Abstract
We investigate the problem of diagnostic lung nodule classification using thoracic Computed Tomography (CT) screening. Unlike traditional studies primarily relying on nodule segmentation for regional analysis, we tackle a more challenging problem on directly modelling raw nodule patches without any prior definition of nodule morphology. We propose a hierarchical learning framework—Multi-scale Convolutional Neural Networks (MCNN)—to capture nodule heterogeneity by extracting discriminative features from alternatingly stacked layers. In particular, to sufficiently quantify nodule characteristics, our framework utilizes multi-scale nodule patches to learn a set of class-specific features simultaneously by concatenating response neuron activations obtained at the last layer from each input scale. We evaluate the proposed method on CT images from Lung Image Database Consortium and Image Database Resource Initiative (LIDC-IDRI), where both lung nodule screening and nodule annotations are provided. Experimental results demonstrate the effectiveness of our method on classifying malignant and benign nodules without nodule segmentation.
W. Shen and M. Zhou—These authors contributed equally.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- Lung nodule classification
- Computed Tomography (CT) Imaging
- Convolutional Neural Networks
- Computer-Aided Diagnoses (CAD)
1 Introduction
Lung cancer is notoriously aggressive with a low long-term survival rate [1]. Quantitative analysis in lung nodules using thoracic Computed Tomography (CT) has been a central focus for early cancer diagnosis, where CT phenotype provides a powerful tool to comprehensively capture nodule characteristics [2].
The importance of diagnostically classifying malignant and benign nodules using CT images is to facilitate radiologists for nodule staging assessment and individual therapeutic planning. Despite different approaches proposed for nodule analysis such as parametric texture feature extraction [4, 8, 13], they are problematic in finding well-suited parameters for robust analysis. It comes without doubt that, technical challenges still remain in defining and extracting quantitative features from clinical images for improving image-guided disease diagnosis. Furthermore, prior studies mostly focused on nodule morphology [5, 19], which may not be able to provide an accurate description of the nodule. For example, the definition of nodule boundaries is obscure and subjective—inter-reader variability from radiologists makes precise nodule delineation a challenging task. In view of these challenges, the following specific questions arise: (a) What should be done to learn discriminative features from heterogeneous nodule data for representing different diagnostic groups? (b) How could one design a robust framework that is capable of extracting quantitative features from original nodule patches—instead of segmented nodules—that is advantageous in completely eliminating onerous preprocessing steps such as a nodule segmentation?
An overview of the MCNN. Our approach first extracts multiple nodule patches to capture the wide range of nodule variability from input CT images. The obtained patches are then fed into the networks simultaneously to compute discriminative features. Finally, our approach applies a classifier to label the input nodule malignancy.
In this paper, we study the problem of lung nodule diagnostic classification based on thoracic CT scans. In contrast to the current methods primarily relying on nodule segmentation and textural feature descriptors for the classification task, we propose a hierarchical learning framework to capture the nodule heterogeneity by utilizing Convolutional Neural Networks (CNN) to extract features (as illustrated in Fig. 1). The learned features can be readily combined with state-of-the-art classifiers (e.g., Support Vector Machine (SVM) and Random Forest (RF)) for related Computer-Aided Diagnoses (CADs). Our method achieves 86.84 % accuracy on nodule classification using only nodule patches. We also observe that the proposed method is robust against noisy corruption—the classification performance is quite stable at different levels of noise inputs, indicating a well generalized property.
Contributions. We introduced an MCNN model to tackle the lung nodule diagnostic classification without delineation on nodule morphology and explored a hierarchical representation from raw patches for lung nodule classification. Our methodological contribution is three-fold:
-
Our MCNN take multi-scale raw nodule patches, rather than the segmented regions, providing evidence that information gained from the raw nodule patches is valuable for lung nodule diagnosis.
-
Our MCNN remove the need of any hand-crafted feature engineering work, such as nodule texture, shape compactness, and nodule sphericity. The MCNN can automatically learn the discriminative features.
-
Although it is challenging to directly deal with noisy data in nodule CT, we show that the proposed MCNN model is effective in capturing nodule characteristics in nodule diagnostic classification even with a high-level noisy corruption.
Related Work. Image-based lung nodule analysis is normally performed with nodule segmentation [5], feature extraction [2], and labelling nodule categories [8, 17, 19]. Way et al. [19] first segmented the nodules and then extracted texture features to train a linear discriminant classifier. El-Baz et al. [5] used shape analysis for diagnosing malignant lung nodules. Han et al. [8] used 3-D texture feature analysis for the diagnosis of pulmonary nodules by considering extended neighbouring structures. However, all of these mentioned methods relied on nodule segmentation as a prerequisite for nodule feature extraction. Notably, automated nodule segmentation can affect classification since segmentation usually depends on initialization, such as region growing and level set methods. Working on these segmented regions may yield inaccurate features that lead to erroneous outputs.
Descriptors of Histogram of Oriented Gradients (HOG) [4] and Local Binary Patterns (LBP) [13] are widely used for feature representation in medical image analysis. However, it is known that they are domain agnostic [15]. In other words, the required hyper-parameters make these approaches sensitive to specific tasks. For example, a repetitious parameter tuning is needed for the neighbourhood points in LBP and the size of the cell window in HOG.
Our work is conceptually similar to the massive training artificial neural network [17], which suggested a feasibility on learning knowledge from artificial neural networks. However, the work was an integrated classifier that required extra support from a 2-D Gaussian distribution for the decision-making, where an image-to-image mapping based on local pixels was learned. Our approach, without knowing any extra distributions, aims at feature extraction globally from the original nodule image space through stacked convolutional operations and max-pooling selections. In contrast to [17], our work is more computationally effective in reducing the feature dimensionality and resulting in highly discriminative features from hierarchical layers.
2 Learning Multi-scale Convolutional Neural Networks
Given a lung nodule CT image, our goal is to discover a set of globally discriminative features using the proposed MCNN model, which captures the essence of class-specific nodule information. The challenge is that the image space is extremely heterogeneous since both healthy tissues and nodules are included. In this work, we make full use of the CNN to learn discriminative features, and build three CNN in parallel to extract multi-scale features from nodules with different sizes. Details are given in this section.
2.1 Convolutional Neural Networks Architecture
Our Convolutional Neural Networks contain two convolutional layers, both of which are followed by a max-pooling layer, and a fully connected layer which represents the final output feature. The detailed structure of the network is shown in Fig. 2. From the input nodule patch to the final feature layer, the sizes of feature maps keep decreasing, which helps remove the potential redundant information in the original nodule patch and obtains discriminative features in nodule classification.
The structure of the Convolutional Neural Networks learned in our work. The numbers along each side of the cuboid indicate the dimensions of the feature maps. The inside cuboid represents the 3D convolution kernel and the inside square stands for the 2D pooling region. The number of the hidden neurons in the final feature layer is marked aside.
The network starts from a convolutional layer, which convolves the input feature map with a number of convolutional kernels and yields a corresponding number of output feature maps. Formally, the convolution operation between an input feature map f and a convolutional kernel h is defined by:
where \(f_c\) and \(h_c\) denote the cth slice from the feature map and that from the convolutional kernel respectively, and b is the bias scalar. \(*\) is the convolution operation. Both h and b are continuously learned in the training process. In order to perform a non-linear transformation from the input to the output space, we adopt the rectified linear unit (ReLu) non-linearity in Eq. 1 for each convolution [11]. It is expressed as \(y=\max (0, x)\), where x is the convolution output.
Following the convolutional layer, a max-pooling layer is introduced to select feature subsets. It is formulated as
where s is the pooling size and x denotes the output of the convolutional layer. An advantage of using the max-pooling layer is its translation invariability which is especially helpful when different nodule images are not well-aligned.
2.2 Multi-scale Nodule Representation
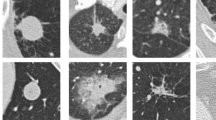
Our idea of the multi-scale sampling strategy is motivated from the clinical fact that nodule sizes vary remarkably, ranging from less than 3 mm to more than 30 mm in the Lung Image Database Consortium and Image Database Resource Initiative (LIDC-IDRI) [3] datasets. In the proposed MCNN architecture, three CNN that take nodule patches from different scales (as shown in Fig. 3) as inputs are assembled in parallel. We briefly refer to the three CNN as \( CNN _{ 0 }\), \( CNN _{ 1 }\), and \( CNN _{ 2 }\). In order to reduce the parameters of the MCNN, we follow the setting in [6] to share parameters among all the CNN. The resulting output of our MCNN is the concatenation of the three CNN outputs, forming the final discriminative feature vector, which will be directly fed to the final classifier without any feature reduction. We also follow the idea of deeply supervised networks (DSN) in [12] to construct our objective function. Unlike the traditional objective function in CNN, DSN introduced “companion objectives” [12] into the final objective function to alleviate the vanishing gradients problem so the training process can be fast and stable. The entire objective function is thus represented as
In our work, \(P(W)={LOSS}(W, w^{(out)})\) is the overall hinge loss function for the concatenated feature layer, and \(Q(W)=\sum _{m=1}^{M}\alpha _m loss(W,w^{(m)})\) is the sum of the companion hinge loss functions from all CNN. \(\alpha _m\) is the coefficient for the mth CNN. W denotes the combination of the weights from all of the CNN, while \(w^{(m)}\) and \(w^{(out)}\) are the weights of the feature layer of the mth CNN and the weights of the final concatenated feature layer respectively. In this way, F(W) keeps each network optimized and also makes the assembly sensible. Figure 4 shows the concatenated features projected into a 2-D subspace. It shows that the proposed MCNN model is able to remove the redundant information in the original images and extract discriminative features.
Feature visualization. The learned features by the MCNN from both training set (a) and test set (b) are illustrated by projecting them into a 2-D subspace with principal component analysis (PCA) [10].
3 Experiments
3.1 Datasets and Setup
We evaluated our method on the LIDC-IDRI datasets [3]. It consists of 1010 patients with lung cancer screening thoracic CT scans as well as mark-up annotated lesions. The nodules are rated from 1 to 5 by four experienced thoracic radiologists, indicating an increasing degree of malignancy (1 denotes low malignancy and 5 is high malignancy). In this study, we included nodules along with their annotated centers from the nodule reportFootnote 1. We chose the averaged malignancy rating for each nodule as in [8]. For those with an average score lower than 3, we labelled them as benign nodules; for those with an average score higher than 3, we labelled them as malignant nodules. We removed nodules with ambiguous IDs and those with an average score of 3. Overall, there were 880 benign nodules and 495 malignant nodules. Since the resolution of the images varied, we resampled those images to set the resolution to a fixed 0.5 mm/pixel along all three axes. Thus, the effect of resolution on the classification performance was removed. Then each nodule patch is cropped from the resampled CT image based on the marked nodule centers. The three scale inputs are \(32 \times 32 \times 32\), \(64 \times 64 \times 64\), and \(96 \times 96 \times 96\) in pixels. Patches are all resampled to \(32 \times 32 \times 32\) so that they can be uniformly fed into each CNN.
3.2 Implementation Details
We used a 5-fold cross validation for evaluating classification performance based on features learned from the MCNN. During each round of validation, there were originally 1100 nodules (704 benign nodules and 396 malignant nodules) in the training set and 275 nodules (176 benign nodules and 99 malignant nodules) in the test set. To enlarge the training samples to train the MCNN, we augmented both benign nodules and malignant nodules by translating the nodule patches along three axes with \(\pm 2\) pixels as in [16]. Thus, each patch was translated 6 times. Such a setting helps capture a range of translation invariant features. Note that the number of benign nodules is almost twice as large as that of malignant nodules. Thus, for the purpose of balancing the datasets, all of the malignant nodules were augmented and only half of the benign nodules were selected for augmentation, resulting in 5588 ((396 + 704/2)\(\,\times \,\)6 + 396 + 704) multi-scale nodules in the training set. Considering that the three CNN share the same parameters, the equivalent number of total augmented nodules can be 16,764. The test set always remained its original number of 275 nodule samples at each validation round.
To systematically evaluate the performance of the MCNN, we covered different network configurations, i.e. different numbers of convolutional kernels of each convolutional layer and that of the hidden neurons in the feature layer. The numbers of the convolutional kernels were \(n_1=\{50, 100\}, n_2=\{50, 100\}\) for the first and second layer and the number of neurons in the hidden layer was \(n_3=\{20, 50\}\). Therefore, there were 8 configurations in total for the MCNN. Note that we set \(\alpha _m = 0.001\) for all m as found best in [12]. The convolutional kernel size is \(5\times 5\times k\) which is quite typical in traditional CNN. k represents the third dimension of the input feature map. The pooling size was fixed to a \(2\times 2\) window. We added L2 norm weight decay during the training process to relieve overfitting. Two classifiers were used in experiments including SVM with a Radial Basis Function (RBF) kernel and RF. The hyper-parameters in both SVM and RF were obtained via a grid search on the training set.
We compared our results with two competing methods including HOG and LBP descriptors. For the HOG descriptor, we included different cell window sizes, \(s_w=\{8, 16, 32\}\) with the number of orientations \(n_o=8\). For the LBP descriptor, the uniform LBP descriptor was computed with different neighbourhood points \(n_{pt}=\{8,16,24\}\). Computation was done on all three scales of nodule patches for both descriptors.
Speaking of time complexity, although training deep networks often takes time, we choose a strategy of off-line training and on-line testing. In other words, once the network is finely trained off-line, it will be fast when a new sample comes in. In our study, using the NVIDIA Tesla K40 GPU, the test time for a single nodule patch was within 0.1 s. The CNN implementation used in this work was the deep learning toolkit CAFFE [9]. The classifiers of SVM and RF were from the scikit-learn package [14]. The HOG descriptor and the uniform LBP descriptor were from the scikit-image package [18].
3.3 Binary Nodule Classification Results
In this section, we evaluated the binary nodule classification. We used the average accuracy to observe the classification performance, i.e. the average ratio of corrected classified nodules from both classes from a 5-fold cross validation. Note that in the test set during each round of cross validation, 176 benign nodules and 99 malignant nodules made a baseline accuracy of 64 % by voting the majority class. From Fig. 5, it was immediately observed that the proposed MCNN showed competitive results above 84 % with different configurations. The highest classification accuracy was obtained with the RF classifier with 86.84 % under the configuration of \(n_1=100\), \(n_2=100\) and \(n_3=50\) (see Fig. 2). The overall performance of both classifiers suggested that our method can achieve promising results. The advantages can be ascribed to a factor that the hierarchical learning strategy selected high-level features, eliminating a number of redundant features. As already proven in [7], convolutional networks can produce useful dimensional reduction that is very helpful for image-related classification.
Accuracy of the HOG and LBP descriptors are shown in Table 1 (numbers in bold denote the best results in columns). It was apparent that the HOG descriptor was quite sensitive to the size of the cell window (\(s_w\)). The results of the HOG descriptor dropped and were even worse than the baseline when \(s_w\) expanded, indicating that the information gained is minimal when the size was becoming larger. For the LBP descriptor, we observed that the number of neighbourhood points (\(n_{pt}\)) was positively related to the performance since sophisticated neighbourhood structures led to better results. However, when comparing best results among different approaches, our MCNN outperformed HOG and LBP descriptors with 10.91 % and 13.17 %, respectively. Overall, our observation confirmed that parametric textural descriptors were sensitive to parameters.
3.4 Robustness to Noise Corruption
We further trained and tested the proposed MCNN on the challenging noisy data. In particular, we imposed a Gaussian noise to the original CT data. Different levels of noises were considered, including a mean value \(\mu =0\) and different standard deviations \(\sigma =\{0.5, 1.0\), \(2.0 \}\) (as shown in Fig. 6). As seen in Table 2, it was surprising that the MCNN still achieved 83.56 % and 83.27 % with \(\sigma = 2.0\), indicating the robustness of the MCNN against noisy inputs. The success could be probably explained by the fact that the max-pooling layers, which use the selective downsampling strategy, “filter out” noisy outliers, rendering the network robust to corrupted information. Therefore, the performance reaffirmed that the MCNN were capable of finding specific patterns that were inherently associated with different nodule classes.
3.5 Discussion
We have shown promising results of the proposed MCNN framework on classifying diagnostic nodule classes. Convolutional network is a powerful tool for image analysis because its capacities can be easily adjusted to a specific task and it makes strong and mostly correct assumptions about the nature of images [11]. In our study, although it suggested a clear need to further investigate the appropriate scales of nodules that lead to improved performance, we experimentally found that even with a single scale, the results remained competitive (for \( CNN _{ 0 }\), \( CNN _{ 1 }\), and \( CNN _{ 2 }\), it achieved 86.12 %, 83.88 %, and 79.00 % respectively). However, using the multi-scale strategy eliminated the careful designing of the patch sizes, which could be a tedious work. Rather than using isotropic kernels, we kept the third dimension of the kernels to be the same with the dimension of the input feature map which is quite common in the intermediate layers of conventional CNN. It also enabled us to directly use CAFFE out of the box.
4 Conclusion
In this paper, we proposed a Multi-scale Convolutional Neural Networks (MCNN) architecture to tackle the challenging problem on learning from lung nodule patches for nodule diagnostic classification. We demonstrated that the learned compact features are able to capture nodule heterogeneity. It is particularly promising that the MCNN model is robust against noisy inputs, which is valuable in the medical image analysis field. Extensive experiments showed that our method achieved 86.84 % for nodule classification and outperformed competing benchmark textural descriptors. In future work, we plan to expand data inclusion for a large-scale evaluation, and we will perform an investigation to seek appropriate scales for improving image-guided nodule analysis.
Notes
References
Aberle, D.R., Adams, A.M., Berg, C.D., Black, W.C., Clapp, J.D., Fagerstrom, R.M., Gareen, I.F., Gatsonis, C., Marcus, P.M., Sicks, J.: Reduced lung-cancer mortality with low-dose computed tomographic screening. N. Engl. J. Med. 365(5), 395–409 (2011)
Aerts, H.J., Velazquez, E.R., Leijenaar, R.T., Parmar, C., Grossmann, P., Cavalho, S., Bussink, J., Monshouwer, R., Haibe-Kains, B., Haibe-Kains, D., Rietveld, D., Hoebers, F., Rietbergen, M.M., Leemans, C.R., Dekker, A., Quackenbush, J., Gillies, R.J., Lambin, P.: Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 5, Article No. 4006 (2014). doi:10.1038/ncomms5006
Armato III, S.G., McLennan, G., Bidaut, L., McNitt-Gray, M.F., Meyer, C.R., Reeves, A.P., Zhao, B., Aberle, D.R., Henschke, C.I., Hoffman, E.A., et al.: The lung image database consortium (LIDC) and image database resource initiative (IDRI): a completed reference database of lung nodules on CT scans. Med. phys. 38(2), 915–931 (2011)
Dalal, N., Triggs, B.: Histograms of oriented gradients for human detection. In: IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR 2005), vol. 1, pp. 886–893 (2005)
El-Baz, A., Nitzken, M., Khalifa, F., Elnakib, A., Gimel’farb, G., Falk, R., El-Ghar, M.A.: 3D shape analysis for early diagnosis of malignant lung nodules. In: Székely, G., Hahn, H.K. (eds.) IPMI 2011. LNCS, vol. 6801, pp. 772–783. Springer, Heidelberg (2011)
Farabet, C., Couprie, C., Najman, L., LeCun, Y.: Learning hierarchical features for scene labeling. IEEE Trans. Pattern Anal. Mach. Intell. 35(8), 1915–1929 (2013)
Hadsell, R., Chopra, S., LeCun, Y.: Dimensionality reduction by learning an invariant mapping. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit. 2, 1735–1742 (2006)
Han, F., Zhang, G., Wang, H., Song, B., Lu, H., Zhao, D., Zhao, H., Liang, Z.: A texture feature analysis for diagnosis of pulmonary nodules using LIDC-IDRI database. In: IEEE International Conference on Medical Imaging Physics and Engineering, pp. 14–18 (2013)
Jia, Y., Shelhamer, E., Donahue, J., Karayev, S., Long, J., Girshick, R., Guadarrama, S., Darrell, T.: Caffe: Convolutional architecture for fast feature embedding (2014). arXiv preprint arXiv:1408.5093
Jolliffe, I.: Principal Component Analysis. Wiley Online Library, Chichester (2005)
Krizhevsky, A., Sutskever, I., Hinton, G.E.: Imagenet classification with deep convolutional neural networks. In: Advances in Neural Information Processing Systems, pp. 1097–1105 (2012)
Lee, C.Y., Xie, S., Gallagher, P., Zhang, Z., Tu, Z.: Deeply-supervised nets (2014). arXiv preprint arXiv:1409.5185
Ojala, T., Pietikainen, M., Maenpaa, T.: Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans. Pattern Anal. Mach. Intell. 24(7), 971–987 (2002)
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V., Thirion, B., Grisel, O., Blondel, M., Prettenhofer, P., Weiss, R., Dubourg, V., Vanderplas, J., Passos, A., Cournapeau, D.: Scikit-learn: machine learning in python. J. Mach. Learn. Res. 12, 2825–2830 (2011)
Prasanna, P., Tiwari, P., Madabhushi, A.: Co-occurrence of Local Anisotropic Gradient Orientations (CoLlAGe): distinguishing tumor confounders and molecular subtypes on MRI. In: Golland, P., Hata, N., Barillot, C., Hornegger, J., Howe, R. (eds.) MICCAI 2014, Part III. LNCS, vol. 8675, pp. 73–80. Springer, Heidelberg (2014)
Roth, H.R., Lu, L., Seff, A., Cherry, K.M., Hoffman, J., Wang, S., Liu, J., Turkbey, E., Summers, R.M.: A new 2.5D representation for lymph node detection using random sets of deep convolutional neural network observations. In: Golland, P., Hata, N., Barillot, C., Hornegger, J., Howe, R. (eds.) MICCAI 2014, Part I. LNCS, vol. 8673, pp. 520–527. Springer, Heidelberg (2014)
Suzuki, K., Li, F., Sone, S., Doi, K.: Computer-aided diagnostic scheme for distinction between benign and malignant nodules in thoracic low-dose ct by use of massive training artificial neural network. IEEE Trans. Med. Imaging 24(9), 1138–1150 (2005)
van der Walt, S., Schönberger, J.L., Nunez-Iglesias, J., Boulogne, F., Warner, J.D., Yager, N., Gouillart, E., Yu, T.: Scikit-image: Image processing in python. Technical report, PeerJ PrePrints (2014)
Way, T.W., Hadjiiski, L.M., Sahiner, B., Chan, H.P., Cascade, P.N., Kazerooni, E.A., Bogot, N., Zhou, C.: Computer-aided diagnosis of pulmonary nodules on CT scans: segmentation and classification using 3D active contours. Med. Phys. 33(7), 2323–2337 (2006)
Acknowledgments
The authors acknowledge the National Cancer Institute and the Foundation for the National Institutes of Health, and their critical role in the creation of the free publicly available LIDC/IDRI Database used in this study. We also gratefully acknowledge the support of NVIDIA Corporation with the donation of the Tesla K40 GPU used for this research. This paper is supported by the Chinese Academy of Sciences Key Deployment Program under Grant No. KGZD-EW-T03, the National Basic Research Program of China (973 Program) under Grant 2011CB707700, the National Natural Science Foundation of China under Grant No. 81227901, 61231004, 81370035, 81230030, 61301002, 61302025, major projects of Biomedicine Department of Shanghai Science and Technology Commission (13411950100), the Chinese Academy of Sciences Fellowship for Young International Scientists under Grant No. 2010Y2GA03, 2013Y1 GA0004, 2013Y1GB0005, the Chinese Academy of Sciences Visiting Professorship for Senior International Scientists under Grant No. 2012T1G0036, 2010T2G 36, 2012T1G0039, 2013T1G0013, the National High Technology Research and Development Program of China (863 Program) under 2012AA021105, the Guangdong Province-Chinese Academy of Sciences comprehensive strategic cooperation program under 2010A090100032 and 2012B090400039, the NSFC-NIH Biomedical collaborative research program under 81261120414, the National Science and Technology Supporting Plan under 2012BAI15B08, the Beijing Natural Science Foundation under Grant No. 4132080, the Fundamental Research Funds for the Central Universities under Grant No. 2013JBZ014.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this paper
Cite this paper
Shen, W., Zhou, M., Yang, F., Yang, C., Tian, J. (2015). Multi-scale Convolutional Neural Networks for Lung Nodule Classification. In: Ourselin, S., Alexander, D., Westin, CF., Cardoso, M. (eds) Information Processing in Medical Imaging. IPMI 2015. Lecture Notes in Computer Science(), vol 9123. Springer, Cham. https://doi.org/10.1007/978-3-319-19992-4_46
Download citation
DOI: https://doi.org/10.1007/978-3-319-19992-4_46
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-19991-7
Online ISBN: 978-3-319-19992-4
eBook Packages: Computer ScienceComputer Science (R0)