Abstract
Assessing prostate cancer grade from whole slide images (WSIs) is a challenging task. While both slide-wise and pixel-wise annotations are available, the latter suffers from noise. Multiple instance learning (MIL) is a widely used method to train deep neural networks using WSI annotations. In this work, we propose a method to enhance MIL performance by deriving weak supervisory signals from pixel-wise annotations to effectively reduce noise while maintaining fine-grained information. This auxiliary signal can be derived in various levels of hierarchy, all of which have been investigated. Comparisons with strong MIL baselines on the PANDA dataset demonstrate the effectiveness of each component to complement MIL performance. For 2,097 test WSIs, accuracy (Acc), the quadratic weighted kappa score (QWK), and Spearman coefficient were increased by 0.71%, 5.77%, and 6.06%, respectively, while the mean absolute error (MAE) was decreased by 14.83%. We believe that the method has great potential for appropriate usage of noisy pixel-wise annotations.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- Multiple instance learning
- Weak supervision
- Noisy labels
- Prostate cancer grade assessment
- Whole slide image
1 Introduction
Prostate cancer is one of the most common cancers in the world [8, 10]. Important prognostic information is inferred from Gleason patterns and grades which are categorized into international society of urological pathology (ISUP) grade groups [5] based on their severity. Assessing prostate cancer grades in whole slide images (WSIs) with giga-scale resolutions is time-consuming and pixel-wise annotations have significant noisiness [1].
Deep neural networks when used to assist diagnosis of cancer must indicate regions where Gleason patterns present for further confirmation. However, pixel-wise Gleason pattern annotations are known to be excessively noisy and its noise levels outweigh its potential benefits. Optimizing patch-wise metrics was insufficient to translate to slide-wise performance, and consequently learning algorithms typically have used pixel-wise annotations have been used only for feature extraction, while the final classifiers have been trained on the less noisy slide-wise annotations [9]. Multiple instance learning (MIL) is a widely used paradigm when classifying histopathological WSIs because slide-wise annotations can be obtained through medical information systems while pixel-wise annotations are not readily available [4]. Attention-based MIL emphasizes regions to locate sparsely-positioned lesions in core needle biopsy tissues but never directly accesses pixel-wise information.
Several attempts to utilize both WSI and pixel-wise annotations are outlined in [2, 9]. Instead of relying on noisy Gleason patterns, the studies use annotations indicating presence of tumor and separate localization from classification. Specifically, Strom and Kartasalo et al. applied boosting on ensembles of detection and grading networks and evaluated their patch-wise performances [9]. Bulten et al. mimicked a clinical setting where a feature extraction network learns to identify tumor positions [2]. Features were then extracted to train a classification model predicting ISUP grade groups. Without training on segmentation masks, [4] ranks of the top-K relevant patches and MIL were utilized. Relevant patches were subjected to recurrent neural network to diagnose malignant or benign tumors.
This work seeks to complement MIL by eliciting useful information from statistical approach in pixel-wise noisy annotations. Our experiments demonstrate that without carefully filtering pixel-wise noise, a combination with MIL amplifies errors in already mis-classified cases, e.g. ISUP grade 3 classified as 2 by a MIL model is classified as 1 by their combination. To allay such issues, we propose to construct weak-supervisory signals from noisy pixel-wise annotations. Annotation abstraction derived from Gleason patterns was shown to enhance spatial attention by reducing pixel-wise noise. Experiments demonstrated how coarse auxiliary signals effectively enhance an attention module’s accuracy and improve ISUP grading of prostate WSIs.
2 Materials and Method
2.1 Data
The Prostate cANcer graDe Assessment (PANDA) dataset containing 10,516 WSIs was used for this study [3]. Data were split into 8,419 and 2,097 WSIs of digitized hematoxylin and eosin (H &E)-stained biopsies for training and test. Slide-wise annotations are provided in the form of Gleason scores and corresponding severity grade ranging 0 to 5 according to the international society of urological pathology (ISUP) standard [5], and endpoints indicating no tumor or malignancy. Mask values in pixel-wise annotation depend on the data provider [3]. Masks acquired from different institutions come with different semantics and are converted to another mask indicating tumor presence. In this work, we excluded slide-wise Gleason scores to focus on the effect of annotation abstraction. The distribution of dataset is detailed in Table 1.
2.2 Architecture
The end-to-end network architecture commonly used throughout this work is described. An ImageNet-pretrained ResNeXt-50 extracted 2,048 channel pre-global average pooling features from a batch of \(b_g=16\) WSI inputs. Each WSI was split into \(b_{s}=32\) patches, with resolution of \(H=W=224\). A learnable global convolution filter followed by sigmoid activation was used to compute attention A and multiplied with the input feature. Post-attention features were fed to the classification layers to predict the ISUP grading group. The classification layer consists of max-pooling, average pooling layer, and fully connected layer(FC layer) as in Fig. 1(a).
2.3 Multiple Instance Learning for Cancer Grade Assessment
Let \(\mathcal {Y} = \left\{ 0, \dots , 5\right\} \) be the set of possible ordinal annotations describing ISUP grades. A classifier is trained to predict slide-wise ordinal annotations \(y \in \mathcal {Y}\). Its softmax prediction is denoted by \(\hat{p}\). Because classes share ordinal relations, the mean variance loss [7] is added to the standard cross entropy loss:
(a) An overview of the proposed method. Attention mechanism for multiple instance learning and additional network layers when using (b) no auxiliary loss, (c) patch-wise auxiliary loss, and (d) slide-wise auxiliary loss. \(b_{g}\): Global batch size, \(b_{s}\): slide batch size, C: Input channel size, W: Input width, H: Input height, \(C_{f}\): Initial feature channel
2.4 Noisy Labels and Weak Supervision
Raw pixel-wise annotations are extremely noisy [3], therefore have often been discarded [1]. Models trained using only MIL often weighed each patch equally because only ISUP grade groups were learnt. Appropriately processed fine-grained annotations can potentially inform the model to utilize local morphological features whose importance should be weighed differently.
The consensus of fine-grained pixel-wise annotations was rarely achievable, so that, the annotations method itself could be major component the noise of pixel-wise annotations. However, their abstraction at the increased coarseness releases pixel-wise noise in WSI annotations. Let \(\gamma _p, \gamma _s\) be ratio of tumor to total tissue area in each patch and slide:
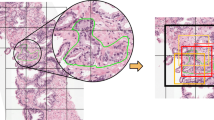
where \(\varOmega _\ell = \left\{ 1, \dots , H_\ell \right\} \times \left\{ 1, \dots , W_\ell \right\} \) is the resolution set of a patch or slide, i.e. its pixel indices, and \(M_\omega \) is the tumor indicator mask. The masks (Fig. 2(a)) are obtained from WSI using Akensert method [1], resolution being 1.0 micron per pixel (mpp). To ensure representation capacity for well-separability, we added a learnable block followed by sigmoid for each coarseness level p, s, shown in Fig. 1(b–d). The auxiliary losses (\(L_{\ell }\)) are then computed as the binary cross entropy \(H_2\) between predictions and the above ratio:
Combining all the losses considered, the total loss is then a convex combination between MIL and the auxiliary loss computed at varying levels of abstraction \(\ell \in \left\{ p, s\right\} \). Here, w is the for the auxiliary loss as follows:
3 Experiments
3.1 Implementation and Evaluation
We compared the performance of three baselines without the auxiliary loss and conducted an ablation study assessing the effectiveness of each auxiliary loss according to abstraction type and its weight (w). All models shared the same ResNeXt-50 (32\(\,\times \,\)4d) encoder. The first baseline is MIL model without both attention and auxiliary loss. This MIL baseline model already achieved high performance by positioning in the top-10 rank in the challenge. [1]. The second baseline model consisted of two stages. In the first stage, a U-Net model with ResNeXt-50 backbone networks was trained on pixel-wise annotations for feature extraction. In second stage, MIL with the freezed ResNeXt-50 in the end of first stage in Fig. 1(a) was trained on only slide-wise annotation based on the first stage’s output as typical methods [2, 9]. The third baseline adds only attention module without abstraction on top of the second baseline.
Ablation study proceeds with increasing levels of abstraction (patch/slide) with various coefficients (w). A coefficient of 0.7 on the auxiliary loss was found to work best via grid search which weighs the abstraction loss during the training of the model. AdamW optimizer [6] with 16 slides in each mini-batch was used with cosine annealing, and the initial learning rate was set to 1e\({-}\)4. Performance for ISUP grade group prediction were evaluated with respect to accuracy, mean absolute error (MAE), quadratic weighted kappa (QWK), and spearman rank correlations.
3.2 Results
As shown in Fig. 3(a), the accuracy of the model trained on pixel-wise noisy annotations was improved with the use of slide-wise annotations. This margin is similar to the gain achieved by adding attention to the MIL baseline (green dotted line in Fig. 3). However, inspecting other criteria (b–d) which penalizes incorrect predictions far from true annotations demonstrates how pixel-wise labels are detrimental in amplifying incorrect predictions. Acc, QWK, and Spearman coefficient were increased by 0.71%, 5.77%, and 6.06%, and MAE was decreased by 14.83% when adding slide-wise label abstraction to the MIL baseline. For such cases, models trained using either patch or slide-wise abstraction predicted ISUP grades closer to true annotations’. The higher levels of abstraction, the more noise filtered naturally, thereby the slide-wise annotations with high noise have achieved benefit. These results support that the use of auxiliary loss using abstracted annotations is more helpful in improving model performance.
We also visualized the distribution of predictions and true ISUP grade groups in Fig. 4. The QWK increased from 0.8190 to 0.8663 when using slide-wise abstractions. Under and over-estimated predictions with margin \(\ge 2\) are highlighted in blue and red triangles, respectively. The implications of under and over-estimates differ: over-estimations (blue) lead to unnecessary costs of care. Under-estimating the severity of cancer (red) is critical because a patient would not receive proper treatment. The cumulative number of upper triangular cases slightly increased by 9 cases (from 64 to 73), but the number of lower triangular cases decreased by 30% from 148 cases to 100 cases. This implies that the potential risk of a patient can be mitigated with the use of our method.
In this study, we tested the effective use of pixel-wise noisy labels in slide-wise inference. It showed a performance improvement in terms of QWK compared to slide-wise classification after attention based on the results of the segmentation model. Compared with the PANDA challenge, the source of the dataset we used, we note that there may be a slight performance difference because the train set and test set used are different from the challenge.
4 Conclusion
We proposed a method to guide a MIL attention network by performing abstraction to filter annotation noise. Our method demonstrated superior performance in comparison with strong baselines. In particular, the performance was improved for samples that were difficult to predict due to noisy annotations, thereby reducing the severity of misdiagnosis. We believe that this study has potential not only for pathology, but also for large-scale environments when fine-grained annotations are contaminated with substantial noise levels.
References
Bulten, W., et al.: Artificial intelligence for diagnosis and Gleason grading of prostate cancer: the panda challenge. Nat. Med. 24, 1–10 (2022)
Bulten, W., et al.: Automated Gleason grading of prostate biopsies using deep learning. arXiv preprint arXiv:1907.07980 (2019)
Bulten, W., Pinckaers, S., Eklund, K., et al.: The PANDA challenge: prostate cancer grade assessment using the Gleason grading system. MICCAI challenge (2020)
Campanella, G., et al.: Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 25(8), 1301–1309 (2019)
Egevad, L., Delahunt, B., Srigley, J.R., Samaratunga, H.: International society of urological pathology (ISUP) grading of prostate cancer-an ISUP consensus on contemporary grading (2016)
Loshchilov, I., Hutter, F.: Decoupled weight decay regularization. arXiv preprint arXiv:1711.05101 (2017)
Pan, H., Han, H., Shan, S., Chen, X.: Mean-variance loss for deep age estimation from a face. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 5285–5294 (2018)
Society, A.C.: About prostate cancer. https://www.cancer.org/cancer/prostate-cancer/about/key-statistics.html
Ström, P., et al.: Pathologist-level grading of prostate biopsies with artificial intelligence. arXiv preprint arXiv:1907.01368 (2019)
UK, P.C.: What is the prostate? https://prostatecanceruk.org/prostate-information/about-prostate-cancer
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Kim, H., Kong, S.T., Lee, H., Kim, K., Jung, KH. (2022). Abstraction in Pixel-wise Noisy Annotations Can Guide Attention to Improve Prostate Cancer Grade Assessment. In: Zamzmi, G., Antani, S., Bagci, U., Linguraru, M.G., Rajaraman, S., Xue, Z. (eds) Medical Image Learning with Limited and Noisy Data. MILLanD 2022. Lecture Notes in Computer Science, vol 13559. Springer, Cham. https://doi.org/10.1007/978-3-031-16760-7_3
Download citation
DOI: https://doi.org/10.1007/978-3-031-16760-7_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-16759-1
Online ISBN: 978-3-031-16760-7
eBook Packages: Computer ScienceComputer Science (R0)