interpolated SH. Furthermore, for the FA map, SH4-net obtains the lowest mean squared errors, thus it is more suitable than autoencoder Avg-b1000-net or the linear interpolation. The opposite trend, i.e. Avg-b1000-net outperforming SH4-net with statistical significance, can be noticed for MD, with exception of the corpus callosum.
Mean squared error of fractional anisotropy (FA) and mean diffusivity (MD) for different methods in three brain regions. See caption Fig. 2 for methods description. (Paired Wilcoxon signed-rank test: **: significant, p<0.028 - t: trending, p \(=\) 0.06 - N.S.: non significant: p>0.06)
3.3 Qualitative Results of FA and MD in Fetuses
The DWIs synthesized by SH4-net using the latent space were visually consistent as they smoothly vary between the adjacent slices. Figure 4 displays the corresponding FA and MD maps for four subjects. We can clearly delineate the smooth transition between the two adjacent slices, especially in late gestational weeks fetuses in which the structures are more visible. For instance, the corpus callosum and the internal capsules of the synthesized slices displayed in FA maps are coherent with respect to their neighbouring slices.
Fractional anisotropy (FA) and mean diffusivity (MD) for four fetal subjects of respectively, from left to right, 4, 5, 4 and 4 mm of slice thickness. The middle row (red frames) illustrates synthesized slices corresponding to the diffusion tensor reconstructed with inferred DWI volumes with SH4-net and b0 with b0-net, using the two neighboring original slices (top and bottom rows). (Color figure online)
4 Conclusion
We have proposed autoencoders for dMRI through-plane slice inference in early brain development. The assessment was performed in both raw signal and spherical harmonics (SH) domains, where the latter proved to be more accurate for DTI-FA maps reconstruction and the former for raw data estimation. We hypothesize that this could be explained by some global bias introduced to the back projected raw signal by the SH trained autoencoder. However, the orientation information (i.e., signal’s shape) was better preserved and hence, FA which is scale invariant, was clearly better depicted by SH autoencoder estimation. Lastly, we have successfully applied our method trained on newborn data to enhance the through-plane resolution of fetal data acquired in a different scanner, with a lower b-value and fewer gradient directions. Inferring missing slices or realistically increasing the through-plane resolution has to potential to translate to more accurate diffusion properties and hence a better uncovering of the underlying brain structure. In future work, we aim to increase the angular resolution in fetal images by using supervised learning to map spherical harmonics coefficients of order 4 (i.e., the maximal order that can be fit with clinical fetal images) to higher orders (6 or 8) using pre-term data.
References
Avants, B.B., Tustison, N., Song, G., et al.: Advanced normalization tools (ants). Insight J. 2(365), 1–35 (2009)
Basser, P.J., Mattiello, J., LeBihan, D.: Mr diffusion tensor spectroscopy and imaging. Biophys. J . 66(1), 259–267 (1994)
Bastiani, M., et al.: Automated processing pipeline for neonatal diffusion mri in the developing human connectome project. Neuroimage 185, 750–763 (2019)
Batalle, D., Edwards, A.D., O’Muircheartaigh, J.: Annual research review: not just a small adult brain: understanding later neurodevelopment through imaging the neonatal brain. J. Child Psychol. Psychiatry 59(4), 350–371 (2018)
Blumberg, S.B., Tanno, R., Kokkinos, I., Alexander, D.C.: Deeper image quality transfer: training low-memory neural networks for 3D images. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11070, pp. 118–125. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00928-1_14
Canales-Rodríguez, E.J., et al.: Sparse wars: a survey and comparative study of spherical deconvolution algorithms for diffusion MRI. Neuroimage 184, 140–160 (2019)
Chatterjee, S., et al.: Shuffleunet: super resolution of diffusion-weighted mris using deep learning. arXiv preprint arXiv:2102.12898 (2021)
Chollet, F., et al.: keras (2015)
Clevert, D.A., Unterthiner, T., Hochreiter, S.: Fast and accurate deep network learning by exponential linear units (elus). arXiv preprint arXiv:1511.07289 (2015)
Dubois, J., Alison, M., Counsell, S.J., Hertz-Pannier, L., Hüppi, P.S., et al.: Mri of the neonatal brain: a review of methodological challenges and neuroscientific advances. J. Magn. Reson. Imaging 53(5), 1318–1343 (2021)
Dyrby, T.B., et al.: Interpolation of diffusion weighted imaging datasets. Neuroimage 103, 202–213 (2014)
Elsaid, N.M., Wu, Y.C.: Super-resolution diffusion tensor imaging using srcnn: a feasibility study. In: 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), pp. 2830–2834. IEEE (2019)
Frank, L.R.: Characterization of anisotropy in high angular resolution diffusion-weighted mri. Magnetic Resonance in Med. Official J. Int. Soc. Magnetic Resonance Med. 47(6), 1083–1099 (2002)
Garyfallidis, E., et al.: Dipy, a library for the analysis of diffusion mri data. Frontiers in neuroinformatics 8 (2014)
Hess, C.P., Mukherjee, P., Han, E.T., Xu, D., Vigneron, D.B.: Q-ball reconstruction of multimodal fiber orientations using the spherical harmonic basis. Magnetic Resonance Med. Official J. Int. Soc. Magnetic Resonance Med. 56(1), 104–117 (2006)
Hüppi, P.S., Dubois, J.: Diffusion tensor imaging of brain development. In: Seminars in Fetal and Neonatal Medicine. vol. 11, pp. 489–497. Elsevier (2006)
Hutter, J., et al.: Time-efficient and flexible design of optimized multishell hardi diffusion. Magn. Reson. Med. 79(3), 1276–1292 (2018)
Jakab, A., et al.: Disrupted developmental organization of the structural connectome in fetuses with corpus callosum agenesis. Neuroimage 111 (2015)
Jakab, A., Tuura, R., Kellenberger, C., Scheer, I.: In utero diffusion tensor imaging of the fetal brain: a reproducibility study. NeuroImage: Clinical 15 (2017)
Jha, R.R., Nigam, A., Bhavsar, A., Pathak, S.K., et al.: Multi-shell d-mri reconstruction via residual learning utilizing encoder-decoder network with attention (msr-net). In: 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), pp. 1709–1713. IEEE (2020)
Kebiri, H., et al.: Through-plane super-resolution with autoencoders in diffusion magnetic resonance imaging of the developing human brain. Frontiers in Neurology 13 (2022)
Kimpton, J., et al.: Diffusion magnetic resonance imaging assessment of regional white matter maturation in preterm neonates. Neuroradiology 63(4), 573–583 (2021)
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980 (2014)
Koppers, S., Haarburger, C., Merhof, D.: Diffusion MRI signal augmentation: from single shell to multi shell with deep learning. In: Fuster, A., Ghosh, A., Kaden, E., Rathi, Y., Reisert, M. (eds.) MICCAI 2016. MV, pp. 61–70. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-54130-3_5
Kuklisova-Murgasova, M., Estrin, G.L., Nunes, R.G., Malik, S.J., Rutherford, M.A., et al.: Distortion correction in fetal epi using non-rigid registration with a laplacian constraint. IEEE Trans. Med. Imaging 37(1) (2017)
Makropoulos, A., et al.: Automatic whole brain mri segmentation of the developing neonatal brain. IEEE Trans. Med. Imaging 33(9), 1818–1831 (2014)
Ning, L., et al.: A joint compressed-sensing and super-resolution approach for very high-resolution diffusion imaging. Neuroimage 125, 386–400 (2016)
Ouyang, M., Dubois, J., Yu, Q., Mukherjee, P., Huang, H.: Delineation of early brain development from fetuses to infants with diffusion mri and beyond. Neuroimage 185, 836–850 (2019)
Ramos-Llordén, G., et al.: High-fidelity, accelerated whole-brain submillimeter in vivo diffusion mri using gslider-spherical ridgelets (gslider-sr). Magn. Reson. Med. 84(4), 1781–1795 (2020)
Sander, J., de Vos, B.D., Išgum, I.: Unsupervised super-resolution: creating high-resolution medical images from low-resolution anisotropic examples. In: Medical Imaging 2021: Image Processing, vol. 11596, p. 115960E. International Society for Optics and Photonics (2021)
Tournier, J.D., Calamante, F., Connelly, A.: Mrtrix: diffusion tractography in crossing fiber regions. Int. J. Imaging Syst. Technol. 22(1), 53–66 (2012)
Tournier, J.D., Yeh, C.H., Calamante, F., Cho, K.H., Connelly, A., Lin, C.P.: Resolving crossing fibres using constrained spherical deconvolution: validation using diffusion-weighted imaging phantom data. Neuroimage 42(2), 617–625 (2008)
Tustison, N.J., Avants, B.B., Cook, P.A., Zheng, Y., et al.: N4itk: improved n3 bias correction. IEEE Trans. Med. Imaging 29(6), 1310–1320 (2010)
Veraart, J., Fieremans, E., Novikov, D.S.: Diffusion mri noise mapping using random matrix theory. Magn. Reson. Med. 76(5), 1582–1593 (2016)
Acknowledgments
This work was supported by the Swiss National Science Foundation (project 205321-182602, grant No. 185897: NCCR-SYNAPSY- “The synaptic bases of mental diseases” and the Ambizione grant PZ00P2_185814). We acknowledge access to the facilities and expertise of the CIBM Center for Biomedical Imaging, a Swiss research center of excellence founded and supported by Lausanne University Hospital (CHUV), University of Lausanne (UNIL), École polytechnique fédérale de Lausanne (EPFL), University of Geneva (UNIGE), Geneva University Hospitals (HUG) and the Leenaards and Jeantet Foundations.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Kebiri, H. et al. (2022). Slice Estimation in Diffusion MRI of Neonatal and Fetal Brains in Image and Spherical Harmonics Domains Using Autoencoders. In: Cetin-Karayumak, S., et al. Computational Diffusion MRI. CDMRI 2022. Lecture Notes in Computer Science, vol 13722. Springer, Cham. https://doi.org/10.1007/978-3-031-21206-2_1
Download citation
DOI: https://doi.org/10.1007/978-3-031-21206-2_1
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-21205-5
Online ISBN: 978-3-031-21206-2
eBook Packages: Computer ScienceComputer Science (R0)