Abstract
The potential application of biological molecules as functional devices has been heralded as the dawn of a new field in biotechnology and medicine. In this regard, molecular motors have attracted the most attention for decades. In the current study, we have studied the bioenergy transfer in a protein chain as a self-powered nanomotor. The effect of different factors on the energy transfer in protein is studied to obtain the best functional condition for the protein machine. The temperature plays the critical role in the control of energy transfer in the system. On the other hand, the external mechanical tension as a vibrator can increase the energy flowing in our system. The chaos theory tools can verify and estimate the results. Generally, one can engineer a self-powered nanomotor based on protein chains and control bioenergy transfer.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Biological molecules, an exciting field with continuous and robust growth for about half a century of existence, encompass the medical, biological, chemical, and material sciences [1]. In the current study, we have studied a protein chain as a biological molecule and its energy transfer. Protein, as the largest macromolecule in the body, is one of the most important components of the life systems that plays vital role in the body. Proteins are composed of amino acid chain sequences that the number of the amino acids can reach several thousand depending on the type of protein [2].
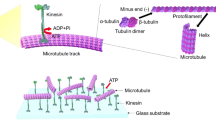
In the current study, we investigate the bioenergy transfer in protein molecules as an automated nanomotor. Automated nanomotor uses the bioenergy in the protein as self-burning energy and converts it into autonomous motion [3]. These nanomotors, which derive their kinetic energy from the biological energy contained in proteins, are called protein nanomotors. Protein motors have potential medical applications and can collect, transport, and release drug carriers of various sizes. Internal order in eukaryotic cells is created by protein motors that transport organs and molecules along the cytoskeletal pathways of self-assembled proteins such as tubulin and shuttle actin. Three known families of cytoskeletal protein motors include kinesin, dynein, and myosin [4]. Kinesins and dyneins, which are the first and second types of protein motors, respectively, move in tubules or microtubules [5]. But Myosins, the third family of protein motors, move on actin filaments and are responsible for muscle contraction in the heart and skeletal muscles [6].
2 Model and Methods
In the current work, the Hamiltonian model used to study the energy transfer in protein molecules is based on the Peng model as follows:
where \(H_{1}\) indicates that a boson-type Frankit exciton is excited in protein molecules using the energy released in ATP hydrolysis written as follows:
\(H_{2}\) defines the harmonic properties of the remaining amino acids:
\(H_{3}\) introduces the interaction between the two modes of motion [7]:
The parameters used in the calculations are shown in Table 1 [8]:
Here, \(a_{n} and a_{n}^{\dag }\) are the creation and annihilation operators for exciton. \(u_{n} and p_{n}\) are the displacement and momentum operators for the amino acid residue at site n, respectively. \(\varepsilon_{0}\) is the energy of the exciton. \(\chi_{1}\) and \(\chi_{2}\) are nonlinear coupling constants. m is the mass of amino acid residue. W is the elasticity constant of the proteins, and J is the dipole–dipole interaction energy between neighboring amino acids [9].
In this work, we have used the classical chaos theory. In this case, the evolution equations for the classical part are derived using the Hamiltonian equation \(\left( {\dot{p}_{n} = - \frac{\partial H}{{\partial q_{n} }}} \right)\). Also, the evolution equations for the quantum part are analyzed using the Heisenberg equation \(\left( {\dot{a}_{n} = - \frac{i}{\hbar }\left[ {a_{n} ,H} \right]} \right)\) as follows:
To study the energy transfer in protein molecules, we have extracted the energy flux. The energy flux obtains using the continuity equation \(\left( {{\text{I}} = - \frac{i}{\hbar }\left[ {a_{n}^{\dag } a_{n} ,H} \right]} \right)\) as follows:
3 Results and Discussion
Different factors effect on energy transfer in biological systems. One of the vital parameters on energy transfer in the protein system is the effect of temperature. We have used the Nosé Hoover thermostat to apply the temperature to the system. The evolution equation of thermostat is written as follows [10]:
where ξ describes a thermodynamic coefficient of friction. T0 is the temperature of the system. KB is the constant of Boltzmann, and M is the thermostat constant.
A biological system can be affected by mechanical shocks. Therefore, we have investigated the effect of mechanical stress on energy transfer in protein molecules. The Hamiltonian of mechanical stress is written as follows [11]:
where \(F_{0}\) and ω are the amplitude and frequency of the external mechanical stress.
We can also examine the best range of parameters affecting energy transfer in protein molecules using multifractal system analysis. Multifractal analysis of the system can confirm the obtained results. In addition, it can classify system parameters to determine the desired results.
In this regard, we use the Renyi dimension spectrum, analogous of the free energy and analogous of the thermodynamic specific heat.
3.1 Simultaneous Effect of Ambient Temperature and the Range of Force Exerting Mechanical Stress
Temperature and external stress as the very influential factors in the performance of biological systems can be considered as the control parameters in the energy transfer in the protein system. We have considered the simultaneous effect of ambient temperature and the range of force exerting mechanical stress on the energy flux in the protein system (Fig. 1). The variation of the ambient temperature from 300 to 330 K and at the same time changing the amplitude of the force from 0 to 2 pN has negligible effect on the amount of energy flux transmitted in the system. In this region, the energy flux fluctuates at a minimum of about 10 to 30 μeV. But, when the temperature rises above 330 K, in the force range of about 1.2 pN, we can see a considerable peak in the energy flux, so that the energy flux reaches about 100 μeV. Similar to this phenomenon is also observed at a temperature of about 345 K and a force range of about 0.8 pN. Therefore, it can be said that temperatures above 330 and the applied stress are two critical parameters in the transfer of bioenergy in our protein system.
3.2 Simultaneous Effect of Temperature and Applied Stress Frequency
The frequency of applied stress simultaneously with the ambient temperature can be another compelling factor in regulating the energy flux through the system. As shown in Fig. 2, at 300–350 K, the energy flux is more than 50 μV. But there are characteristic frequencies that can significantly increase the energy flux through the system. A frequency of about 0.09 THz at 310 K causes the energy flux of the system to reach about over 200 μeV, and this shows that energy transport in the protein system is highly sensitive to several frequencies that can be used to optimize the performance of the biological system.
3.3 Multifractal Analysis
We have studied the multifractal spectrum of the energy flux through the protein system by using Renyi dimension. In this regard, we consider the d dimensional phase space of system and divide it into cubes of size land calculate the Renyi dimension (Dq) as follows [12]:
Figure 3 shows the Renyi dimension spectrum for four different frequencies of applied stress.\(D_{q}\) shows the descending behavior by increasing q which indicates that the system is multifractal. If we go back to the previous results, we observe that the temperature of 300 K for a force amplitude of 3 pN, the frequencies ω = 0.01 THz, ω = 0.03 THz, ω = 0.05 THz are in the blue region where the energy flux crossing the system is minimal (Fig. 2). Also, the frequency ω = 0.09 THz is in the red region where the energy flux passing through the system is maximum. As shown in Fig. 3, for negative q, the Renyi dimensions at 300 K and the force amplitude of 3 pN corresponds to the frequencies ω = 0.01THz, ω = 0.03THz, ω = 0.05THz are coincident with almost the same amount. But the Renyi dimension of the frequency ω = 0.09THz shows a greater value for negative values of q. The Renyi dimension distinguishes the regions with the highest energy flux from the regions with the lowest energy flux.
To have a similarity between a multifractal system and a thermodynamic state, relations are usually equated with their thermodynamic equivalents. We consider τ(q) as analogous the thermodynamic free energy of the system, and check the multifractality of the system. The analogous of the free energy is written as follows [13]:
The system is a homogenous fractal when τ(q) have a linear dependence to q. On the other hand, τ(q) shows a deviation from the linear state when the system is multifractal and the higher deviation from the linear state shows the more multifractality of the system. According the Fig. 4, τ(q) is deviated from the linear state in terms of q for all frequencies at the point q = 0, and this confirms the multifractality of the system. On the other hand, for negative q, τ(q) is the same for the frequencies in the blue region. But the frequency ω = 0.09 THz, which is in the red area, has the highest deviation from linear behavior. Thus, the analogous of the thermodynamic free energy shows the distinct regions for negative q at 300 K and for the force range of 3 pN.
On the other hand, the analogous of the specific heat C(q) which is obtained from a second-order derivative of the analogous of free energy concerning q can be analyze the multifractal behavior of system through the following equation:
Analogous of specific heat is one of the measures to check the multifractality of a system. As shown in Fig. 5, the frequencies of the blue region overlap in the peak area. The frequency ω = 0.09 THz, which is in the red region, shows a higher peak. Therefore, a analogous of thermodynamic specific heat also shows the distinct regions and confirms the previous results.
4 Conclusions
We have studied the bioenergy transfer in a protein system to design a nanomotor. In this regard, we have investigated the effect of various parameters such as temperature and mechanical stress on the energy transfer in protein nanomotors. We have analyzed the multifractal nature of the system using the multifractal analysis methods. Using the multifractal analysis, we are able to confirm the previous results and show the distinct areas.
References
I. Kulinets, Biomaterials and their applications in medicine, in Regulatory Affairs for Biomaterials and Medical Devices (2015) (Woodhead Publishing), pp. 1–10
D.S. Miller, P.R. Payne, A theory of protein metabolism. J. Theor. Biol. 5(3), 398–411 (1963)
F. Zha, T. Wang, M. Luo, J. Guan, Tubular micro/nanomotors: Propulsion mechanisms, fabrication techniques and applications. Micromachines 9(2), 78 (2018)
P.D. Vogel, Nature’s design of nanomotors. Eur. J. Pharm. Biopharm. 60(2), 267–277 (2005)
R.D. Vale, T.S. Reese, M.P. Sheetz, Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell 42(1), 39–50 (1985)
M.E. Brown, P.C. Bridgman, Myosin function in nervous and sensory systems. J. Neurobiol. 58(1), 118–130
X.F. Pang, M.J. Liu, The influences of temperature and chain-chain interaction on features of solitons excited in Α-helix protein molecules with three channels. Int. J. Mod. Phys. B 23(10), 2303–2322 (2009)
X.F. Pang, M.J. Liu, The effects of damping and temperature of medium on the soliton excited in α-helix protein molecules with three channels. Mod. Phys. Lett. B 23(01), 71–88 (2009)
X.F. Pang, H.W. Zhang, Y.H. Luo, The influence of the heat bath and structural disorder in protein molecules on soliton transported bio-energy in an improved model. J. Phys. Condens. Matter 18(2), 613 (2005)
M. Peyrard, A.R. Bishop, Statistical mechanics of a nonlinear model for DNA denaturation. Phys. Rev. Lett. 62(23), 2755 (1989)
S. Fathizadeh, S. Behnia, Control of a DNA based piezoelectric biosensor. J. Phys. Soc. Jpn. 89(2), 024004 (2020)
S. Behnia, S. Fathizadeh, E. Javanshour, F. Nemati, Light-driven modulation of electrical current through DNA sequences: Engineering of a molecular optical switch. J. Phys. Chem. B 124(16), 3261–3270 (2020)
S. Fathizadeh, Behnia, Control of a DNA based piezoelectric biosensor. J. Phys. Soc. Jpn. 89(2), 024004 (2020)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Sefidkar, N., Fathizadeh, S., Nemati, F. (2022). Multifractal Analysis of Bioenergy Transport in a Protein Nanomotor. In: Skiadas, C.H., Dimotikalis, Y. (eds) 14th Chaotic Modeling and Simulation International Conference. CHAOS 2021. Springer Proceedings in Complexity. Springer, Cham. https://doi.org/10.1007/978-3-030-96964-6_28
Download citation
DOI: https://doi.org/10.1007/978-3-030-96964-6_28
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-96963-9
Online ISBN: 978-3-030-96964-6
eBook Packages: Physics and AstronomyPhysics and Astronomy (R0)