Abstract
This manuscript presents the evolution of the computational technologies and image-based algorithms applied to skull prosthesis modeling in recent years. Medical diagnoses and surgical planning are characterized as processes that engineering tools can support. The scope is about 3D modeling evaluation of the complex geometry of virtual prosthesis pieces built from medical images. The scenario has a relationship with the improvement of automated manufacturing processes, addressed by the engineering viewpoint. Advances in manufacturing technologies, and integration with computer-aided design techniques, using CAD/CAM (Computer-Aided Design, Computer-Aided Manufacturing) and/or 3D Printing tools, has allowed prosthesis manufacturing to be customized according to the individual’s morphological characteristics. Among ongoing technologies, those all dealing with the manipulation of digital images have gained support within the IoHT (Internet of Health Things) approach in the context of process automation. The literature was reviewed, summarizing the recent studies in this theme in a total of 34 articles. This review enabled us to improve the techniques explored. As a result, we present improvements on previous methods addressed by an efficient strategy based on the Variational AutoEncoder (VAE) neural network to generate each layer of the reconstructed prosthesis. Thus, the purpose of this study is to provide short feedback on AI-based applications in the context of bone modeling and serve as a roadmap for future developments.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
1 Introduction
The recent evolutions in the Artificial Intelligence (AI) field have introduced innovative approaches in the many medicals processes. Nowadays, the same pillars of industry 4.0 revolution, as the Internet of Things (IoT) or IoHT to “health things” in the medical area), Cloud Computing and Cyber-Physical Systems (CPS) have made some relevant contributions to solve the problems in medicine with a focus on processes optimization. Inside these innovative concepts, the data based on medical images analysis are essential to support different processes.
From images, those processes that require 3D reconstruction depend on a wide set of data, and from the engineering's viewpoint, require a high computational processing level.
Medical images from computed tomography (CT) or magnetic resonance imaging (MRI) are frequently used to model the existing bones structures, improve visualization and clinical interpretation, and used in the surgical preparation of implants, construction of molds, and virtual models for prosthetics pieces.
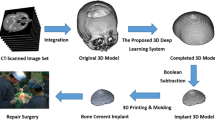
Virtual bone modeling is a challenging process due to the complexity of the geometry and because sometimes we do not have enough information to build a model. In this context, the skull prosthesis modeling has the same challenge, mainly for non-symmetric fractures, because mirroring techniques are ineffective, for example, as in Fig. 11.1a (VISUALDATA 2021). The conceptual basis for a personalized reconstruction is to look for the image parameters from knowing data. In this way, as in Fig. 11.1b, a virtual model can rebuild a complete skull, and an estimative of a prosthesis piece can be calculated to fill the gaps in the bone. The region of interest (ROI) addressed in this research is the calvaria region.
Following the concept presented in Fig. 11.1, some techniques were experimented by the PPGEPS (Industrial and Systems Engineering Program) team during last years based on optimization techniques and AI tools and made some different approaches to virtual model creation as (i) Adjusted Ellipses and Super-Ellipses concept, (ii) PSO (Particle Swarm Optimization), (iii) Splines and Bezier Curves, (iv) Data Mining, (v) Content-Based Retrieval, (vi) Neural Networks, (vii) CNNs (Convolutional Neural Networks) and Deep learning, and (viii) Semantic Segmentation; all of them aligned with virtual design systems integration. The unfolding from these initial experiments using more advanced approaches as Variational AutoEncoder (VAE) has been analyzed here. Thus, the objective is to show the advances in prosthesis modeling by investigating the most recent research studies and propose a suitable solution for the anatomic prosthesis.
2 The State of the Art for the Skull Modelling
Several approaches have been used over the years to address the task of skull reconstruction. We reviewed these approaches by searching for articles related to skull reconstruction in PubMed, Science Direct, and Google Academic as in the method of (Reche et al. 2020). We considered all articles, with no restriction towards the publication year. After our article selection step, we selected 34 articles that are summarized and analyzed over this section. All selected articles are summarized in Table 11.1; we provide information regarding the dataset, the approach, the dataset size, and the obtained results. Some of the articles evaluated their results visually, others with knowing metrics, for instance, applying the Dice Similarity Coefficient (DSC) and Hausdorff distance (HD).
Among the methods for skull modeling, there are approaches based on mirroring. Mirroring uses the reflection of the healthy half of the skull onto the defective one, using the patient's data as a template for the implant (Marzola et al. 2019). Unilateral defects can be solved with mirroring, either with minimal manual effort or automatically (Mainprize et al. 2020).
Preliminary studies from Lee et al. (2002) and Hieu et al. (2003) from the 2000s. Both used mirroring and an additional manual adjustment step. The approaches from Gall et al. (2016) and Egger et al. (2017) also used mirroring and manual adjustment. The adjustment was based on Laplacian smoothing followed by a Delaunay triangulation. The mirroring provided an initial design, while the smoothing and triangulation provided an aesthetic-looking and well-fitting outcome (Egger et al. 2017). The approach from Rudek et al. (2015b) also applied symmetric mirroring. Further, Chen et al. (2017) developed a mirroring, contour clipping, and surface fitting method. Marzola et al. (2019) developed a mirroring and surface interpolation method for unilateral and quasi-unilateral defects.
However, just mirroring may not be enough for an implant, as human heads are generally too asymmetric (Gall et al. 2016). Further, mirroring can only be applied to unilateral defects, not to defects that cross the symmetry plane (Shi and Chen 2020; Marzola et al. 2019).
An alternative is using slice-based techniques, where 2D images (generally CT images) are used to fit curves to the bone contours (Marzola et al. 2019). The information from the healthy part is usually used as a boundary to the curve optimization algorithms and functions.
Among the techniques, Rudek et al. (2018), Rudek et al. (2016), and Rudek et al. (2015a) generated control points for each CT slice with Cubic Bezier Curves and the ABC algorithm to optimize the curve. Afterward, they compared control points of the defective slices with healthy slices, selecting the healthy slice with the smallest error to fulfill the missing part. Mohamed et al. (2015) also used Bezier Curves; they combined C1 Rational Bezier Curves with Harmony Search (HS) to complete the missing piece on every slice with a defect.
The approaches of Lin et al. (2017) and Chang and Cheng (2018) used methods based on active contour models. Lin et al. (2017) developed a self-adjusting method based on active contour models, optimizing the shape for every slice and then combining the results for each slice. While Chang and Cheng (2018) developed a self-adjusting method based on active contour models, using the Adaptive Balloon Force Active Surface Model.
Further, Rudek et al. (2013) and Lin et al. (2016) used methods based on Superellipses. Rudek et al. (2013) adjusted Superellipses to the missing part. The Superllipses parameters were optimized by Particle Swarm Optimization (PSO). Lin et al. (2016) developed a self-adjusting method based on Superellipse combined with Differential Evolution for the Superellipse parameter optimization.
The downside of Curve optimization methods is that they work only at slice-level and only consider information about that specific slice for which the curve is being optimized. Hence, the lack of information about the missing area (all slices that have a defect) could affect the reconstruction (Marzola et al. 2019). Further, methods trained over several images have a better chance of “learning” the characteristics of missing parts in general and better designing more realistic implants.
Over time deep learning models have become an alternative to traditional machine algorithms traditionally used in medical imaging (Singh et al. 2020). Convolution neural networks (CNNs) are widely used in medical imaging, considering 2D or 3D images. Additionally, classification problems involving 3D images, such as CT images, can always be downgraded to 2D-level. The drawbacks of developing deep learning for 3D images are the limited available data to the algorithms and computational cost (Singh et al. 2020). However, data augmentation techniques and more powerful GPUs mitigate these issues (Singh et al. 2020).
Over the literature, we verified a predominance of publications involving CNN-based models in the latest years, especially in 2020 and 2021. Most of the publications were related to the AutoImplant 2020 shared task,Footnote 1 with 210 complete skulls from the CQ500 datasetFootnote 2 with their corresponding defective skulls and the implants (Li and Egger 2020a, b).
The approaches used for the AutoImplant 2020 challenge are detailed below. Pimentel et al. (2020) adjusted a 3D statistical shape model (SSM) to locate and correct the defect, followed by a 2D generative adversarial network (GAN) to make corrections and better fit the design. Shi and Chen (2020) decomposed into slices for each axis (2D level), applying three 2D CNNs (one for each axis) and then combining the results from all axes. Further, Bayat et al. (2020) approached was based on CNNs, with a 3D encoder-decoder to complete the downsampled defected followed by a 2D upsampler to upsample the shape. Downsampling turned the approach more feasible for commonly available GPUs. Li et al. (2020) developed a baseline approach for the AutoImplant 2020 shared task. The approach was based on an encoder-decoder network that predicted a downsampled coarse implant; afterward, the data is upsampled, a bounding box is applied to locate the defected region on the high-resolution volume, and the data passes by another encoder/decoder network that generated an implant from the bounded region.
Several of the papers used models based on U-net (Ronneberger et al. 2015). As highlighted by (Ronneberger et al. 2015), the U-net model has the advantage of being trained end-to-end from very few images, achieving excellent results while the training is fast.
The papers that used models based on U-net are detailed further below. Ellis and Aizenberg (2020) augmented the dataset with data transformations that added different shapes and orientations (9903 additional images—significant augmentation) and used a U-Net model with residual connections. Kodym et al. (2020a)’s approach was based on skull alignment with landmark detection with 3D CNN and two 3D U-net models for the reconstruction. They also applied shape pros-processing steps. Mainprize et al. (2020) predicted the skull with a U-Net model and data augmentation, subtracted from the original skull to obtain the prosthesis, and added further post-processing. The data augmentation occurred by adding cubic and spherical defects. Eder et al. (2020)’s framework used 2 U-Net models, the first one was used to reconstruct the skull with low-resolution data, and the other was used to up-sample the low-resolution data. Also, they applied some post-processing filters to provide specific corrections. Kodym et al. (2020b) predicted the implant with two U-net models, testing discriminative and generative models. Additionally, provided tests with synthetic data.
Further, Matzkin et al. (2020b) adapted the model from Matzkin et al. (2020a) to consider flaps similar to the ones for the challenge dataset, tested two methods: (1) using a 3D DE-UNet model and (2) DE-UnET model with data augmentation from additional input from shape priors. Wang et al. (2020) predicted the complete skull with a Residual Dense U-net (RDU-Net) model with the encoder of the Variational Auto-encoder (VAE) model being added as the U-net regulation term, later subtracted the defective skull to delimiter the implant. Jin et al. (2020) approach was based on a V-net model, an adaptation of the U-Net model proposed by Milletari et al. (2016). To address the problem of high-resolution input images to neural networks tested two methods: resizing the images and partitioning the images.
Among the papers related to the AutoImplant 2020 shared task, we highlight the approach of Ellis and Aizenberg (2020), in which the augmentation of training data with registration directly improved the classifier results. Further, for Matzkin et al. (2020b), data augmentation was helpful for out-of-distribution cases, in which the defects did not follow the distribution from the training data, where the network tended to fail. Thus, data augmentation can provide additional data for the network’s training with the cost of computational power. Generally, deep learning models tend to achieve superior performance by providing additional data.
Outside of the AutoImplant 2020 shared task dataset, some papers used neural networks. Hsu and Tseng (2000) and Hsu and Tseng (2001) are preliminary studies that addressed that used models based on 3D orthogonal neural networks. Further, Rocha et al. (2020) used a Generative Adversarial Network (GAN) model to predict the missing part of each slice. Additionally, the Morais et al. (2019) approach was based on the Volumetric Convolutional Denoising Autoencoder model that Sharma et al. (2016) proposed with some adaptions. Further, they used MRI images, not CT images. Fuessinger et al. (2018)'s approach used a statistical shape model (SSM) based on 131 CT scans combined with geometric morphometrics (GM) methods, comparing the results with the mirroring technique (only for unilateral defects).
Further, (Chang et al. 2021) used a CNN-based deep learning network with 12 layers and data augmentation to complete the defect, subtracting the completed skull from the incomplete skull to obtain the implant. In the approach proposed by Matzkin et al. (2020a), the images passed by registration, resampling, and thresholding being fed into a U-Net that directly predicted the implant. Tested several architectures and reconstruction strategies (direct estimation and reconstruct and subtract), achieving superior results with U-Net and direct estimation of the implant.
Hence, in general, we verified that most of the preliminary studies addressed the reconstruction task with mirroring or slice-based techniques. With the advances in deep learning models, especially CNNs and their variants (U-net), deep learning-based models became the primary approach. Further, techniques that relieve the computational cost of the training, such as the usage of downsampling combined with upsampling techniques and data augmentation techniques that provide additional images to the training step, are relevant to the research theme.
Among the techniques that were used to address skull reconstruction, VAE models have addressed the task well. Hence, in the next section, we detail some of our experiments of skull reconstruction with a VAE-based model. The following section provides details about our proposed method with its results. Additionally, we provide details about standard evaluation metrics and several images to give the viewer a better understanding of the whole process.
3 Background
3.1 The VAE Neural Network
The Variational AutoEncoder (VAE), as presented in Fig. 11.2, is a neural network that encodes and decodes data. In the encoding part, the data is compressed into a regularized latent space. In the decoding part, the data is decompressed from this space. The regularization of the latent space is made by minimizing the Kullback and Leibler Divergence (KL divergence), this makes the VAE a generative model.
Figure 11.2 represents the VAE architecture where x represents the input data, h represents the coded data, σ represents the measure of the standard deviation of h, µ represents the measure of the mean of h, z represents the sampled data of the probability distribution µ + p(0, 1)*σ2 and ŷ represents the classification of decoding data.
3.2 Evaluation Criteria
The VAE neural network (Patterson and Gibson 2017) is a generative model that encodes the input into a latent space close to the uniform distribution using KL loss and decodes the latent space into a classification problem measured using reconstruction loss, additional methods like dice similarity coefficient and Hausdorff distance are used to compare results between proposed method and results in (Li and Egger 2020a, b), assuming y is the removed part of skull and ỹ is 1 where ŷ > 0.5 and 0 otherwise. The measuring methods are described below.
3.2.1 The Dice Similarity Coefficient
As presented by Taha and Habury (2015), the Dice Similarity Coefficient (DSC) measures the difference between an y and an ỹ. Both images are required to have the same size and pixels values should be 0 or 1. The formula uses the sum of pixels and the intersection between both images, according to Eq. 11.1.
3.2.2 The Hausdorff Distance
According to Taha and Habury (2015), the Hausdorff Distance (HD) is a way of measuring the difference between two images based on the maximum distance between a pixel in y and a pixel in ỹ, according to Eq. 11.2.
3.2.3 The Kullback and Leibler Divergence
From Bonaccorso (2018), the Kullback and Leibler divergence (KL divergence) is used to compare two distributions, where the KL loss is the comparison between a p(µ, σ) distribution and a normal distribution p(0,1), with µ the measured mean and σ the measured standard deviation, according to Eq. 11.3.
3.2.4 The Reconstruction Loss
As presented in Bonaccorso (2018), the Reconstruction loss measures a classification using a reference binary image and a classification image with values between 0 and 1, according to Eq. 11.4.
4 Proposed Method
The method uses a Variational AutoEncoder (VAE) neural network (Patterson and Gibson 2017) to generate each layer of the prosthesis. The method was separated into four main steps: (i) image selection, (ii) image segmentation, (iii) neural network training, and (iv) reconstruction, as in Fig. 11.3.
4.1 Step 1—Image Selection
The creation of the training, validation and test sets separated some files of skulls without fracture identification. The fracture identification was performed by three specialists and noted in an auxiliary dataset as in Table 11.2. The skulls without any indication of fracture were selected. The images size is 512 × 512 pixels, with voxel dimensions from 0.441 × 0.441 × 0.625 mm to 0.488 × 0.488 × 0.625 mm. Also, each exam having from 233 to 256 image slices of the upper skull region (calvaria region). The selection resulted in 123 skulls, where 80% of them were used for training, 10% for validation, and 10% for the test.
Table 11.3 represents the voxel height statistics considering those images without fracture, with a dimension of 512 × 512 pixels and a voxel depth of 0.625 mm.
Table 11.4 shows the statistics of the number of layers after voxel selection between 25 and 75%.
The region of interest was selected as 40% of the upper layers, as presented in Fig. 11.4. We can simulate a frontal defect area in this region by removing the data from a one-by-one 2D CT slicing.
4.2 Step 2—Image Segmentation
Bone tissue was identified using the Hounsfield scale in the image sets. The value of 500HU was found based on segmentation tests and histogram comparison. After, for each skull, a volume was created with the CT slices overlapping operation. Finally, identifying connected pixels and selection of the largest connected group, removing unwanted objects presented in Fig. 11.5.
4.3 Step 3—Neural Network Training
The training set and the validation set of CT images were used to train and validate the neural network. The total number of skull images was augmented by changes in position and size to increase the number of data samples. From each skull file, the image is transformed by: a vertical inversion with a 50% change, rotated between ‒45° and 45°, scaled between 90 and 110%, and finally translocated between ‒16 and 16 pixels. After the transformation, up to 25% of the image is removed and used as the neural network’s output. Further, the image complement is used as the input of the neural network. Figure 11.6 represents an example of each type of transformation.
The VAE neural network architecture was used with input and output of 512 × 512 × 1, six layers of convolution and deconvolution with 2layer filters, latent space with two layers of 4096 and samples among the latent space. The ReLu was used as the activation function in the convolution and deconvolution layers. The Linear activation function was applied in the latent space layers, and the Sigmoid activation function was used in the output of the neural network. Figure 11.7 represents the generation of a prosthesis layer, going through the stages of convolution, parameter estimation, sampling, and deconvolution.
The sum of Reconstruction Loss with KL Loss was used as the loss function. The Reconstruction Loss was calculated between ŷ and y. The KL Loss was calculated between µ and σ. The DSC and HD metrics were calculated between y and ỹ, these metrics were used only for visualization and did not interfere with learning. The ADAM optimizer (Bonaccorso 2018) was used for training. Figure 11.8 represent the metrics in respective scale.
4.4 Step 4—Reconstruction
The test set without transformation and with removed skull parts was used to create the three-dimensional models, where the interference between x and ỹ was removed. The marching cubes (Lorensen and Cline 1987) method was used to obtain the vertices and faces of the triangles obtained on the surface of the prosthesis (superimposition of layers), using the voxel dimensions as a reference. The Laplacian smoothing (Vollmer et al. 1999) method was used to reduce the sharp surface differences. Figure 11.9 represents the reconstruction process.
5 Application Example
The dataset provided by the Center for Advanced Research in Imaging, Neurosciences, and Genomics (CARING) (VISUALDATA 2021) was used in the simulations. It contains 491 skulls with and without fractures, and each skull is composed of different data collections where the number of files and the dimensions of the voxel (Height, Width, Depth) may vary. The absolute path, study name, fracture identification, image size, image position, and voxel dimensions (HxWxD) were mapped to each file. A machine with AMD Ryzen 5 3600X Hexa-core processor, NVIDIA Graphics Card, was used for neural network training and 3D mesh generation. An RTX 2060 super 8 GB VRAM, 32 GB RAM, and Windows 11 operating system. Python programming language was used for software development.
Figure 11.10 shows the synthetic cut artificially created in (a) by removing 25% of the skull and the generated prosthesis in (b). The colors scale in (b) represents the distance between the prosthesis and the original removed part of the skull. See Fig. 11.11 about color scale and respective metrics.
Figure 11.10b shows differences in the prosthesis boundary connected with the original removed piece, with measures differences around 2 mm as indicated by the green representation. Table 11.5 represents the metrics obtained from removed part of the skull (piece cut as in Fig. 11.10a). The (+) and (‒) signals represent if the difference is on outer or inner surface respectively by comparing with the real bone.
The other view of the generated prosthesis is presented in Fig. 11.11. From colored scale, in green the distances are between ‒2 and 2 mm; in red, the distances are greater than 2 mm; and in blue, the distances are smaller than -2 mm.
6 Conclusion
The research presented the advances to skull prosthesis modeling based on a literature review and respective content analysis. A set of 35 papers were selected to demonstrate the techniques addressed in recent years. The CNN-based techniques are proved as the best way to find missing information on CT/MRI images with efficiency validated by the metrics Mean DSC and Mean HD.
We observed that the virtual skull repairing based on Variational Auto-encoder (VAE) model presented in literature looks like promisor and we investigated its application for own method. As presented in example, the best result for our method was 0.792 to Mean DSC and 5.587 to Mean HD.
The performance achieved by the proposed method according to the metrics: Mean HD and Mean DSC, present important improvement by comparison of the previous explored methods if compared with PSO, Superellipse, ABC and other previously studied by the authors. However, the metrics indicates that we do not get similar results as found in literature.
The next step is improving HD and DSC by testing addicting 3D CNN, more filters in convolution/deconvolution or more neurons in latent space.
References
Bayat A, Shit S, Kilian A et al (2020) Cranial implant prediction using low-resolution 3D shape completion and high-resolution 2D refinement In: Cranial implant design challenge, pp 77–84
Bonaccorso G (2018) Mastering machine learning algorithms: expert techniques to implement popular machine learning algorithms and fine-tune your models. Packt Publishing Ltd
Chang CC, Cheng CY (2018) The adaptive balloon forces for active surface models in skull repair technique. In: 2018 International conference on engineering, applied sciences, and technology (ICEAST), pp 1–5
Chang YZ, Wu CT, Yang YH (2021) Three-dimensional deep learning to automatically generate cranial implant geometry
Chen X, Xu L, Li X et al (2017) Computer-aided implant design for the restoration of cranial defects. Sci Rep 7(1):1–10
Eder M, Li J, Egger J (2020) Learning volumetric shape super-resolution for cranial implant design. In: Cranial implant design challenge, pp 104–113
Egger J, Gall M, Tax A et al (2017) Interactive reconstructions of cranial 3D implants under MeVisLab as an alternative to commercial planning software. PLoS One 12(3):e0172694
Ellis DG, Aizenberg MR (2020) Deep learning using augmentation via registration: 1st place solution to the autoimplant 2020 challenge. In: Cranial implant design challenge, pp 47–55
Fuessinger MA, Schwarz S, Cornelius CP et al (2018) Planning of skull reconstruction based on a statistical shape model combined with geometric morphometrics. Int J Comput Assist Radiol Surg 13(4):519–529
Gall M, Li X, Chen X et al (2016) Computer-aided planning and reconstruction of cranial 3D implants. In: 2016 38th annual international conference of the IEEE engineering in medicine and biology society (EMBC), pp 1179–1183
Hieu LC, Bohez E, Vander et al (2003) Design for medical rapid prototyping of cranioplasty implants. Rapid Prototyping J 9(3):175–186
Hsu JH, Tseng CS (2000) Application of orthogonal neural network to craniomaxillary reconstruction. J Med Eng Technol 24(6):262–266
Hsu JH, Tseng CS (2001) Application of three-dimensional orthogonal neural network to craniomaxillary reconstruction. Comput Med Imaging Graph 25(6):477–482
Jin Y, Li J, Egger J (2020) High-resolution cranial implant prediction via patch-wise training. In: Cranial implant design challenge, pp 94–103
Kodym O, Spanel M, Herout A (2020a) Cranial defect reconstruction using cascaded CNN with alignment. In: Cranial implant design challenge, pp 56–64
Kodym O, Spanel M, Herout A (2020b) Skull shape reconstruction using cascaded convolutional networks. Comput Biol Med 123:103886
Lee MY, Chang CC, Lin CC et al (2002) Custom implant design for patients with cranial defects. IEEE Eng Med Biol Mag 21(2):38–44
Li J, Egger J (2020a) Dataset descriptor for the autoimplant cranial implant design challenge. In: Cranial implant design challenge, pp 10–15
Li J, Egger J (2020b) Towards the automatization of cranial implant design in cranioplasty. Springer, Cham
Li J, Pepe A, Gsaxner C et al (2020) A baseline approach for autoimplant: the miccai 2020 cranial implant design challenge. In: Multimodal learning for clinical decision support and clinical image-based procedures, pp 75–84
Lin Y, Cheng C, Cheng Y et al (2017) Skull repair using active contour models. Procedia Manufact 11:2164–2169
Lin YC, Cheng CY, Cheng YW et al (2016) Using differential evolution in skull prosthesis modelling by superellipse
Lorensen WE, Cline HE (1987) Marching cubes: a high resolution 3D surface construction algorithm. Comput Graph 21(4)
Mainprize JG, Fishman Z, Hardisty MR (2020) Shape completion by U-Net: an approach to the autoimplant MICCAI cranial implant design challenge. In: Cranial implant design challenge, pp 65–76
Marzola A, Governi L, Genitori L et al (2019) A semi-automatic hybrid approach for defective skulls reconstruction. Comput-Aided Des Appl 17:190–204
Matzkin F, Newcombe V, Glocker B et al (2020b) Cranial implant design via virtual craniectomy with shape priors. In: Cranial implant design challenge, pp 37–46
Matzkin F, Newcome V, Stevenson S et al (2020a) Self-supervised skull reconstruction in brain CT images with decompressive craniectomy. In: International conference on medical image computing and computer-assisted intervention, pp 390–399
Millletari F, Navab N, Ahmadi AS (2016) V-Net: fully convolutional neural networks for volumetric medical image segmentation. In: 2016 fourth international conference on 3D vision (3DV), pp 565–571
Mohamed N, Majid AA, Piah ARM et al (2015) Designing of skull defect implants using C1 rational cubic Bezier and offset curves. In: AIP conference proceedings, p 050003
Morais A, Egger J, Alves V (2019) Automated computer-aided design of cranial implants using a deep volumetric convolutional denoising autoencoder. In: World conference on information systems and technologies, pp 151–160
Patterson J, Gibson A (2017) Deep learning: a practitioner’s approach. O’Reilly Media, Inc.
Pimentel P, Szengel A, Ehlke M et al (2020) Automated virtual reconstruction of large skull defects using statistical shape models and generative adversarial networks. In: Cranial implant design challenge, pp 16–27
Reche AYU, Canciglieri Junior O, Estorilio CCA et al (2020) Integrated product development process and green supply chain management: contributions, limitations and applications. J Clean Prod 249:119429–1194459
Rocha LGS, Rudek JVL, Rudek M (2020) Extraction of geometric attributes based on GAN for anatomic prosthesis modeling. In: ICIST 2020 proceedings, pp 64–67
Ronneberger O, Fischer P, Brox T (2015) U-Net: convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention, pp 234–241
Rudek M, Gumiel YB, Canciglieri Junior O et al (2018) A cad-based conceptual method for skull prosthesis modelling. Facta Univ Ser: Mech Eng 16(3):285–296
Rudek M, Canciglieri Junior O, Jahnen A et al (2013) CT slice retrieval by shape ellipses descriptors for skull repairing. In: 2013 IEEE international conference on image processing, pp 761–764
Rudek M, Gumiel YB, Canciglieri Junior O et al (2015a) Optimized CT skull slices retrieval based on cubic bezier curves descriptors. In: CIE45—the 45th international conference on computers & industrial engineering
Rudek M, Mendes GC, Canciglieri Junior O et al (2015b) Skull failure-correction modelling method by symmetry mirroring. In: CIE45—the 45th international conference on computers & industrial engineering
Rudek M, Gumiel YB, Canciglieri Junior O et al (2016) Optimized CT skull slices retrieval based on cubic Bezier curves descriptors. In: 6th international conference on information society and technology ICIST 2016, pp 75–79
Sharma A, Grau O, Fritz M (2016) Vconv-dae: Deep volumetric shape learning without object labels In: European conference on computer vision, pp 236–250
Shi H, Chen X (2020) Cranial implant design through multiaxial slice inpainting using deep learning. In: Cranial implant design challenge, pp 28–36
Singh SP, Wang L, Gupta S et al (2020) 3D deep learning on medical images: a review. Sensors 20(18):5097
Taha AA, Habury A (2015) Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool. BMC Med Imaging 15(1):1–28
VISUALDATA (2021) CQ500. A dataset of head CT scans. http://headctstudy.qure.ai/#dataset
Vollmer J, Mencl R, Mueller H (1999) Improved laplacian smoothing of noisy surface meshes. In: Computer graphics forum. Blackwell Publishers Ltd, Oxford, UK and Boston, USA, pp 131–138
Wang B, Liu Z, Li Y et al (2020) Cranial implant design using a deep learning method with anatomical regularization. In: Cranial implant design challenge, pp 85–93
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
da Rocha, L.G.S., Gumiel, Y.B., Rudek, M. (2022). Modelling of the Personalized Skull Prosthesis Based on Artificial Intelligence. In: Canciglieri Junior, O., Trajanovic, M.D. (eds) Personalized Orthopedics. Springer, Cham. https://doi.org/10.1007/978-3-030-98279-9_11
Download citation
DOI: https://doi.org/10.1007/978-3-030-98279-9_11
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-98278-2
Online ISBN: 978-3-030-98279-9
eBook Packages: EngineeringEngineering (R0)