Abstract
Purpose: Lung cancer is a life-threatening disease that affects both men and women . Accurate identification of lung cancer has been a challenging task for decades. The aim of this work is to perform a bi-level classification of lung cancer nodules. In Level-1, candidates are classified into nodules and non-nodules, and in Level-2, the detected nodules are further classified into benign and malignant.
Methods: A new preprocessing method, named, Boosted Bilateral Histogram Equalization (BBHE) is applied to the input scans prior to feeding the input to the neural networks. A novel Cauchy Black Widow Optimization-based Convolutional Neural Network (CBWO-CNN) is introduced for Level-1 classification. The weight updation in the CBWO-CNN is performed using Cauchy mutation, and the error rate is minimized, which in turn improved the accuracy with less computation time. A novel hybrid Convolutional Neural Network (CNN) model with shared parameters is introduced for performing Level-2 classification. The second model proposed in this work is a fusion of Squeeze-and-Excitation Network (SE-Net) and Xception, abbreviated as “SE-Xception”. The weight parameters are shared for the SE-Xception model trained from CBWO-CNN, i.e., a knowledge transfer approach is adapted.
Results: The recognition accuracy obtained from CBWO-CNN for Level-1 classification is 96.37% with a reduced False Positive Rate (FPR) of 0.033. SE-Xception model achieved a sensitivity of 96.14%, an accuracy of 94.75%, and a specificity of 92.83%, respectively, for Level-2 classification.
Conclusion: The proposed method’s performance is better than existing deep learning architectures and outperformed individual SE-Net and Xception with fewer parameters.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- Lung cancer
- Cauchy Black Widow Optimization based Convolutional Neural Network (CBWO-CNN)
- Shared network parameters
- SE-Xception
1 Introduction
Cancer is one of the world’s deadliest illnesses, with a high death rate in men and women. Cancer is formed by abnormal cell growth in any tissue, which leads to the formation of tissue lumps, masses, or nodules. Lung cancer is one of the most life-threatening cancers, accounting for the majority of cancer-related deaths. There has been increasing interest in research in the early identification of lung cancer by investigating lung nodules. Some Computer-Aided Diagnosis (CAD) systems have been previously developed, but there is still no accurate CAD system designed to identify and classify lung nodules. As most patients get lung cancer diagnosed in the middle or advanced stages, CAD systems will help in providing a second opinion for radiologists to proceed with further invasive tests [19]. Nodules in lung cancer may be characterized into two categories, namely, cancerous and non-cancerous. Malignant nodules are cancerous, while benign nodules are not.
The vital information is captured using images. These images can be acquired in different forms, such as Magnetic Resonance Imaging (MRI), Computed Tomography (CT) scans, radiographs (X-rays), Positron Emission Tomography (PET) scans, etc. CT scans proved to be more effective than other techniques and is preferable to the image mentioned above acquisition techniques. CT scanning can be used to identify lung mass tissue as it can detect minor irregularities that suggest lung cancer [1]. A bi-level lung cancer classification with shared network parameters is performed in this work. A novel Cauchy Black Widow based Convolutional Neural Network (CBWO-CNN) is used to classify nodules and non-nodules. A second model, which is a hybrid of SE-Net and Xception, is developed in this work. These two architectures have not been proposed as far as our knowledge is concerned. A knowledge transfer approach is used to train the SE-Xception model. The proposed method outperformed most state-of-the-art deep learning architectures in terms of performance.
1.1 Contribution of the Work
-
A novel preprocessing technique named Boosted Bilateral Histogram Equalization (BBHE) is adapted in this work to improve the quality of CT scans.
-
Level-1 classification is performed using the novel CBWO-CNN, in which Cauchy mutation is used to choose the best weights.
-
A novel SE-Xception CNN model with shared network parameters is proposed for performing Level-2 classification of lung cancer in CT scans.
-
The results obtained from the models are verified and recommended for deploying in real-time analysis from an expert pulmonologist.
The rest of the paper is organized as follows: The details of existing CAD systems are presented in Sect. 2. The dataset used in the proposed work is discussed in detail in Sect. 3. Section 4 provides detailed information on the proposed methodology. Section 5 presents a discussion of current architectures as well as the findings of the proposed approach. Section 6 provides the conclusion of the work done in this paper.
1.2 Related Work
In recent research, deep learning has captured many researchers’ attention in developing CAD systems for detecting and classifying nodules in lung cancer. An enhanced CNN was used to classify nodule and non-nodule on CT scans [9]. Deep Neural Networks (DNNs) usually are computationally expensive as they consist of many layers. In contrast to this, there are light-weight CNNs that provide comparatively accurate results close to the results of a DNN. A multi-section light-weight CNN for classification of nodules was proposed in [14]. Two such architectures, namely AlexNet and LeNet, were used in [21]. An optimal DNN was trained using a modified gravitational search algorithm used in [10], where these features were provided to Linear Discriminant Analysis (LDA) classifier. A deep convolutional network for lung nodule detection and classification was proposed in [12]. A combination of dense convolution network and a binary tree network, DenseBTNet, was proposed in [11]. A CNN architecture was proposed in [20] to perform lung cancer classification. Three different deep learning architectures were used in their work, namely CNN, DNN, and Stacked AutoEncoder. To classify lung cancer nodules, authors from [17] suggested a multi-scale CNN model. Hence, DNN usage can improve the CAD system’s performance.
2 Dataset
The dataset utilized for performing lung cancer nodules classification is LUng Nodule Analysis (LUNA), released in the year 2016. The LUNA16 challenge is an open challenge, where the reference standards and the images are made publicly available. LUNA16 dataset is a curated version of the LIDC-IDRI dataset [2], which is also a public-access dataset. Four expert radiologists have provided the annotations [15]. The main aim of the challenge was to develop large-scale automated nodule detection algorithms for the LIDC-IDRI dataset. Details related to the LUNA dataset are provided in Table 1.
3 Proposed Methodology
3.1 Overview
The proposed method is divided into four main stages: data preparation, Level-1 classification, transfer-learned knowledge, and Level-2 classification. The block diagram of the proposed methodology is shown in Fig. 1.
In the first stage, data preparation involves the preprocessing of input CT scans. The input CT needs to be preprocessed before providing the image data for the Level-1 classification task. BBHE filtering technique is used to preprocess input CT scans. The second stage is to differentiate between nodules and non-nodules. The dataset consists of very few positive nodules as compared to non-nodules. To mitigate this issue, positive lung nodule data is augmented only for training and validation dataset splits. This process is described in the data augmentation section. The level-1 classification task is performed using the proposed CBWO-CNN. The third stage is the transferring of learned knowledge from the CBWO-CNN model to the proposed SE-Xception model. The reason for adapting the transfer learning approach is that the size of the data used for classifying benign and malignant nodules is significantly less. Deep learning models perform well on large datasets. Therefore, to address this issue, a weight-sharing scheme is adapted in this work. The best pre-trained weights obtained from the trained CBWO-CNN model for nodule and non-nodule images are used as initialization weights for training the SE-Xception model for benign and malignant nodules. The classification of lung nodules into benign and malignant nodules is the fourth stage of the proposed method. A novel architecture named “SE-Xception” is proposed in this work. A detailed description of all the methods is provided in the below sections.
3.2 Image Preprocessing
Preprocessing provides the quality enhanced image to locate small particles in the scanned image. CT scans are stored in the image format of the ‘raw’ and ‘mhd’ extension. These CT scans are loaded using the python tool SimpleITK. The location of candidates is provided in X, Y, and Z coordinates. The candidate location is chosen based on the center of the nodule or non-nodule. A 32\(\,\times \,\)32 region is cropped out from each candidate location and saved into two classes, positive and negative, i.e., nodules and non-nodules. These classes are decided based on the annotations provided by the radiologists.
Various preprocessing methodologies have been developed, but still, quality remains to be a challenge. A Boosted Bilateral Histogram Equalization algorithm has been developed to improve the image quality so that small parts can be seen clearly. The detailed illustration of the BBHE is as follows:
Initially, the histogram is generated for the image \(\hbar \left( \chi _j \right) = n_j^p\). Then the histogram is smoothed by bilateral filtering to preserve the edges in an image given by Eq. 1.
where, \(\chi _j\) represents the \(j^{th}\) intensity level, \(n_j^p\) denotes the numbers of pixels having intensity level \(\chi _j\) and \(\psi \) denotes the bilateral filtering. Thereafter, boosting is applied on the edge preserved histogram given by Eq. 2.
where, m(k) denotes the peak histogram’s smoothed value, \(P_{min}\) and \(P_{max}\) denotes the local minimum and maximum pixel values, \(\alpha \) boosts the minor regions which is given by \(\alpha = log(P_{max}-P_{min})/log(m(k)-P_{min})\). Now, mapping is done using HE and the contrast enhance image is obtained (\(\chi '\)). \(\delta \) is multiplied by the gain (\(\varGamma _g\)) and noise reduction (\(\varGamma _n\)) functions to improve detail. The detail gain function is given as Eq. 3.
where, \(\varGamma _{g}\left( i,j \right) = 1/\left\{ \mathbb {R}.\left[ \chi _s(i,j)+1.0 \right] ^p \right\} \) and where \(\varGamma _{n}\left( i,j \right) = N_{offset} + \left\{ N_{band}/\left[ 1+e^{-\delta (i,j)-E} \right] \right\} \).
Finally, by multiplying the details with the detail function (\(\varGamma _{detail}\)) using Eq. 4, the improved detail image \(\delta '\) is obtained.
Equation 5 is used to integrate the \(\chi '\) and \(\delta '\) in the final step.
where, the final enhanced image is \(\chi ''{}\left( i,j\right) \), and w is a weighted function. The value of w can be anywhere between 0.0 and 1.0.
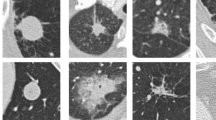
An illustration of nodules, non-nodules, benign and malignant nodules is shown in Fig. 2. There is a considerable difference in number of nodules and non-nodules in this dataset. To avoid the above-mentioned skewness in the data, the non-nodule data images are sub-sampled, i.e., the images are randomly selected from all the subsets of the dataset. Nodules must be further classified into benign and malignant nodules after the nodule, and non-nodule categories have been established.
3.3 Data Augmentation
In the above step, there is a clear data imbalance in the data of the two classes. To overcome this problem, the augmentation of positive nodules is performed. The images are augmented by performing some image operations such as modifying brightness, contrast, random rotation of the image to 90 \(^\circ \), transposing, scaling the images, and flipping the images horizontally and vertically. Figure 3 illustrates some of the augmented images of a nodules’ CT scan. The augmentation images shown in the figure are taken only from one CT scan.
3.4 Level-1 Classification: CBWO-CNN
Classifying candidates in a lung CT scan into nodules and non-nodules is critical because some nodules resemble tissues or organs in that area, making it difficult to classify the nodule properly. The training should be done properly to avoid high bias and low variance as well as low bias and high variance.

The first step in the algorithm is feature extraction \(F'_{EXT}\) from the input images. These features are fed convolutional layers \(\wp _2^{conv}\). The input image is filtered, and the convolution operation learns the same feature across the image. The pooling layer is responsible for reducing the Convolved Feature’s spatial size \(\wp _3^{pooling}\). This is to reduce the computing power needed by dimensionality reduction to process the data. The convolutional layer and the pooling layer form the \(i^{th}\) layer of a convolutional neural network together. The number of such layers can be increased to capture low-level information much more depending on the complexity of the images. Thus, the output from the convolutional layer is flattened \(\wp _3^{pooling}\left( \zeta _{flatten} \right) \). The output of the convolutional layer is distorted and fed to the fully connected layer as the input to the fully connected layer \(\wp _4^{FC}\). In order to minimize the loss Back Propagation is done which minimize the loss between the observed output and actual output. CBWO provides with selecting the best weights that is \( W_K^+ = \left\{ w_1^+, w_2^+,w_3^+,w_4^+,...w_n^+ \right\} \) and improving the accuracy within less computation time. The outline of the proposed technique that is CBWO-CNN, is elaborated in the form of pseudo-code in Algorithm 1.
3.5 Level-2 Classification: SE-Xception
Lung cancer nodule classification is a challenging task. A novel CNN, SE-Xception, a combination of two best performing and popular deep learning architectures, SE-Net, and Xception is introduced in this work. SE-Net architecture consists of a block named as Squeeze-and-Excitation (SE) block. In this block, the channel-wise feature responses are adaptively recalibrated using modeling channel inter-dependency explicitly. The SE block illustration can be shown in Fig. 4(a).
In the SE block diagram, \(F_{tr}\) represents a convolution operation where the input X is transformed to U. The \(F_{tr}\) in the proposed work is an Xception block. In previous works, inception and residual blocks are used as convolution operations. \(F_{tr}\) is represented in Eq. 6.
The notation \(V = [v_1, v_2, . . ., v_C]\) is used to illustrate a set of learned filter kernels, where \(v_C\) denotes the parameters of filter kernel. The outputs can be written as \( U = [u_1, u_2, . . . , u_C]\), where \(u_C\) is given in Eq. 7.
The features that are obtained after performing \(F_{tr}\) are U. The first operation that is carried out in the network is the passing of features through the squeeze operation (\(F_{sq}\)). A channel descriptor is produced from the feature maps when passed through the squeeze operation. The feature maps are aggregated across the spatial dimensions (H x W). An embedding of the global distribution of feature responses is generated channel-wise from this descriptor. It makes the information from the network’s global receptive area that all its layers are to use. The squeeze operation can be shown in Eq. 8. By shrinking U by its spatial dimensions H x W, a statistic \(z \epsilon R \) is generated such that \(c^{th}\) is calculated.
Followed by the squeeze operation, to capture the aggregated information of the channel descriptors, an excitation operation is performed. This operation fully captures the channel-wise dependencies, where it learns the non-linear and non-mutually-exclusive relationship of the channels. This operation is represented in Eq. 9, where Rectified Linear Unit (ReLU) activation function is denoted using \(\delta \) notation, \(W_1 = \mathbb {R}^{\frac{C}{r}X C}\), and \(W_2 = \mathbb {R}^{\frac{C}{r}X C}\).
Using a reduction ratio r, a bottleneck with two completely connected (FC) layers is created with dimensionality reduction. This block’s final output is given by rescaling U with s activations, which can be expressed in Eq. 10.
where \(X_c= x_1, x_2, . . . , x_c\), \(F_{scale}(u_c,s_c)\) represents channel-wise multiplication of scalar \(s_c\) and the feature map \(u_c \epsilon \mathbb {R} ^ {(H x W) } \).
Xception network architecture is completely based on depth-wise separable convolution blocks. The working of the Xception model is illustrated in Fig. 4(b). The hypothesis includes calculating spatial correlations, so it is possible to decouple cross-channel correlations in the CNN feature maps fully. The name Xception means “Extreme Inception”. It is named Xception because it has a more robust hypothesis than the underlying architecture of InceptionV3 [3]. Xception architecture consists of 36 convolution layers. These convolution layers are organized into 14 different modules. Every module consists of linear residual connections except for the first and last layers. In brief, the Xception network can be said as the stacking of depth-wise separable convolution layers in a linear fashion, which consists of residual connections. The input is data is mapped to spatial correlations for each output channel separately, and then 1\(\,\times \,\)1 depthwise convolution operation is performed. This operation captures the cross-channel correlation. These correlations can be pictured as a 2D+1D mapping instead of a 3D mapping. Here, the 2D space correlations are performed first, and then 1D space correlation is performed. Xception proved to provide slightly better results as compared to InceptionV3 on the LUNA16 dataset.
The proposed SE-Xception model is a combination of SE-Net and Xception. SE-Net consists of a squeeze-and-excitation block, which performs operations mentioned in the above sections. The addition of these modules in the Xception reduces the parameters of the model. Figure 4(c) shows the graphical representation of the proposed methodology. The figure represents the operations performed in the SE-Xception model. Only one module operation is illustrated. Input X is first passed to an Xception module. The input is given to a convolution filter with a filter size of 1, and then it is passed to a convolution filter with a filter size of 3. These two operations are concatenated, which is known as depth-wise separable convolution. This operation is followed by a SE-block where the squeeze and excitation operation is performed. Initially, to generate a channel descriptor, a global pooling operation is performed on the Xception module’s output. The pooling operation is followed by a Fully Connected (FC) layer with the ReLU as the activation function. The sigmoid activation function acts as a simple gating mechanism. This operation is known as the excitation operation. Once this step is completed, the output is scaled, represented as \(\widetilde{X}\).
4 Results and Discussion
4.1 Level-1 Classification
This section describes the performance of the proposed models. As the data is skewed, the non-nodules are selected by performing sub-sampling, where a set of non-nodule images are chosen from each subset to balance the data. The data imbalance may be the reason for overfitting the model, which affects the model’s performance. The model is validated on 10% of the total data and tested on 20% of the data. Adam optimizer used in both the networks. The loss function used is binary cross-entropy. The total number of epochs are set to 200. An early-stopping criterion is used while training, in which if no improvement is found in the validation loss after 20 epochs, the training is terminated. CBWO-CNN model is initially evaluated on four different activation functions namely, Exponential Linear Unit (ELU) [4], Tanh [13], LeakyReLU [5], and Rectified Linear Unit (ReLU) [8]. The model performance of various activation functions on the CBWO-CNN model is demonstrated in Fig. 5.
Considering ELU as an activation function in the networks, its convergence is faster than other activation functions. The difference between tanh and sigmoid lies in the range of the output value it returns. Tanh output value ranges from −1 to 1, whereas the sigmoid output value ranges from 0 to 1. However, the tanh function’s performance in the models’ hidden layers is relatively poor compared to other activation functions. LeakyReLU activation function is an extension of ReLU. The alpha value is added to the function to solve the issue of “dying ReLU" in this activation function. The results obtained from the LeakyReLU activation function in the hidden layer are second-best among the other activation functions used to evaluate the model. ReLU provide faster and more accurate results as it removes the negative values to pass to the next layer. The model performed best with the ReLU activation function in the hidden layer.
The performance metrics used to evaluate the CBWO-CNN model are accuracy, specificity, sensitivity, precision, recall, F1-score, and FPR. The corresponding results achieved from these confusion matrices are illustrated in Table 2. Average results obtained from the 5-fold validation of the model are depicted in the table. The table represents the performance measures stated above with their corresponding results.
Several popular deep learning architectures are used to evaluate the lung cancer classification system. The architectures chosen for the evaluation of the Level-1 classification system are CNN, VGG-19, Inception-V3, Deep Belief Network, ResNet, Recurrent Neural Network, and proposed CBWO-CNN. Accuracy, sensitivity, and specificity are the performance metrics that are evaluated. The results obtained are presented in Table 3. Hence, it can be noted that the proposed CBWO-CNN model performed better for Level-1 classification.
4.2 Level-2 Classification
The task of classifying positive lung nodules into benign and malignant nodules is known as lung nodule classification. In this work, the lung cancer nodule classification is performed using the proposed SE-Xception model with shared parameters from the CBWO-CNN model trained in Level-1 classification. To get the best performing model, the proposed model is evaluated on four activation functions in the model’s hidden layer. The activation functions used are ELU, Tanh, LeakyReLU, and ReLU. The network proved to provide better performance for the ReLU activation function. The four activation functions’ performance has been assessed for three models, SE-Net, Xception, and proposed SE-Xception is demonstrated in Fig. 6.
The proposed SE-Xception is evaluated with performance metrics such as accuracy, sensitivity, specificity, precision, F1-score, and FPR. The results achieved are presented in Table 4. The results are presented without shared network parameters and with shared network parameters for the SE-Xception model. The model trained without shared parameters resulted in overfitting of the model due to fewer training images. The accuracy achieved for the proposed SE-Xception is 82.29%. The improvement in the result is performed using pre-trained weights from Level-1 classification. This improved accuracy by almost 12%. The accuracy achieved for the proposed SE-Xception was 94.76%.
The proposed SE-Xception model is compared with the previous works performing lung cancer nodule classification. The results achieved from the proposed model outperformed the previous works. Performance comparison of the previous works is illustrated in Table 5.
The effect of the proposed SE-Xception method is visually demonstrated in Fig. 7. The images given in the green box are malignant nodules correctly classified as malignant nodules, and the images presented in the red box are benign nodules misclassified as malignant nodules. The reason for some of this misclassification is the indistinguishable similarity in the anatomical structure of the nodules. Even though the FPR of the proposed method is significantly less, the method still has the scope of improvement, as minimal misclassification is also not acceptable in medical applications.
5 Conclusion
Lung cancer classification has been in the interest of researchers for three decades. Several traditional and deep learning methods have been proposed for classifying and detecting lung cancer nodules. In this work, a bi-level classification model is proposed for the classification of the lung cancer nodules. To enhance the CT scan images, a novel BBHE technique is used. Level-1 classification performs bifurcation among nodules and non-nodules found in the candidates based on the locations provided by the radiologists. Level-2 classification performs classification of benign and malignant lung cancer nodules from the nodules identified in the Level-1 classification.
A novel deep learning architecture, CBWO-CNN, is introduced to perform Level-1 classification and a novel SE-Xception network is introduced to perform Level-2 classification. The proposed SE-Xception model uses the shared network parameters from the CBWO-CNN model used for training nodules and non-nodules. This hybrid network is designed and developed using recently proposed and best performing models, namely, SE-Net and Xception. SE-Net is made use to obtain a lesser number of parameters. The Xception network is an extreme version of the InceptionV3 network. CBWO-CNN and SE-Xception models are the contributions of this paper. The proposed network’s performance outperformed recently introduced deep learning models. In the future, a 3D model can be built and trained using 3D image input, where the model can learn volumetric information.
References
Aggarwal, T., Furqan, A., Kalra, K.: Feature extraction and LDA based classification of lung nodules in chest CT scan images. In: 2015 International Conference on Advances in Computing, Communications and Informatics (ICACCI), pp. 1189–1193. IEEE (2015)
Armato III, S., et al.: The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): a completed reference database of lung nodules on CT scans. Med. Phys. 38, 915–931 (2011)
Chollet, F.: Xception: deep learning with depthwise separable convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1251–1258 (2017)
Clevert, D.A., Unterthiner, T., Hochreiter, S.: Fast and accurate deep network learning by exponential linear units (ELUS). arXiv preprint arXiv:1511.07289 (2015)
Glorot, X., Bordes, A., Bengio, Y.: Deep sparse rectifier neural networks. In: Proceedings of the Fourteenth International Conference on Artificial Intelligence and Statistics, pp. 315–323 (2011)
Gong, J., Liu, J.Y., Wang, L.J., Sun, X.W., Zheng, B., Nie, S.D.: Automatic detection of pulmonary nodules in CT images by incorporating 3d tensor filtering with local image feature analysis. Physica Medica 46, 124–133 (2018)
Gupta, A., Das, S., Khurana, T., Suri, K.: Prediction of lung cancer from low-resolution nodules in CT-scan images by using deep features. In: International Conference on Advances in Computing, Communications and Informatics (ICACCI), pp. 531–537 (2018)
He, K., Zhang, X., Ren, S., Sun, J.: Delving deep into rectifiers: surpassing human-level performance on ImageNet classification. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 1026–1034 (2015)
Kasinathan, G., Jayakumar, S., Gandomi, A.H., Ramachandran, S.J., Patan, R.: Automated 3-D lung tumor detection and classification by an active contour model and CNN classifier. Expert Syst. Appl. 134, 112–119 (2019)
Lakshmanaprabu, S., Mohanty, S.N., Shankar, K., Arunkumar, N., Ramirez, G.: Optimal deep learning model for classification of lung cancer on CT images. Futur. Gener. Comput. Syst. 92, 374–382 (2019)
Liu, Y., Hao, P., Zhang, P., Xu, X., Wu, J., Chen, W.: Dense convolutional binary-tree networks for lung nodule classification. IEEE Access 6, 49080–49088 (2018)
Mendoza, J., Pedrini, H.: Detection and classification of lung nodules in chest x-ray images using deep convolutional neural networks. Comput. Intell. 36(2), 370–401 (2020)
Nwankpa, C., Ijomah, W., Gachagan, A., Marshall, S.: Activation functions: Comparison of trends in practice and research for deep learning. arXiv preprint arXiv:1811.03378 (2018)
Sahu, P., Yu, D., Dasari, M., Hou, F., Qin, H.: A lightweight multi-section CNN for lung nodule classification and malignancy estimation. IEEE J. Biomed. Health Inform. 23(3), 960–968 (2018)
Setio, A.A.A., et al.: Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: the LUNA16 challenge. Med. Image Anal. 42, 1–13 (2017)
Shaukat, F., Raja, G., Ashraf, R., Khalid, S., Ahmad, M., Ali, A.: Artificial neural network based classification of lung nodules in CT images using intensity, shape and texture features. J. Ambient. Intell. Humaniz. Comput. 10(10), 4135–4149 (2019)
Shen, W., Zhou, M., Yang, F., Yang, C., Tian, J.: Multi-scale convolutional neural networks for lung nodule classification. In: Ourselin, S., Alexander, D.C., Westin, C.-F., Cardoso, M.J. (eds.) IPMI 2015. LNCS, vol. 9123, pp. 588–599. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-19992-4_46
Silva, G.L.F.D., Filho, A.O.D.C., Silva, A.C., Paiva, A.C.D., Gattass, M.: Taxonomic indexes for differentiating malignancy of lung nodules on CT images. Res. Biomed. Eng. 32(3), 263–272 (2016)
Sinha, T.: Tumors: benign and malignant. Cancer Therapy Oncol. Int. J. 10(3), 555790 (2018)
Song, Q., Zhao, et al.: Using deep learning for classification of lung nodules on computed tomography images. J. Healthc. Eng. 1–7 (2017)
Zhao, X., Liu, L., Qi, S., Teng, Y., Li, J., Qian, W.: Agile convolutional neural network for pulmonary nodule classification using CT images. Int. J. Comput. Assist. Radiol. Surg. 13(4), 585–595 (2018). https://doi.org/10.1007/s11548-017-1696-0
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Dodia, S., Annappa, B., Padukudru, M.A. (2022). A Novel Bi-level Lung Cancer Classification System on CT Scans. In: Yang, G., Aviles-Rivero, A., Roberts, M., Schönlieb, CB. (eds) Medical Image Understanding and Analysis. MIUA 2022. Lecture Notes in Computer Science, vol 13413. Springer, Cham. https://doi.org/10.1007/978-3-031-12053-4_43
Download citation
DOI: https://doi.org/10.1007/978-3-031-12053-4_43
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-12052-7
Online ISBN: 978-3-031-12053-4
eBook Packages: Computer ScienceComputer Science (R0)