Abstract
Omics data are now inexpensive to collect in vast quantities, across a wide variety of not only multiple data platform, but also distinct functional units. These bioinformatic datasets can enable scientific analysis of system-level cellular processes, including complex diseases such as cancers. Recent experimental research has found significant interactions between non-coding microRNAs (miRNAs) and genes. We propose an integrated, graphical regression model to endogenize the directed miRNA–gene target interactions and control for their effects in signaling pathway disturbance. We identify prominent miRNA–gene interactions and propose a graphical representation of the targeting. We merge this network with signaling pathway networks to obtain a cross-functional graph representation of regulatory relationships between genes and miRNAs. We integrate gene expression and miRNA expression, in tandem with graphical integration of epigenetic and transcriptomic data types, and estimate a statistical model. We find that our integration approach improves the statistical power, using a simulation study. We demonstrate our integrated model with an application to disturbance of the BRAF signaling pathway across 9 cancers. We find that integration of miRNA–gene targets clarifies the differential activity between healthy and tumor tissues, which in turn reflects different roles for the pathway across the different cancers.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
The widespread availability of genomic data has dramatically increased the scope of quantitative research into biology at the molecular, genomic, and systems levels. The diversity of data available for study improves the detail available to characterize the functional processes of the genome. Significantly, these data may provide valuable new insight into the drivers of complex diseases. Multi-view datasets are now routinely collected in multiple modalities across separate biological structures, and large-scale research studies coordinate to improve the quality and quantity of data available to advance knowledge, treatment, and prevention.
To analyze these high-resolution omics data, robust methods are essential to ensure scientific rigor and validity. New experimental techniques should be complemented by statistical methods that reflect the biology in a sophisticated way. Increasingly, data is collected for genomic entities other than the gene, such as non-coding microRNAs (miRNAs).
Notably, miRNA research is fundamentally integrative in nature. Individual miRNAs are believed to target genes in a functional manner (Lewis et al. 2005), and it is often the case that single miRNAs target multiple genes. To model correlated gene activity due to a shared miRNA parent, miRNA–gene target interactions must be known and available to researchers. Early miRNA–gene target research validated individual targets experimentally, but the combinatoric problems introduced by large numbers of genes and miRNAs motivated meta-analytic and computational approaches. One such study of miRNA–gene targets was miRTarBase (Hsu et al. 2010), which identified gene targets for fewer than 700 miRNAs by manual aggregation of experimental evidence. Modern informatic methods permit large-scale analyses to identify miRNA targets. Frameworks such as miRTarBase and DIANA-miRPath (Vlachos et al. 2015) utilize web interfaces to access and explore association analyses between miRNAs and genes. DIANA-miRPath adapts methods originally applied to gene expression. In addition to Fisher’s exact test, they also test for differential activity in miRNAs using the enrichment analysis method of Bleazard et al. (2015). Computational approaches have been used to identify targets, too. Hsu et al. (2011) proposed miRTar, a successor to miRTarBase that used gene set enrichment analysis for significance testing of differential activity. These are often applied to specific phenomena or systems. Other databases include DIANA-TarBase (Karagkouni et al. 2017) and TargetScan (Agarwal et al. 2015). Coll et al. (2015) used correlation analysis to identify miRNA–gene targets related to cirrhosis of liver tissue.
Integrative analysis unifies multiple data types into a single whole. However, real-world analysis is often highly restrictive in its assumptions and the sophistication of its representation of biological systems. In many cases, “integrated” analysis refers to qualitative aggregation of separate marginal analyses on different data types, as well as correlation analysis between data types. Early statistical analyses of miRNAs were characterized by straightforward statistical methods. The CORNA method of Wu and Watson (2009) applied hypergeometric and Fisher’s exact tests to assess differential activity in miRNA–gene interactions and networks. Du and Zhang (2015) integrated methylation in a small-sample analysis of lung cancer that also included expression in genes and miRNAs. They used miRNA–gene target databases, but the interactions were excluded from gene enrichment analysis. Godard and van Eyll (2015) performed pathway analysis of miRNA in the context of Alzheimer’s disease, also using a hypergeometric enrichment test. Their procedure also treated pathways as simple gene sets, thereby ignoring known structural information about the signaling pathways. Miao et al. (2017) analyzed the relationship between miRNAs and DNA methylation in sheep. They identified gene–miRNA networks on the basis of a correlation analysis but only applied a basic t-test for differential activity across gene sets, ignoring network topology. Moreover, their integration was largely restricted to correlation analysis to cluster genes targeted by the same miRNAs. Volinia and Croce (2013) analyzed gene expression and miRNA expression for a breast cancer dataset. Their analysis focused on survival outcomes, and the extent of their data integration was to include both genes and miRNAs as covariates, rather than a structural or model-based integration. Cava et al. (2014) considered copy number as well as gene and miRNA expression. But, genes and miRNAs were only heuristically integrated, by performing separate marginal analyses, as well as comparing up- and downregulation across the different data types.
This lack of a single coherent integration scheme is also found in miRNA analyses applied to cancer datasets. Enerly et al. (2011) studied miRNA suppression in a novel miRNA and gene expression breast tumor dataset. But, their integration was limited to correlation analysis and separate studies on each data type. Likewise, Yu et al. (2019) identified specific biomarkers with differential survival outcomes in lung cancer, and Li et al. (2018) used differential correlation analysis between miRNAs and genes in cancer, both using correlation analysis.
We address this lack of technical statistical methods for joint integrative analysis of data observed on genes and miRNAs. We identify prominent miRNA–gene interactions and construct a graphical model to represent the targets. We merge this network with signaling pathways to estimate pathway activity while controlling network effects and coexpression of genes due to the miRNAs. We extend the NetGSA regression model for analysis of signaling pathways, which was restricted to gene-level measurements, originally only gene expression (E) in Shojaie and Michailidis (2009) and Shojaie and Michailidis (2010). Zhang et al. (2017) extended the signaling pathway network to include gene methylation (M) and copy number (C), and we use their EMC-NetGSA model to integrate gene-level omics observations. In this chapter, we also incorporate miRNAs into the statistical model. Furthermore, we also introduce a semi-parametric bootstrap procedure to assess the robustness of the statistical inference.
This chapter proceeds as follows. In Sect. 2, we give an overview of the omics datasets we use for integrative analysis. In Sect. 3, we give details of the network integration and pathway model. In Sect. 4, we first perform simulation studies to examine the statistical level and power of the proposed method in Sect. 4.1 and then conduct a data analysis of pathway disturbance in the BRAF signaling pathway in 9 cancers in Sect. 4.2. Finally, we conclude our paper with discussion in Sect. 5.
2 Data
We consider a multi-platform omics dataset assembled from observations published by The Cancer Genome Atlas (TCGA). TCGA is an ongoing, international study funded by the National Cancer Institute (NCI) that collects tumorous tissue samples in patients with more than 30 distinct cancers (Tomczak et al. 2015). For each cancer, we obtained measurements of gene expression, copy number variation (CNV), and methylation, as well as miRNA expression. In order to analyze differential activity by cancer, we downloaded omics data for all tumor samples, as well as matched healthy control tissue samples. We describe in Sect. 4.2 our steps to aggregate methylation and copy number at the level of individual genes.
Unlike gene-level integration of methylation and copy number features, no standard, direct mapping exists between genes and miRNAs. Instead, we use resources on functional miRNA–gene targets to construct an integrated statistical model. Substantial work has been done to identify miRNA–gene targets. One resource that quantifies the degree of experimental evidence in support of a given miRNA–gene target interaction is mirDIP. Tokar et al. (2017) compiled the database as a meta-analysis to integrate predicted miRNA–gene targets from 30 separate sources of experimentally validated interactions. It includes information on the degree to which the source databases overlap in their conclusion.
3 Methods
We integrate the omics data described in the previous section through a statistical model across the four data types collected across miRNAs and genes. At a high level, we start with a known genetic signaling pathway, specified as a directed graph on vertices representing genes. We identify likely miRNA–gene targets and integrate these with directed edges. We use a similar approach to integrate gene methylation and copy number within each gene. Finally, the graph adjacency matrix of the fully integrated omics network is used to form a design matrix for a mixed linear model. This enables hypothesis testing for differential pathway activity between two populations.
We introduce our integrated graphical network constructively. We start with a genetic signaling pathway specifying known functional relationships between genes. We define a directed graph \(\mathcal {G}=\lbrace \mathcal {V}, \mathcal {E}\rbrace \). Initially, \(\mathcal {V}\) contains p graph vertices, and \(\mathcal {E}\) contains the directed edges between elements of the genes \(\mathcal {V}\) that comprise the signaling pathway. In general, the graph vertices in \(\mathcal {V}\) represent biological features, while the edges in \(\mathcal {E}\) represent the functional interactions.
Graph \(\mathcal {G}\) can be represented by a p × p adjacency matrix, \({\mathbf {A}}^{\star }_{\text{E}}\). The subscript “E” emphasizes that the graph relations in \({\mathbf {A}}^{\star }_{\text{E}}\) specify relationships between vertices for gene expression. The element α jk of \({\mathbf {A}}^{\star }_{\text{E}}\) is an indicator for the presence of a directed edge from vertex k to vertex j, for all j, k = 1, …, p. α jk is nonzero when gene j is conditionally dependent on gene k. For each gene, we observe gene expression as a vector y i1 of p elements, where i = 1, …, N indexes tissue samples.
Suppose the p pathway genes are targeted by g miRNAs, and for each sample, we observe a vector of g elements y i2, the values of which measure miRNA expression. We add g vertices to \(\mathcal {V}\) integrated the miRNA–gene target interactions, and we construct the p × g graph adjacency matrix \({\mathbf {A}}^{\star }_{\text{mi}}\). The element \(\tau _{j\ell }\in {\mathbf {A}}^{\star }_{\text{mi}}\) is an indicator value for miRNA ℓ targeting gene j, j = 1, …, p, ℓ = 1, …, g. Each miRNA–gene target is represented by a directed edge which we add to \(\mathcal {E}\).
We construct the (p + g) × (p + g) integrated adjacency matrix \({\mathbf {A}}^{\star }_{\text{miE}}\) that spans both the genetic pathway and the miRNA–gene targets and contains elements of 0 and 1:
where O m×n is a m × n matrix of zeros.
Moreover, for each gene in y i1, we also observe copy number and methylation, contained in the vectors y i3 and y i4, respectively. We adopt the EMC-NetGSA model (Zhang et al. 2017) to integrate {y i1, y i3, y i4} by adding 2p vertices to \(\mathcal {V}\), one for each gene for copy number and methylation, and 2p directed edges to \(\mathcal {E}\), from the copy number and methylation vertices to their counterpart gene expression vertices. This produces a fully integrated adjacency matrix, \({\mathbf {A}}_{\text{miEMC}}^{\star }\):
In real-world datasets, individual elements of y i2, y i3, and y i4 may be missing across all N samples. An advantage of our unidirectional integration, which does not model directed interactions from genes to the other omics features, is that we may simply omit the columns and rows for the corresponding miRNA, copy number, and methylation features in \({\mathbf {A}}_{\text{miEMC}}^{\star }\) prior to the pathway analysis. Without loss of generality, we consider the full (g + 3p) × (g + 3p) adjacency matrix \({\mathbf {A}}_{\text{miEMC}}^{\star }\), with the knowledge that its true dimension q is such that q ≤ (g + 3p).
The network in Eq. 2 composes three distinct network layers: (1) the primary signaling network on elements of y i1; (2) the miRNA integration layer of directed relationships, possibly many-to-one, from miRNAs in y i2 to genes in y i1; and (3) a within-gene layer integrating copy number in y i3 and methylation in y i4. Therefore, the graph simultaneously provides for causal relationships between genes, allows correlation between genes, and reduces noise by controlling for epigenetic and transcriptional effects. Each of the three components is supported by scientific knowledge of the complex underlying biological processes.
For expositional clarity, define m = (g + 3p), \({\mathbf {y}}_{i}=({\mathbf {y}}_{i1}^{\prime },{\mathbf {y}}_{i2}^{\prime },{\mathbf {y}}_{i3}^{\prime },{\mathbf {y}}_{i4}^{\prime })'\), \({\mathbf {A}}^{\star }\equiv {\mathbf {A}}^{\star }_{\text{miEMC}}\), and the elements of A ⋆ by δ jk, j, k = 1, …, m.
Gaussian graphical models formalize the conditional dependence of vertex j on vertex k as the partial correlation ρ jk between gene-level random variables Y j, Y k, controlling for the effects of the remaining (m − 2) vertices in \(\mathcal {V}\). Writing the random variables for the remaining (m − 2) vertices by \(\mathcal {Z}\), \(\rho _{jk}=\text{corr}(Y_{j\setminus {\mathcal {Z}}}, Y_{k\setminus {\mathcal {Z}}})\), where \(Y_{j\setminus {\mathcal {Z}}} = Y_{j}-\mathcal {P}_{\mathcal {Z}}Y_{j}\) is the orthogonal complement of Y j with respect to \(\mathcal {Z}\), and \(\mathcal {P}_{\mathcal {Z}}\) is a projection onto \(\mathcal {Z}\) (Krämer et al. 2009).
We estimate ρ jk with the sample partial correlation r jk. We first estimate two separate regressions, one of Y j on \(\mathcal {Z}\) and the other of Y k on \(\mathcal {Z}\). Then, we estimate r jk by the Pearson correlation coefficient between the residuals of the two regressions (Kim 2015). Finally, we form a weighted adjacency matrix A with elements a jk = r jk α jk, j, k = 1, …, m. Elements of A thus take either the value 0, when no interaction exists, or a value in the interval (−1, 1) corresponding to the strength of conditional association between two vertices with a functional interaction.
The effect of coexpression due to the graph topology \(\mathcal {G}\) can be summarized by a transformation Λ of A, called the influence matrix. As detailed in Shojaie and Michailidis (2009), in the special case of directed acyclic graphs (DAGs), it can be shown that Λ = (I m −A)−1. This definition extends to all graphs for which the adjacency matrix has eigenvalues all of which are smaller than 1 in magnitude. Shojaie and Michailidis (2010) extended the definition to non-DAG, non-substochastic graphs. They used a limit approximation to induce in arbitrary directed graphs the necessary eigenvalue properties of A.
The NetGSA statistical model uses Λ to structure the mean of y i, by setting E y i = Λ β for an unknown vector of m regression coefficients β. β is the network-adjusted activity parameter, giving the mean values for the m observed elements of y i, controlling for pass-through network effect due to \(\mathcal {G}\). The influence matrix also structures the covariance of y i. The NetGSA model parameterizes variability in individuals’ mean expression via a mixed effects linear regression model: y i = Λ β + Λ γ i + 𝜖 i, γ i ∼ N m(\(\boldsymbol {0_{m}}, \sigma ^{2}_{\gamma } {\mathbf {I}}_{m}\)), 𝜖 i ∼ N m(\(\boldsymbol 0_{m}, \sigma ^{2}_{\epsilon }{\mathbf {I}}_{m}\)), where γ i is a sample-level random effect, and i = 1, …, N.
In proposing the NetGSA model, Shojaie and Michailidis (2009) also proposed a hypothesis test for difference in mean vectors between two populations. Denote the population label for sample i by c i, where c i ∈{C, T}, “C” corresponds to control, and “T” corresponds to treatment. In our pathway analysis of cancerous tumors, we assign healthy tissue samples the label of “control,” and tumor tissues the label “treatment.” We estimate separate weighted adjacency matrices A C, A T, yielding distinct influence matrices Λ C, Λ T, and population-specific mean parameters β C, β T.
We test for differential activity in subsets of the pathway features, corresponding to elements in β C, β T, using an indicator vector b for the omics features of interest. The NetGSA network contrast is ℓ = (−b ⋅b Λ C, b ⋅b Λ T), and this yields a test statistic T ∝ ℓ β, \(\boldsymbol \beta =(\boldsymbol \beta _{\text{C}'}, \boldsymbol \beta _{\text{T}'})'\). T follows a Student’s t distribution with degrees of freedom estimated using Satterthwaite’s approximation.
The above inference depends upon both the assumption of normality and the composition of the control population. In practice, the size of the healthy sample population is very small, and we may wish to assess the robustness of the inference to the specific control samples. Therefore, we propose a semi-parametric bootstrap test based on the principles discussed by MacKinnon (2009).
Specifically, we generate B pairwise bootstrap replicates under the null hypothesis of no difference in the network-adjusted mean parameters in the two populations. We randomly select pairs of population labels and omics observations, with replacement. If a sample contains fewer than 10 observations in the control (healthy) population, we re-generate the bootstrap sample. Because the distribution of the NetGSA test statistic is approximated using a function of the estimated variance, the distributions of the test statistic are not comparable between bootstrap replicates, as the degrees of freedom vary substantially. Instead, when the empirical p-value is nominally significant, we calculate a 95% bootstrap upper confidence bound. Inversely, for a p-value that is nominally not significant, we calculate a 5% bootstrap lower bound.
This bootstrap procedure offers a semi-parametric criterion for assessing the robustness of the outcome of a hypothesis test. The procedure still depends upon the underlying assumptions of the mixed linear model but instead addresses the robustness of the procedure to small sample sizes in the control population.
4 Results
First, we use a simulation study to demonstrate the improved power of our method due to the integration of miRNA–gene target information. We then apply our method to the TCGA dataset introduced in Sect. 2 for pathway analysis of the BRAF pathway.
4.1 Simulations
Our simulation study borrows the ideas on simulation designs in the EMC-NetGSA paper (Zhang et al. 2017). Broadly, we construct a pathway composed of a binary tree signaling pathway; we model miRNA–gene targets that drive correlated gene expression, and we integrate the methylation and copy number within genes. This supplies three separate layers of network information that may contribute to gene expression, and we examine the relationship between omics integration and statistical power.
We modeled a signaling pathway consisting of a five-level binary tree containing 31 genes. We integrated miRNA with directed edges from miRNA to corresponding genes. For every gene, we added three distinct miRNA vertices, i.e., each had out-degree 1. We then partitioned the genes in \(\mathcal {V}\) into disjoint sets of two, proceeding from the root node. We assigned to each pair of genes one shared miRNA, i.e., with out-degree 2. Finally, we repeated this procedure for sets of three genes. We assigned each triplet one miRNA with out-degree 3. The network topology for the control population signaling pathway used the same integrated binary tree structure, with all edges in the tree’s left branch removed. For EMC-NetGSA integration, we add \(\mathcal {G}\) directed edges into each gene from two vertices representing methylation and copy number.
We set the correlation between expression vertices to 0.8 in the top third (two levels) of the tree; association is 0.5 in the middle third (third level); and association 0.2 in the final third (final level). We set the magnitude of the association strength between miRNA and their gene targets to 0.4. In the TCGA dataset, we found that the partial correlation coefficients between miRNA and expression were generally symmetric. Therefore, we assigned alternating edges from miRNAs to genes to have positive and negative association, respectively. This may be understood as simulating cases where miRNAs with multiple gene targets have the same sign for association, as well as different associations with different genes for a single miRNA. We set the association 0.5 between copy number and gene expression and −0.25 between methylation and expression.
We generated observation vectors y i, i = 1, …, N, from the NetGSA linear mixed model, where N = N C + N T. The number of control samples was N C = 50, and the number of treatment samples was N T = 150. This reflects the imbalanced sample sizes in the real cancer datasets. We set the variance parameters as \(\sigma _{\gamma }^{2}=5\) and \(\sigma _{\epsilon }^{2}=0.5\).
Denoting the mean vectors for gene expression, miRNA expression, gene copy number, and gene methylation by β c1, β c2, β c3, β c4, we simulated two scenarios for the network-adjusted mean parameter β. Here, c ∈{C, T} indexes the control and treatment populations. In the first scenario, we assigned β cj = 0, c ∈{C, T}, j = 1, 2, 3, 4. In the second mean scenario, we held β Cj = 0 for all j. For the top two-thirds levels of the binary tree, we set (β T1, β T2, β T3, β T4) = (0.25, 0.5, 1.0, 0.5). In the bottom third of the binary tree, we maintained (β T1, β T2, β T3, β T4) = 0, as in the first scenario.
For each simulated dataset, we tested four gene sets for differential activation: (1) the full binary tree; (2) the top third of the tree; (3) the top two-thirds of the tree; and (4) the bottom third of the tree. We estimated the miEMC-NetGSA model for the entire simulated dataset, as well as the NetGSA variants with the adjacency matrices A miE, A EMC, and A E, as well as the corresponding hypothesis test for each network. We ran 1000 replicates of the simulation. We calculated the power for each method by the proportion of hypothesis test p-values that were significant at the α = 0.05 level, i.e., the proportion of replicates for which we reject the null hypothesis of no difference in pathway-adjusted mean parameters.
Figure 1 shows boxplots of the −log10 p-values from the significance tests. The left-hand panel shows the results of the first mean scenario, in which no features are differentially expressed. The right-hand panel shows the second mean scenario, in which the top two-thirds of the binary tree signaling pathway are differentially expressed. We compare the performance of miEMC-NetGSA with the other integrated models described above.
Boxplots of −log10 p-values from simulation study. The left-hand panel shows significance tests for four gene sets of interest, under the first mean scenario in which no omics features exhibit differential activation. The right-hand panel shows the second mean scenario, in which the top two-thirds of the simulated binary tree signaling pathway is differentially activated, but the final third is equal in the control and treatment populations. miEMC-NetGSA is shown in green, miE-NetGSA in yellow, EMC-NetGSA in blue, and NetGSA (“E”) on expression only in white
The first mean scenario permits assessment of the false positive rate under different omics integration schemes. We observe that in all gene sets that we tested, all four NetGSA-based methods have low false positive rates. Most importantly, integration of miRNA with expression alone (“miE”) does not cause an elevated false positive rate over the original NetGSA method. Likewise, although the false positive rate is somewhat elevated in miEMC-NetGSA, we observe that it is not elevated significantly over the existing EMC-NetGSA method. Therefore, we do not attribute to the miRNA integration a meaningful increase of the type I error rate.
Further, we note that the number of miRNAs is large relative to the number of genes. This causes the type I error rate to be lower for the methods that integrate miRNA–gene targets, shown in the test set of the top 1/3 genes. The reason is that the larger overall number of features provides increased accuracy to miEMC-NetGSA and miE integration, so more substantial information is available to the inference procedure than the methods with only gene-level network features.
The second mean scenario provides an assessment of the method’s power. We observe that in the three test sets that contain differentially activated features, the power of models that integrate miRNA–gene targets dominates the gene-only analyses. In fact, an integrated model of miRNA and expression has power comparable to EMC-NetGSA integration of methylation and copy number, although miRNA–gene expression remains under-powered relative to EMC-NetGSA.
Also prominent is the increased power of the new method, which integrates both the miRNA–gene targets and gene copy number and methylation. Integration of miRNA reduces the type II error rate of the EMC-NetGSA model; equivalently, it increases the analytic power. Finally, the miRNA methods continue to exhibit low rates of type I errors for the gene set that is not differentially activated.
Taken in tandem, the results of the two simulation scenarios confirm the value of pathway analysis that integrates miRNA–gene targets. We find miRNA contributes to pathway analyses improved statistical power, relative to analyses conducted solely at the level of individual genes. At the same time, we find that miRNA integration does not artificially elevate the false positive rate. Finally, although the addition of miRNA to an expression-only analysis improves power, the increase is marginally less substantial than is provided by integrating copy number and methylation. But, our composition of these two integration schemes achieves the highest statistical power and does not noticeably increase the type I error rate.
4.2 Data Analysis
Prior to analysis, we formatted the dataset described in Sect. 2. We downloaded level 3 TCGA data for 33 cancers from the NCI Genomic Data Commons (Grossman et al. 2016), using the R package TCGA-Assembler, version 2.0.0 (Zhu et al. 2014; Wei et al. 2017).
Starting with gene expression data measured using RNASeqV2, we used a normalization of the read counts provided by TCGA, fragments per kilobase of transcript per million mapped reads upper quartile (FPKM-UQ) (Grossman et al. 2016). We further took a log2 transformation of the normalized read counts. We used CNV data with common germ-line copy number variants removed and averaged gene-level CNV across the corresponding DNA regions. We aggregated observed methylation beta values across CpG sites by gene and took the mean.
Similar to gene expression, miRNA expression data are available from TCGA in two formats: raw read counts and normalized reads per million (RPM). Raw read counts were collected on the miRNASeq platform, and the TCGA processing pipeline outlined by Chu et al. (2015) is consistent with procedures in comparable projects, such as ENCODE (ENCODE Project Consortium et al. 2012). After alignment and read trimming, a library of approximately 22 base pairs of mature strands was used with an insert length of approximately 22.
Typical miRNASeq analyses use methods traditionally developed for RNASeq. For example, Stokowy et al. (2014) employed RPM normalization and cited its original definition from Mortazavi et al. (2008), in the context of gene expression. They cited other work that applies RPM normalization to miRNASeq data, including Chen et al. (2013). Following RPM normalization but prior to the primary analysis, those authors applied a log2 transformation. Han et al. (2018) integrated gene and miRNA expression. They first applied FPKM-UQ normalization RNASeq gene expression values, then calculated RPM for miRNASeq values, and transformed logarithmically. TCGA provides RPM-normalized transformation of the data, so for our integrative analysis, we applied a log2 transformation to the RPM-normalized values. Empirically, we observed that this normalization was comparable to FPKM-UQ applied to the raw read counts.
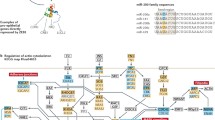
To demonstrate our procedure, we performed data analysis of the BRAF pathway, a genetic signaling pathway previously studied by Zhang et al. (2017). The left-hand panel of Fig. 2 shows the network topology of the BRAF pathway. It consists of 10 genes—AKT1, BRAF, MAP2K1, MAP2K2, MAPK1, MTOR, NRAS, PIK3CA, PTEN, and RAF1—which are connected by 12 directed edges. The BRAF pathway is a DAG, and this property is preserved under integration with miRNA, copy number, and methylation.
Network diagrams for the BRAF signaling pathway. The left-hand network is the directed graph representing the BRAF genetic signaling pathway, consisting of 10 genes with 12 directed edges between. The right-hand network contains the same 10 gene vertices, but edges represent a shared miRNA parent. In other words, two genes that share an edge are both targets of a single miRNA and possibly several. The graph contains 25 such edges. miRNA–gene targets are chosen as those entries in the mirDIP database with an “very high” confidence score. Neither the NRAS nor PTEN genes are targeted by any other genes. MAP2K1, MAP2K2, MAPK1, and PIK3CA each have in-degree 2 from other genes. The remaining genes in the network each in-degree 1 from genes. The density of miRNA–gene targets is much higher: in-degree from miRNAs ranges from 10–50 (AKT1, MAP2K1, MTOR, PIK3CA, RAF1) to 143 (PTEN), and BRAF and MAP2K2 are not targeted by any miRNAs
The mirDIP database compiled by Tokar et al. (2017) aggregates predicted miRNA–gene target relationships from several experimentally validated sources. The database assigns each miRNA–gene pair found across any of the 30 sources a composite integrative score. The score, valued on the interval [0, 1], quantifies the strength of experimental evidence that supports the existence of the interaction. The scores are stratified by the so-called confidence classes, expressed in the labels “very high,” “high,” “medium”, and “low” confidence. These classes, respectively, represent the top 1% of scores (very high), the next 4% of scores (high), remainder of top 33% of scores (medium), and all other scores (low). The classes offer a discrete criterion for determining whether to include in \({\mathbf {A}}_{\text{miEMC}}^{\star }\) a specific miRNA–gene interaction. We downloaded their mirDIP unidirectional database, version 4.1, and we considered both 3’ and 5’ UTR miRNA entries for miRNAs that were also present in the TCGA dataset.
Figure 3 shows a scatter plot of the miRNA–gene target scores from mirDIP for all genes in the BRAF pathway. For a given gene, we identified all miRNAs in the database that target that gene. In the figure, the scores are grouped by gene and colored by confidence class: blue represents very high confidence, green is high confidence, yellow is medium confidence, and red is low confidence.
miRNA–gene interaction scores for all miRNA in TCGA dataset that target any of the 10 genes that comprise the BRAF pathway. Scores are grouped by gene and colored by confidence class: very high, high, medium, and low confidence are colored blue, green, yellow, and red, respectively. For our data analysis, we included only miRNA–gene target interactions with “very high” confidence that the interaction exists, i.e., the blue scores
In our analysis, we used the subset of miRNA–gene targets for which the scores in Fig. 3 belong to the “very high” confidence class. We can construct the secondary graph formed between genes that are both targeted by a mutual miRNA parent. Compared with the genetic signaling pathway, this secondary network has a far denser edge set: whereas the BRAF pathway contains 12 edges, the miRNA co-target graph contains 25 edges.
More generally, this larger edge set due to miRNA–gene targets indicates that miRNA integration substantially complexifies the network structure used as input for the pathway analysis. This contrasts with the underlying simplicity of the original graph: whereas the signaling pathway consists of 10 genes, miRNA integration introduces to the network 238 vertices for miRNA observations. Nearly, half of these miRNAs target multiple genes in the BRAF pathway.
The miRNA–gene target subnetwork, corresponding to the unweighted adjacency matrix \({\mathbf {A}}^{\star }_{\text{mi}}\) in Eq. 2, is shown in Fig. 4. This graph shows the subnetwork produced by the directed edges from miRNA vertices to genes, based on miRNA–gene targets. The number of miRNAs targeting a given gene varies substantially, from as many as 143 miRNAs targeting PTEN to as few as to 0. These in-degrees from miRNA vertices are given in the caption to Fig. 2.
Topology of subnetwork of the integrated BRAF signaling pathway corresponding to the unweighted adjacency matrix \({\mathbf {A}}^{\star }_{\text{mi}}\) in Eq. 2, namely, the graph formed by miRNA–gene targets. All edges are directed from the miRNA vertex to the gene vertex. The miRNA node colors, edge widths, and edge colors correspond to the degree of the miRNA vertex, that is, the number of genes in the BRAF pathway targeted by a given miRNA. Gray graph nodes are genes, while colored graph nodes are miRNAs, targeting 1 gene (red), 2 genes (yellow), 3 genes (green), 4 genes (blue), and 5 genes (purple). The genes BRAF and MAP2K2 are not targeted by any genes. Darker edge colors correspond to higher out-degree of the associated miRNA
Although most miRNAs in the TCGA dataset target a single gene in the BRAF pathway, 47% of the miRNAs target two or more genes. In the network diagram in Fig. 4, miRNAs and their edges are colored according to the degree of the miRNA node, that is, the number of genes in the BRAF pathway targeted by the miRNA. Although there are 128 miRNAs that target only a single gene, the remaining 110 target multiple genes. This intricate structure is not balanced, in neither the in-degree of gene nodes nor the out-degree of miRNA nodes. The number of unique miRNAs that target each gene varies from 58 (PTEN) to 4 (PIK3CA); while one miRNA targets 5 genes, there are 19 that target 4, 31 that target 3, and 59 that target 2.
We obtained observations of gene expression, miRNA expression, methylation, and copy number from TCGA, as described in Sect. 2. Among the TCGA cancer studies, we restricted our analysis to the subset of cancers for which all 10 BRAF pathway genes were observed, and for which more than 10 samples were available in both the tumor and normal tissue sample populations. This yielded nine cancers for integrative pathway analysis of the BRAF pathway: bladder, breast, head and neck squamous cell, kidney renal clear cell and papillary cell, liver hepatocellular, thyroid, and uterine corpus endometrial carcinomas, and prostate adenocarcinoma. Sample sizes and proportional representation of the healthy samples were consistent across the nine cancers. For example, the bladder urothelial carcinoma dataset contained 341 samples of tumor tissue and 15 samples of healthy tissue. This corresponds to 356 total samples, 4% of which are healthy controls.
Within each cancer, we performed the same four NetGSA-based pathway analyses as in the simulation study: full integration of gene expression, miRNA expression, and gene-level methylation and copy number (miEMC-NetGSA); integration of gene and miRNA expression (“miE”); integration of gene expression, methylation, and copy number (EMC-NetGSA); and the original NetGSA for expression only (“E”). To correct for the multiple comparison problem, we adjusted p-values within each cancer using the method of Benjamini and Hochberg (1995) (BH).
The adjusted p-values are plotted in the top panel of Fig. 5. The pathway is significantly disturbed at the α = 0.05 level across all cancers for both methods that integrate miRNA. However, the cancers diverge in their decision outcomes when only gene-level features are included. In head and neck squamous cell carcinoma (HNSC) and prostate adenocarcinoma (PRAD), both methods that integrate only gene-level features, namely, EMC-NetGSA and expression-only NetGSA, fail to reject the null hypothesis of no pathway disturbance, but miRNA integration confirms pathway disturbance. In bladder urothelial carcinoma (BLCA), thyroid carcinoma (THCA), and uterine corpus endometrial carcinoma (UCEC), the expression-only analysis does not reject the null hypothesis, while integration of any features beyond gene expression leads to the conclusion of pathway disturbance. In these cancers, integration of miRNA features leads to differential effects: in the bladder and thyroid cancers, miRNA integration causes a large increase in significance. Likewise, although the pathway is significantly disturbed in kidney renal clear cell carcinoma (KIRC) and liver hepatocellular carcinoma (LIHC), miRNA integration substantially increases the significance of the hypothesis test. The same is largely true of breast invasive carcinoma (BRCA). On the other hand, in the uterine cancer, the change in significance is less pronounced between gene-only and miRNA–gene integration. Similarly, kidney renal papillary cell carcinoma (KIRP) exhibits a lesser degree of differentiation between the significance of the three integrative methods.
(Top) Results of significance tests for pathway disturbance in the BRAF pathway, across 9 TCGA cancers. Barplots give values of −log10 p-values, after Benjamini–Hochberg adjustment for multiple comparisons. Full gene expression miRNA expression, and gene-level methylation and copy number are shown in green (“miEMC”); integration of gene and miRNA expression is in yellow (“miE”); integration of gene expression and gene-level methylation and copy number are in blue (“EMC”); and the original, expression-only NetGSA results are in white (“E”). The horizontal dotted line gives the significance threshold at the α = 0.05 level. (Bottom) Test statistics for pathway disturbance in 9 TCGA cancers, for the BRAF pathway. Bar plot heights give the test statistic value. miEMC-NetGSA is in green, miE-NetGSA in yellow, EMC-NetGSA in blue, and NetGSA (“E”) in white
Figure 5 also shows bar plots of the test statistics corresponding to the p-values. For most cancers with substantial increases in significance due to miRNA integration, bladder, breast, kidney renal clear cell, prostate, and thyroid correspond to test statistics with the same sign and distinctly greater magnitude than the gene-level analyses. The test statistics of kidney renal papillary cell carcinoma display a similar pattern in the relative magnitudes of the test statistics, despite the lesser differences in statistical significance between the EMC-NetGSA and the miRNA-NetGSA analyses. The liver and head and neck cancers have test statistics with the opposite sign from those six cancers, in conjunction with sign switches in the test statistics before and after miRNA integration. In contrast, the test statistics for the uterine cancer display test statistics with consistent signs, though the magnitude increases with miRNA integration.
Across the nine cancer types, we did not observe any apparent pattern in the relation between the test statistic signs and the relative significance of the tests. Relatively weaker significance in gene-level analyses sometimes corresponds to small test statistics with the same sign, as in breast and kidney renal clear cell cancers. But it also corresponds to sign changes in the test statistics, as in bladder, head and neck, and thyroid cancers. The head and neck, liver, and uterine cancers are notable for their inversion of the test statistic’s sign in the miRNA analyses, relative to the other cancers. Despite the similarity between kidney renal papillary cell carcinoma and uterine corpus endometrial carcinoma in terms of p-values and relative significances of the four analyses, the kidney cancer test statistic sign is more comparable to the other kidney cancer, among others. The large magnitude of the prostate cancer test statistics under miRNA integration matches the sharp increase in significance.
Finally, we assessed the robustness of our inference to the assumptions, especially symmetric errors. Overdispersion can be a characteristic of RNASeq count data (Zhou et al. 2011), but this typically arises in the context of count data and the choice between Poisson and negative binomial distributions. Empirically, the transformations we apply to the raw counts induce symmetry, and the assumption of normality provides separate parameters for location and dispersion.
Furthermore, we applied the bootstrap method described in the previous section, with B = 9999, and found that the inference for the full miEMC-NetGSA integration and the partial miRNA–gene expression integration were both robust in all cancers, and the semi-parametric decision outcome matches that of the parametric test. On the other hand, the EMC-NetGSA tests were not robust in any cancers except the kidney cancers, and the expression-only tests for bladder and thyroid cancers were likewise inconclusive. Therefore, we conclude that the inferences we draw are robust to the model assumptions, and the evidence for pathway disturbance in the miRNA-integrated analyses is valid in the semi-parametric setting, as well.
These results indicate statistically meaningful contributions of the miRNA features to the pathway analysis. Moreover, they suggest the effect of miRNA integration does not lead deterministically to a specific decision outcome. In some cancers, miRNA integration reinforces the conclusions of pathway analysis using existing methods based solely on gene-level features. In others, the miRNAs identify a significant disturbance that is less apparent when the BRAF pathway is considered using only gene-level features. This may be due to the reduction in noise at the level of gene expression features using the augmented network, thereby accentuating the differential expression in the pathway. Finally, in all the cancers, the reduction of gene-level noise that is accomplished by the miRNA–gene target network effect clarifies the expression of pathway genes.
We have made publicly available the code to produce the results of our analysis at:
https://github.com/Zhang-Data-Science-Research-Lab/miEMC-NetGSA-BRAF
5 Discussion
In this chapter, we highlighted the importance of integrating miRNA measurements into network analysis of genetic signaling pathways. We proposed a statistical modeling approach that incorporates recent biological research on the functional relationships between protein-coding and non-coding RNAs. We identified significant miRNA–gene targets and constructed a graphical model of these interactions. We combined this graph with a signaling pathway graph to account for correlated coexpression in genes through a biological mechanism external to the signaling pathway of interest. We used a simulation study to show that an integrative model of miRNA–gene targets has higher statistical power than analysis that focuses only on gene-level features. At the same time, we demonstrated that our method does not increase the false discovery rate, relative to the existing methods. Our miEMC-NetGSA model offers a statistical framework for data integration and pathway analysis of multiple omics data types, obtains high power, and is grounded in current biological research.
Finally, we applied the miRNA–gene analysis to the BRAF signaling pathway on a large-scale cancer study. Although the genetic pathway itself consisted of only 12 edges on 10 vertices, we found that the fully integrated network of gene expression, miRNA expression, gene copy number, and gene methylation observations consisted of 268 vertices and 452 directed edges. The decision outcome in a hypothesis test for differential expression varied across the 9 cancers under consideration. In some cancers, integration of either miRNA or gene-level copy number and methylation led to higher significance, but the composition of all integrative features did not necessarily strengthen the significance. In other cancers, integration of miRNA–gene targets with expression confirmed the result of a gene expression-only analysis; while the integration of gene expression with methylation and copy number indicated strong significance, the further addition miRNA integration was in consensus with the expression-only and miRNA–gene analyses.
References
Agarwal, V., Bell, G.W., Nam, J.-W., & Bartel, D.P. (2015). Predicting effective microRNA target sites in mammalian mRNAs. elife, 4:e05005.
Benjamini, Y., & Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B (Methodological), 57(1), 289–300.
Bleazard, T., Lamb, J.A., & Griffiths-Jones, S. (2015). Bias in microRNA functional enrichment analysis. Bioinformatics, 31(10), 1592–1598.
Cava, C., Bertoli, G., Ripamonti, M., Mauri, G., Zoppis, I., Della Rosa, P. A., Gilardi, M. C., & Castiglioni, I. (2014). Integration of mRNA expression profile, copy number alterations, and microRNA expression levels in breast cancer to improve grade definition. PLoS ONE, 9(5), e97681.
Chen, M., Zhang, X., Liu, J., & Storey, K. B. (2013). High-throughput sequencing reveals differential expression of miRNAs in intestine from sea cucumber during aestivation. PLoS One, 8(10), e76120.
Chu, A., Robertson, G., Brooks, D., Mungall, A. J., Birol, I., Coope, R., Ma, Y., Jones, S., & Marra, M. A. (2015). Large-scale profiling of microRNAs for the cancer genome atlas. Nucleic Acids Research, 44(1), e3–e3.
Coll, M., El Taghdouini, A., Perea, L., Mannaerts, I., Vila-Casadesús, M., Blaya, D., Rodrigo-Torres, D., Affò, S., Morales-Ibanez, O., Graupera, I., et al. (2015). Integrative miRNA and gene expression profiling analysis of human quiescent hepatic stellate cells. Scientific Reports, 5, 11549.
Du, J., & Zhang, L. (2015). Integrated analysis of DNA methylation and microRNA regulation of the lung adenocarcinoma transcriptome. Oncology Reports, 34(2), 585–594.
ENCODE Project Consortium et al. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature, 489(7414),57.
Enerly, E., Steinfeld, I., Kleivi, K., Leivonen, S.-K., Aure, M. R., Russnes, H. G., Rønneberg, J. A., Johnsen, H., Navon, R., Rødland, E., et al. (2011). miRNA-mRNA integrated analysis reveals roles for miRNAs in primary breast tumors. PLoS One, 6(2), e16915.
Godard, P., & van Eyll, J. (2015). Pathway analysis from lists of microRNAs: common pitfalls and alternative strategy. Nucleic Acids Research, 43(7), 3490–3497.
Grossman, R. L., Heath, A. P., Ferretti, V., Varmus, H. E., Lowy, D. R., Kibbe, W. A., & Staudt, L. M. (2016). Toward a shared vision for cancer genomic data. New England Journal of Medicine, 375(12), 1109–1112.
Han, S., Kim, D., Shivakumar, M., Lee, Y.-J., Garg, T., Miller, J. E., Kim, J. H., Kim, D., & Lee, Y. (2018). The effects of alternative splicing on miRNA binding sites in bladder cancer. PLoS One, 13(1):e0190708.
Hsu, S.-D., Lin, F.-M., Wu, W.-Y., Liang, C., Huang, W.-C., Chan, W.-L., Tsai, W.-T., Chen, G.-Z., Lee, C.-J., Chiu, C.-M., et al. (2010). miRTarBase: a database curates experimentally validated microRNA–target interactions. Nucleic Acids Research, 39(suppl_1), D163–D169.
Hsu, J. B. K., Chiu, C. M., Hsu, S. D., Huang, W. Y., Chien, C. H., Lee, T. Y., & Huang, H. D. (2011). miRTar: an integrated system for identifying miRNA-target interactions in human. BMC Bioinformatics, 12(1), 300.
Karagkouni, D., Paraskevopoulou, M. D., Chatzopoulos, S., Vlachos, I. S., Tastsoglou, S., Kanellos, I., Papadimitriou, D., Kavakiotis, I., Maniou, S., Skoufos, G., et al. (2017). DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA–gene interactions. Nucleic Acids Research, 46(D1), D239–D245.
Kim, S. (2015). ppcor: an R package for a fast calculation to semi-partial correlation coefficients. Communications for Statistical Applications and Methods, 22(6), 665.
Krämer, N., Schäfer, J., & Boulesteix, A.-L. (2009). Regularized estimation of large-scale gene association networks using graphical Gaussian models. BMC Bioinformatics, 10(1), 384.
Lewis, B. P., Burge, C. B., & Bartel, D. P. (2005). Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell, 120(1), 15–20.
Li, X., Yu, X., He, Y., Meng, Y., Liang, J., Huang, L., Du, H., Wang, X., & Liu, W. (2018). Integrated analysis of microRNA (miRNA) and mRNA profiles reveals reduced correlation between microRNA and target gene in cancer. BioMed Research International, 2018.
MacKinnon, J. G. (2009). Bootstrap hypothesis testing. Handbook of Computational Econometrics, 183, 213.
Miao, X., Luo, Q., Zhao, H., & Qin, X. (2017). An integrated analysis of miRNAs and methylated genes encoding mRNAs and lncRNAs in sheep breeds with different fecundity. Frontiers in Physiology, 8, 1049.
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., & Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-seq. Nature Methods, 5(7), 621.
Shojaie, A., & Michailidis, G. (2009). Analysis of gene sets based on the underlying regulatory network. Journal of Computational Biology, 16(3), 407–426.
Shojaie, A., & Michailidis, G. (2010). Network enrichment analysis in complex experiments. Statistical Applications in Genetics and Molecular Biology, 9(1).
Stokowy, T., Eszlinger, M., Świerniak, M., Fujarewicz, K., Jarząb, B., Paschke, R., & Krohn, K. (2014). Analysis options for high-throughput sequencing in miRNA expression profiling. BMC Research Notes, 7(1), 144.
Tokar, T., Pastrello, C., Rossos, A. E. M., Abovsky, M., Hauschild, A.-C., Tsay, M., Lu, R., & Jurisica, I. (2017). mirDIP 4.1–integrative database of human microRNA target predictions. Nucleic Acids Research, 46(D1), D360–D370.
Tomczak, K., Czerwińska, P., & Wiznerowicz, M. (2015). The cancer genome atlas (TCGA): an immeasurable source of knowledge. Contemporary Oncology, 19(1A), A68.
Vlachos, I. S., Zagganas, K., Paraskevopoulou, M. D., Georgakilas, G., Karagkouni, D., Vergoulis, T., Dalamagas, T., & Hatzigeorgiou, A. G. (2015). DIANA-miRPath v3. 0: deciphering microRNA function with experimental support. Nucleic Acids Research, 43(W1), W460–W466.
Volinia, S., & Croce, C. M. (2013). Prognostic microRNA/mRNA signature from the integrated analysis of patients with invasive breast cancer. Proceedings of the National Academy of Sciences, 110(18), 7413–7417.
Wei, L., Jin, Z., Yang, S., Xu, Y., Zhu, Y., & Ji, Y. (2017). TCGA-assembler 2: software pipeline for retrieval and processing of TCGA/CPTAC data. Bioinformatics, 34(9), 1615–1617.
Wu, X., & Watson, M. (2009). CORNA: testing gene lists for regulation by microRNAs. Bioinformatics, 25(6), 832–833.
Yu, N., Yong, S., Kim, H. K., Choi, Y.-L., Jung, Y., Kim, D., Seo, J., Lee, Y. E., Baek, D., Lee, J., et al. (2019). Identification of tumor suppressor miRNAs by integrative miRNA and mRNA sequencing of matched tumor–normal samples in lung adenocarcinoma. Molecular Oncology, 13(6), 1356.
Zhang, Y., Linder, M. H., Shojaie, A., Ouyang, Z., Shen, R., Baggerly, K. A., Baladandayuthapani, V., & Zhao, H. (2017). Dissecting pathway disturbances using network topology and multi-platform genomics data. Statistics in Biosciences, 1–21.
Zhou, Y.-H., Xia, K., & Wright, F. A. (2011). A powerful and flexible approach to the analysis of RNA sequence count data. Bioinformatics, 27(19), 2672–2678.
Zhu, Y., Qiu, P., & Ji, Y. (2014). TCGA-assembler: open-source software for retrieving and processing TCGA data. Nature Methods, 11(6), 599.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Linder, H., Zhang, Y. (2022). MiRNA–Gene Activity Interaction Networks (miGAIN): Integrated Joint Models of miRNA–Gene Targeting and Disturbance in Signaling Pathways. In: He, W., Wang, L., Chen, J., Lin, C.D. (eds) Advances and Innovations in Statistics and Data Science. ICSA Book Series in Statistics. Springer, Cham. https://doi.org/10.1007/978-3-031-08329-7_1
Download citation
DOI: https://doi.org/10.1007/978-3-031-08329-7_1
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-08328-0
Online ISBN: 978-3-031-08329-7
eBook Packages: Mathematics and StatisticsMathematics and Statistics (R0)