Abstract
The nuclear receptor superfamily is a group of transcriptional regulators that orchestrate multiple vital processes such as inflammation, metabolism, and cell proliferation. In recent years, it has become clear that some nuclear receptors form condensates in living cells. These condensates contain high concentrations of proteins and can contain millions of molecules. At these sites, high concentrations of nuclear receptors and co-factors potentially contribute to efficient transcription. While condensate formation has been observed for some nuclear receptors, the majority have unknown condensate formation abilities. Condensate formation abilities for these NRs would implicate an additional layer of regulation for the entire nuclear receptor family. Here, we consider the nuclear receptor superfamily, the current evidence for condensate formation of some of its members and the potential of the whole superfamily to form condensates. Insights into the regulation of assembly or disassembly of nuclear receptor condensates and our considerations for the understudied family members imply that condensate biology might be an important aspect of nuclear receptor-regulated gene transcription.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Transcriptional regulation
- Nuclear receptors
- Condensate formation
- intrinsically disordered regions
- Biomolecular condensates
- Liquid-liquid phase separation
1 The Nuclear Receptor Superfamily
The nuclear receptor (NR) superfamily is a group of transcription factors (TFs), of which many members are activated by different ligands such as steroid and thyroid hormones [1, 2]. There is a lot of interest in these receptors because ligand-activated NRs regulate multiple essential processes such as inflammation, metabolism, and cell proliferation. NRs regulate these processes by recruiting co-factors to specific promoter or enhancer sites, which results in transcriptional activation or repression [2,3,4,5,6]. Dysregulation of several NRs is implicated in cancer, atherosclerosis, diabetes, and other pathologies [2,3,4,5,6].
Currently, 48 members of the NR family are known in humans [6, 7]. These differ in their ligand and DNA binding domains, which is well summarized elsewhere [7, 8]. In general, NRs have a standard protein structure that consists of multiple domains in a specific order: the N-terminal domain (NTD), DNA binding domain (DBD), hinge, ligand binding domain (LBD), and the C-terminal domain [3, 5, 9, 10]. The NTD contains an activator function-1 region (AF1), which is responsible for interactions with co-factors, and this region is also important for transcriptional activation [3, 5, 9, 11]. The DBD is the most conserved region in the NR, comprises two zinc fingers and is responsible for targeting the NR to a specific DNA sequence. The hinge region is not only described to function as a connecter between the DBD and the LBD involved in nuclear translocation, but is in many cases also described as a region that is post-translationally modified, and this influences NR transactivation and ligand sensitivity [3, 5, 7]. The LBD is formed by 12 conserved α-helical regions numbered from H1 to H12 that undergo allosteric changes after ligand binding, leading to the activation of the NR. The activator function-2 region (AF-2) is a part of the LBD, which is responsible for transcriptional activation by recruitment of coregulator proteins and the transcription complex [3, 11,12,13].
In addition to the similarities in structure between the NRs, the transcriptional activation and repression by NRs are also regulated in a common way. Some NRs behave as transcriptional repressors when their ligand is absent. This repression is mediated by the recruitment of co-repressors such as NCoR1 (nuclear receptor co-repressor) in the unliganded state. This leads, for example, to the mobilization of histone deacetylases, and the resulting deacetylated histones lead to a more condensed chromatin structure. This prohibits RNA polymerase II binding, preventing transcriptional activation [7, 14,15,16]. Upon ligand binding and the resulting conformational change in the NR, the co-repressor complex is released [7, 15, 17]. Other NRs are localized in the cytosol in the unliganded state. Upon binding of the ligand, the NR translocates to the nucleus and binds to the DNA.
Subsequently, co-activators are recruited to the NR. Over 350 NR co-activators are known, of which some co-activators have histone acetyltransferase (HAT) activity which, in contrast to the histone deacetylases, leads to histone acetylation and decondensation of the chromatin [7, 18, 19]. Co-activators also support initiation of transcription by catalyzing the assembly of the transcription preinitiation complex at promoters [7, 18, 19].
Activated NRs bind as monomer to a specific DNA hexameric sequence, or as a homodimer or heterodimer to a dual hexameric repeat, which can be positioned in an inverted, everted, or direct orientation [3, 5, 7, 9, 12, 17]. Ultimately, NR-cofactor binding to these so-called hormone responsive elements (HRE) results in the recruitment of RNA polymerase II and the activation of transcription.
For transcriptional activation, it is vital that ligand, co-factors, RNA polymerase II and NR find each other in the crowded environment of the cell at the appropriate time and place to regulate transcription successfully [20,21,22,23]. As we will highlight in the next paragraphs, it is thought that NR condensation is the means by which this intricate process is achieved.
2 A Condensate Model for NR Transcriptional Regulation
Compartmentalization of the necessary proteins into organelles with membranes (e.g., nucleus, endoplasmic reticulum, lysosomes) is one of the methods by which a cell regulates the spatial and temporal localization of proteins required to assert certain functions. The second way this localization is regulated is the formation of membraneless organelles [20, 22, 24].
These membraneless organelles, also called biomolecular condensates, are compartments within a cell in which biomolecules such as proteins and nucleic acids assemble, and are typified by their droplet-like structure [20,21,22,23,24,25]. Although consensus around the biomolecular condensate term is recent, these structures have been observed since the late 1800s, when E.B. Wilsons described a liquid droplet-like organization in protoplasm using simple light microscopy [20, 26]. Recently, it has been demonstrated that several TFs, including some NRs (e.g., estrogen receptor alpha (ERα), glucocorticoid receptor (GR)), can form condensates within the nucleus [16, 20,21,22,23,24,25, 27, 28]. Moreover, these condensates have been observed to play a role in efficient transcriptional regulation [16, 20,21,22,23,24,25]. Condensates are likely to be formed through a biophysical process of phase separation. In phase separation, part of a homogenous solution de-mixes into two phases, a dense phase and a dilute phase [20, 23, 26, 29]. In cells, a specific type of phase separation can drive condensate formation by forming a liquid compartment in a liquid environment; this process is called liquid-liquid phase separation (LLPS) [20, 23, 26].
Currently, the evidence for LLPS underlying condensate formation is incomplete, mainly because some of the characteristics of LLPS are difficult to demonstrate in a cellular context [23]. However, there are now numerous examples that show that many different proteins can form condensates, which result in cellular compartments with high concentrations of these proteins. Among these examples are several NRs and co-factors, indicating that condensate biology likely plays a role in NR function.
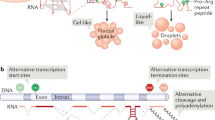
Formation of protein condensates is driven by weak multivalent interactions between proteins, and is dependent on protein concentration [20,21,22,23,24,25]. These weak multivalent interactions often involve prion-like domains or, more broadly, intrinsically disordered regions (IDRs). Prion-like domains in proteins are defined by the ability to assume multiple conformational states, and one of these states enabling binding to other copies of the same protein, which is favorable for condensate formation [30, 31]. An IDR is characterized by a low number of hydrophobic amino acids and enrichment in polar, charged and aromatic residues [21, 22, 25, 32]. These IDR properties also result in lack of a fixed 3D structure and these amino acids facilitate multivalent interactions that can potentially drive condensate formation (Fig. 14.1) [21, 25, 32].
The importance of these domains in condensate formation was demonstrated using OCT4 (octamer-binding transcription factor 4). OCT4 induced the formation of condensates that included the essential co-factor complex Mediator [33]. Disrupting the IDR of OCT4 blocked the formation of condensates, demonstrating the dependency of condensate formation on the IDR of OCT4. Interestingly, the lack of condensate formation was accompanied by a lower transcriptional output of OCT4 target genes [20, 33, 34]. Besides showing the importance of certain IDRs for condensate formation, this example suggests that condensate formation is involved in increasing the activation potential of TFs, which may include NRs.
Besides IDRs, important factors that influence condensate formation of TFs are the DNA accessibility and density of TF binding motifs. Digestion of DNA in situ disrupts condensate formation and adjusting the density of DNA elements controls condensate nucleation, indicating a role for DNA in nucleating condensates (Fig. 14.1) [22, 35, 36]. In addition to DNA, environmental factors such as temperature, ionic strength, protein and RNA concentrations, osmolarity, and pH levels have been suggested to play a role in condensate formation [20, 25, 36,37,38] (Fig. 14.1). The capacity of condensates to integrate so many biological signals, together with their effect on transcriptional output, suggests condensate formation is an additional layer of regulation of TFs, including NRs.
3 Evidence of NR Condensate Formation
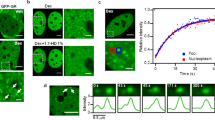
Since condensate formation results in the assembly of TFs, transcription complexes and co-factors, thereby increasing the transcription of target genes, this suggests an additional regulatory mechanism for NR-mediated transcriptional regulation (Fig. 14.1). Recently, condensate formation has been established for some NRs, mainly the group of steroid receptors. One of these steroid receptors is the glucocorticoid receptor (GR), which is activated upon glucocorticoid binding in the cytoplasm. GR subsequently translocates to the nucleus and binds to its HRE [39]. The existence of GR condensates has been shown via expression of a GFP-fusion construct in several cell lines [35, 36, 40, 41].
Several GR domains have been implicated to be essential for its condensation. Deletion of either the DBD or LBD reduced the number of GR condensates in cells [36, 42]. Moreover, mutation of a single amino acid (phenylalanine at position 623) decreased ligand binding, which reduced the number of condensates compared to wild-type GR, suggesting that ligand binding is essential for GR-condensate formation [42, 43]. Interestingly, deletion of the NTD did reduce the number of condensates in vitro, but not in cell culture experiments [36, 41, 42]. This is remarkable because the NTD contains an IDR, which are often found to be crucial in condensate formation [20, 33, 34].
Nevertheless, the NTD has a function in the formation of condensates under certain environmental circumstances [36, 41]. An increase in NaCl, osmolarity, or temperature induces a rise in the number of condensates for wild-type GR [36, 41] and is reversible, suggesting that the formed condensates are not the result of abnormal aggregate development [36]. However, NaCl treatment could not induce an increase in condensate formation of GR lacking the NTD [36]. This suggests that the NTD in GR is involved in condensate formation upon specific environmental cues. Contrarily, the LBD and DBD are essential for condensate formation independent of environmental cues [36, 41, 42]. This shows that specific domains within one protein can have different effects on condensate regulation.
To investigate the potential role of DNA density in the formation of GR condensates, Stortz et al. performed a GR condensate formation assay [36]. This demonstrated that GR condensates are formed independently of a particular chromatin state, which is in contrast to other TFs [35, 36, 41]. In addition, the condensate formation assay revealed that stimulation with a GR agonist results in GR condensate development at specific locations in the nucleus. Moreover, a lack of specific GR DNA binding motifs leads to a decrease in GR condensate formation [41]. Overall, these results indicate that specific GR binding DNA sequences are necessary for condensate formation. In these condensates, co-factors, such as Mediator, G9a, and SRC (steroid receptor co-activator), colocalize with GR [35, 41, 44].
The GR is not the only NR for which condensate formation has been described. Condensates containing ERα [28, 33, 45,46,47,48], mineralocorticoid receptor (MR) [49, 50], progesterone receptor (PR) [51, 52], and androgen receptor (AR) have also been demonstrated [46, 53, 54]. Similar to the GR, the LBD and DBD are essential in condensate formation of these NRs [50, 51, 54]. These NRs do not have prion-like domains, but do contain an IDR located in the NTD [11, 31, 41]. Interestingly, only the NTD of the AR has been found to have a crucial role in condensate formation, independent of environmental cues or other domains [50, 51, 54]. The contribution to condensate formation of the NTD is still debated for other NRs [45, 48, 53, 54].
Like GR, the condensates of ERα, PR, and AR colocalize with Mediator, but also with other NR-specific cofactors [45, 51, 53, 54]. For example, condensates of ERα together with MegaTrans components were observed at enhancer clusters together with estrogen responsive genes upon estrogen stimulation [28]. Interestingly, knockdown of Mediator decreased AR condensate formation and transcriptional output, which was not the case for GR (Fig. 14.1) [54]. This indirectly suggests that AR-condensate formation influences transcription. Another way of investigating the effect of NR condensate formation on target gene expression is with the aliphatic alcohol 1,6-hexanediol (HD) [36, 54, 55]. HD disrupts hydrophobic interactions between proteins and is used to target condensate formation. Despite pleiotropic effects of HD, it was shown that HD treatment disrupts ERα and AR condensates, which led to decreased gene activity of their target genes [28, 45, 47, 48, 54]. While these methods to determine transcriptional effects of condensate formation do not demonstrate a direct effect, Wei et al. showed a direct link between TF condensate formation and transcription by establishing that nascent RNA is enriched in the condensates, compared to an even distribution of RNA when these condensates are absent [56]. These results suggest that also for NRs there can be a direct link between condensate formation and transcriptional regulation.
In conclusion, condensate formation has been demonstrated for multiple NRs. The exact role of the different NR structures, such as the IDR, and the precise mechanism behind condensate formation are still unknown or might differ for each NR. However, the expectation is that NR condensate formation can influence transcription.
4 Potential Condensate Formation of the NR Superfamily
Currently, condensate formation has been described for five NRs (ERα, PR, AR, GR, and MR). These five NRs have characteristics typically associated with condensate formation, such as an IDR and interaction with Mediator (Fig. 14.1). To estimate the relevance of condensate biology for the NR family, we here predict the potential to form condensates for the other NR family members based on the characteristics of these five NRs.
To gain insight into the ability of the other NRs to form condensates, we have used different phase separation prediction tools. The characteristics of the condensate forming GR [35, 36, 40, 41, 57, 58], AR [54], MR [49, 50], PRs [51, 52], and ERα [47] were used to set a baseline for predicting condensate formation.
Typically, phase-separation prediction tools use one aspect that is important for phase separation, such as the presence of an IDR, a prion-like domain, or charged amino acids. However, dSCOPE (Detecting Sequence Critical fOr Phase sEparation) uses a combination of these factors to predict if a protein has a phase separation domain, a domain that has a combination of factors favorable for phase separation including IDRs, charged amino acids, low complexity, hydropathy, polarity, and a prion-like domains [59]. Therefore, dSCOPE was used to predict the presence of a “phase separation domain” in all 48 NRs (Table 14.1). dSCOPE predicted a phase separation domain in 22 out of the 48 human NRs. However, only three out of the five described condensate-forming NRs (ERα, PR, AR, GR, and MR) were predicted to have a phase separation domain by dSCOPE. To avoid relying on a single algorithm, two other common prediction programs were investigated for their ability to predict condensate formation.
Firstly, PondR (Predictor Of Naturally Disordered Regions) with predictor VSL2 was used (Table 14.1 and Fig. 14.2) [60, 61]. PondR provides a disorder score for each amino acid in a particular protein and when applied to the NR family it showed an IDR in most of the 48 NR family members, including each of the five benchmark NRs.
Secondly, PLAAC (Prion-Like Amino Acid Composition) was used to predict a prion-like domain in NRs. PLAAC predicts a prion-like domain in only two out of the five NRs known to form condensates, so the prion-like domain is unfit to predict condensate formation on its own. These results illustrate the difficulty of accurately predicting condensate formation. However, by comparing the results of the three prediction programs for these five NRs, some features seem to be common and potentially required for the formation of these condensates and could be used as criteria to estimate the likelihood of NR condensate formation.
Firstly, the presence of an IDR and the presence of either phase separation or prion-like domain(s) correlate with a higher likelihood to form condensates. A second, data-informed, factor is interaction with the Mediator complex which has been demonstrated to influence condensate formation not only for some NRs but also for other TFs [47, 51]. We have ranked the 48 NR factors according to likelihood of condensate formation based on the information above (Table 14.1). Based on the different prediction methods, all of the NRs have some hallmarks associated with condensate formation and we estimate that it is likely that a large portion of the family will indeed form condensates in vivo.
Future studies will validate whether the NRs indeed form condensates. Overexpression studies should be interpreted with care and at least be validated with endogenous NR expression (for example, by generating knock-in of an endogenously expressed NR-mEGFP fusion). Next, condensate formation upon ligand addition can be determined by confocal microscopy [36, 62]. To exclude aggregate formation, fluorescence recovery after photobleaching (FRAP) should be performed, to establish that the NR condensates are dynamic structures that exchange molecules with their surroundings [36, 62]. Together, such experiments will validate whether NRs can form liquid-like condensates in cells. Subsequently, essential NR domains can be investigated by means of deletion mutants or inactivating point mutations. Lastly, the effect on transcription should be demonstrated by nascent RNA labeling in the presence and absence of NR condensates [63]. These studies will provide insight in the function of condensate formation as a mechanism of transcriptional regulation for NR target genes.
Detailed understanding of NR condensates will be crucial to identify new methods to manipulate transcriptional output. In conclusion, based on the chosen prediction tools, many more NRs outside the five for which experimental evidence is available likely have the ability to form protein condensates.
5 Conclusion and Future Perspectives
The past few years have provided a lot of new insights into NR condensate formation suggesting that nuclear protein condensates partly regulate NR function [70, 71]. We used three different prediction programs to predict NR condensation for the complete NR family based on the characteristics of the five NRs (ERα, PR, AR, GR, MR) for which condensate formation has been established [59, 61, 72]. This showed that NR condensate formation is likely to be much more common than current experimental data has shown, potentially affecting a broad swathe if not all of the NR family.
Current knowledge of NR condensates and the implications for transcription is based on experiments using HD to disrupt interactions or by knocking out co-factors [45, 47, 48, 54], which need to be carefully interpreted because the pleiotropic effects of HD or potential indirect effects of co-factor knock-out. Therefore, detailed studies of NR condensate formation and its influence on transcription are necessary to provide more insight into how NR transcription is regulated.
Other transcriptional regulators, such as TAF15 (TATA-binding protein-associated factor 15) and p300, can form condensates that enhance transcriptional output and gene activation [56, 70]. This demonstrates that these transcriptional regulators influence transcription, implicating that NR condensates can potentially also directly influence transcriptional output. The different prediction tools showed that TAF15 has a predicted percentage disordered of 93%, a phase separation domain and prion-like domain [56, 59, 61, 72]. This supports our suggestion that a high fraction of disordered protein combined with a phase separation domain, and a prion-like domain enhance the chance of condensate formation.
However, there is an important difference between NRs and other TF such as TAF15. TF activation is complex and can involve different intracellular signal transduction pathways, while NRs are directly activated by lipophilic ligands [73]. The five described NRs form condensates only in the presence of their ligand, suggesting a role for ligands in NR condensate formation [39]. NR ligands thereby add to the complexity of condensate regulation.
Further detailed studies on the underlying forces of NR condensate formation and the influence of condensate formation on transcription will provide a better understanding of how NR condensate formation can influence transcription, and could potentially be exploited to manipulate this therapeutically.
References
Robinson-Rechavi M, Garcia HE, Laudet V (2003) The nuclear receptor superfamily. J Cell Sci 116(4):585–586
Sever R, Glass CK (2013) Signaling by nuclear receptors. Cold Spring Harb Perspect Biol 5(3):a016709
Weikum ER, Liu X, Ortlund EA (2018) The nuclear receptor superfamily: a structural perspective. Protein Sci 27(11):1876–1892
Mangelsdorf DJ, Thummel C, Beato M et al (1995) The nuclear receptor superfamily: the second decade. Cell 83(6):835
Olefsky JM (2001) Nuclear receptor minireview series. J Biol Chem 276(40):36863–36864
Kumar R, McEwan IJ (2012) Allosteric modulators of steroid hormone receptors: structural dynamics and gene regulation. Endocr Rev 33(2):271–299
Pawlak M, Lefebvre P, Staels B (2012) General molecular biology and architecture of nuclear receptors. Curr Top Med Chem 12(6):486–504
Moras D, Gronemeyer H (1998) The nuclear receptor ligand-binding domain: structure and function. Curr Opin Cell Biol 10(3):384–391
Mukha A, Kalkhoven E, van Mil SW (2021) Splice variants of metabolic nuclear receptors: Relevance for metabolic disease and therapeutic targeting. Biochim Biophys Acta (BBA) - Mol Basis Dis 2021:166183
Mazaira GI, Zgajnar NR, Lotufo CM et al (2018) The nuclear receptor field: a historical overview and future challenges. Nucl Receptor Res 5:101320
Simons SS Jr, Edwards DP, Kumar R (2014) Minireview: dynamic structures of nuclear hormone receptors: new promises and challenges. Mol Endocrinol 28(2):173–182
Porter BA, Ortiz MA, Bratslavsky G, Kotula L (2019) Structure and function of the nuclear receptor superfamily and current targeted therapies of prostate cancer. Cancers 11(12):1852
Wärnmark A, Treuter E, Wright AP, Gustafsson J (2003) Activation functions 1 and 2 of nuclear receptors: molecular strategies for transcriptional activation. Mol Endocrinol 17(10):1901–1909
Yoon H, Chan DW, Huang Z et al (2003) Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J 22(6):1336–1346
Sladek FM (2011) What are nuclear receptor ligands? Mol Cell Endocrinol 334(1–2):3–13
Cho W, Spille J, Hecht M et al (2018) Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361(6400):412–415
Holzer G, Markov GV, Laudet V (2017) Evolution of nuclear receptors and ligand signaling: toward a soft key–lock model? Curr Top Dev Biol 125:1–38
Han SJ, Lonard DM, O’Malley BW (2009) Multi-modulation of nuclear receptor coactivators through posttranslational modifications. Trends Endocrinol Metab 20(1):8–15
McKenna NJ, Lanz RB, O’Malley BW (1999) Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev 20(3):321–344
Peng L, Li E, Xu L (2020) From start to end: Phase separation and transcriptional regulation. Biochim Biophys Acta, Gene Regul Mech 2020:194641
Banani SF, Lee HO, Hyman AA, Rosen MK (2017) Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18(5):285–298
Sabari BR, Dall’Agnese A, Young RA (2020) Biomolecular condensates in the nucleus. Trends Biochem Sci
Bhat P, Honson D, Guttman M (2021) Nuclear compartmentalization as a mechanism of quantitative control of gene expression. Nat Rev Mol Cell Biol:1–18
Alberti S, Hyman AA (2021) Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat Rev Mol Cell Biol 22(3):196–213
Shin Y, Brangwynne CP (2017) Liquid phase condensation in cell physiology and disease. Science 357(6357)
Woringer M, Darzacq X (2018) Protein motion in the nucleus: from anomalous diffusion to weak interactions. Biochem Soc Trans 46(4):945–956
Sabari BR, Dall’Agnese A, Boija A et al (2018) Coactivator condensation at super-enhancers links phase separation and gene control. Science 361(6400)
Nair SJ, Yang L, Meluzzi D et al (2019) Phase separation of ligand-activated enhancers licenses cooperative chromosomal enhancer assembly. Nat Struct Mol Biol 26(3):193–203
Boeynaems S, Alberti S, Fawzi NL et al (2018) Protein phase separation: a new phase in cell biology. Trends Cell Biol 28(6):420–435
Gotor NL, Armaos A, Calloni G et al (2020) RNA-binding and prion domains: the yin and yang of phase separation. Nucleic Acids Res 48(17):9491–9504
Li L, McGinnis JP, Si K (2018) Translational control by prion-like proteins. Trends Cell Biol 28(6):494–505
Dignon GL, Best RB, Mittal J (2020) Biomolecular phase separation: from molecular driving forces to macroscopic properties. Annu Rev Phys Chem 71:53–75
Boija A, Klein IA, Sabari BR et al (2018) Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175(7):1842-1855. e16
Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA (2017) A phase separation model for transcriptional control. Cell 169(1):13–23
Stortz M, Presman DM, Bruno L et al (2017) Mapping the dynamics of the glucocorticoid receptor within the nuclear landscape. Sci Rep 7(1):1–14
Stortz M, Pecci A, Presman DM, Levi V (2020) Unraveling the molecular interactions involved in phase separation of glucocorticoid receptor. BMC Biol 18:1–20
King JT, Shakya A (2021) Phase separation of DNA: from past to present. Biophys J
Strom AR, Brangwynne CP (2019) The liquid nucleome–phase transitions in the nucleus at a glance. J Cell Sci 132(22):jcs235093
Stortz M, Presman DM, Pecci A, Levi V (2021) Phasing the intranuclear organization of steroid hormone receptors. Biochem J 478(2):443–461
Htun H, Barsony J, Renyi I, Gould DL, Hager GL (1996) Visualization of glucocorticoid receptor translocation and intranuclear organization in living cells with a green fluorescent protein chimera. Proc Natl Acad Sci 93(10):4845–4850
Frank F, Liu X, Ortlund EA (2021) Glucocorticoid receptor condensates link DNA-dependent receptor dimerization and transcriptional transactivation. Proc Natl Acad Sci 118(30)
Schaaf MJ, Lewis-Tuffin LJ, Cidlowski JA (2005) Ligand-selective targeting of the glucocorticoid receptor to nuclear subdomains is associated with decreased receptor mobility. Mol Endocrinol 19(6):1501–1515
Kauppi B, Jakob C, Färnegårdh M et al (2003) The three-dimensional structures of antagonistic and agonistic forms of the glucocorticoid receptor ligand-binding domain: RU-486 induces a transconformation that leads to active antagonism. J Biol Chem 278(25):22748–22754
Voegel JJ, Heine MJ, Zechel C, Chambon P, Gronemeyer H (1996) TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J 15(14):3667–3675
Stenoien DL, Mancini MG, Patel K, Allegretto EA, Smith CL, Mancini MA (2000) Subnuclear trafficking of estrogen receptor-α and steroid receptor coactivator-1. Mol Endocrinol 14(4):518–534
Ahmed J, Meszaros A, Lazar T, Tompa P (2021) DNA-binding domain as the minimal region driving RNA-dependent liquid–liquid phase separation of androgen receptor. Protein Sci 30(7):1380–1392
Saravanan B, Soota D, Islam Z et al (2020) Ligand dependent gene regulation by transient ERα clustered enhancers. PLoS Genet 16(1):e1008516
Tanida T, Matsuda KI, Yamada S, Hashimoto T, Kawata M (2015) Estrogen-related receptor β reduces the subnuclear mobility of estrogen receptor α and suppresses estrogen-dependent cellular function. J Biol Chem 290(19):12332–12345
Fejes-Tóth G, Pearce D, Náray-Fejes-Tóth A (1998) Subcellular localization of mineralocorticoid receptors in living cells: effects of receptor agonists and antagonists. Proc Natl Acad Sci 95(6):2973–2978
Pearce D, Náray-Fejes-Tóth A, Fejes-Tóth G (2002) Determinants of subnuclear organization of mineralocorticoid receptor characterized through analysis of wild type and mutant receptors. J Biol Chem 277(2):1451–1456
Arnett-Mansfield RL, Graham JD, Hanson AR et al (2007) Focal subnuclear distribution of progesterone receptor is ligand dependent and associated with transcriptional activity. Mol Endocrinol 21(1):14–29
Muñoz-Gil G, Romero C, Mateos N et al (2020) Phase separation of tunable biomolecular condensates predicted by an interacting particle model. bioRxiv
Saitoh M, Takayanagi R, Goto K et al (2002) The presence of both the amino-and carboxyl-terminal domains in the AR is essential for the completion of a transcriptionally active form with coactivators and intranuclear compartmentalization common to the steroid hormone receptors: a three-dimensional imaging study. Mol Endocrinol 16(4):694–706
Zhang F, Wong S, Lee J et al (2021) Dynamic phase separation of the androgen receptor and its coactivators to regulate gene expression. bioRxiv
Düster R, Kaltheuner IH, Schmitz M, Geyer M (2021) 1, 6-hexanediol, commonly used to dissolve liquid–liquid phase separated condensates, directly impairs kinase and phosphatase activities. J Biol Chem 296
Wei M, Chang Y, Shimobayashi SF, Shin Y, Strom AR, Brangwynne CP (2020) Nucleated transcriptional condensates amplify gene expression. Nat Cell Biol 22(10):1187–1196
Weikum ER, Knuesel MT, Ortlund EA, Yamamoto KR (2017) Glucocorticoid receptor control of transcription: precision and plasticity via allostery. Nat Rev Mol Cell Biol 18(3):159–174
van Steensel B, Brink M, van der Meulen K et al (1995) Localization of the glucocorticoid receptor in discrete clusters in the cell nucleus. J Cell Sci 108(9):3003–3011
Li S, Yu K, Zhang Q et al (2021) dSCOPE: A software to detect sequences critical for liquid-liquid phase separation. bioRxiv
Shen B, Chen Z, Yu C, Chen T, Shi M, Li T (2021) Computational screening of biological phase-separating proteins. Genomics Proteomics Bioinformatics
Xue B, Dunbrack RL, Williams RW, Dunker AK, Uversky VN (2010) PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim Biophys Acta Protein Proteomics 1804(4):996–1010
Alberti S, Gladfelter A, Mittag T (2019) Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 176(3):419–434
McSwiggen DT, Hansen AS, Teves SS et al (2019) Evidence for DNA-mediated nuclear compartmentalization distinct from phase separation. elife 8:e47098
Chen W, Roeder RG (2011) Mediator-dependent nuclear receptor function. Semin Cell Dev Biol 22(7):749–758
Ranjan A, Ansari SA (2018) Therapeutic potential of mediator complex subunits in metabolic diseases. Biochimie 144:41–49
Torra IP, Freedman LP, Garabedian MJ (2004) Identification of DRIP205 as a coactivator for the farnesoid X receptor. J Biol Chem 279(35):36184–36191
Wang S, Lai K, Moy FJ, Bhat A, Hartman HB, Evans MJ (2006) The nuclear hormone receptor farnesoid X receptor (FXR) is activated by androsterone. Endocrinology 147(9):4025–4033
Cornelison JL, Cato ML, Johnson AM et al (2020) Development of a new class of liver receptor homolog-1 (LRH-1) agonists by photoredox conjugate addition. Bioorg Med Chem Lett 30(16):127293
Atkins GB, Hu X, Guenther MG, Rachez C, Freedman LP, Lazar MA (1999) Coactivators for the orphan nuclear receptor RORα. Mol Endocrinol 13(9):1550–1557
Wagh K, Garcia DA, Upadhyaya A (2021) Phase separation in transcription factor dynamics and chromatin organization. Curr Opin Struct Biol 71:148–155
Klosin A, Oltsch F, Harmon T et al (2020) Phase separation provides a mechanism to reduce noise in cells. Science 367(6476):464–468
Lancaster AK, Nutter-Upham A, Lindquist S, King OD (2014) PLAAC: a web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics 30(17):2501–2502
Barnes PJ, Drazen JM, Rennard SI, Thomson NC (2009) Asthma and COPD: basic mechanisms and clinical management. Elsevier
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Appelman, M.D., Hollaar, E.E., Schuijers, J., van Mil, S.W.C. (2022). Protein Condensation in the Nuclear Receptor Family; Implications for Transcriptional Output. In: Campbell, M.J., Bevan, C.L. (eds) Nuclear Receptors in Human Health and Disease. Advances in Experimental Medicine and Biology, vol 1390. Springer, Cham. https://doi.org/10.1007/978-3-031-11836-4_14
Download citation
DOI: https://doi.org/10.1007/978-3-031-11836-4_14
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-11835-7
Online ISBN: 978-3-031-11836-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)