Abstract
Locating the hip joint center (HJC) from X-ray images is frequently required for the evaluation of hip dysplasia. Existing state-of-the-art methods focus on developing functional methods or regression equations with some radiographic landmarks. Such developments employ shallow networks or single equations to locate the HJC, and little attention has been given to deep stacked networks. In addition, existing methods ignore the connections between static and dynamic landmarks, and their prediction capacity is limited. This paper proposes an innovative hybrid framework for HJC identification. The proposed method is based on fast deep stacked network (FDSN) and dynamic registration graph with four improvements: (1) an anatomical landmark extraction module obtains comprehensive prominent bony landmarks from multipose X-ray images; (2) an attribute optimization module based on grey relational analysis (GRA) guides the network to focus on useful external anatomical landmarks; (3) a multiverse optimizer (MVO) module appended to the framework automatically and efficiently determines the optimal model parameters; and (4) the dynamic fitting and two-step registration approach are integrated into the model to further improve the accuracy of HJC localization. By integrating the above improvements in series, the models’ performances are gradually enhanced. Experimental results show that our model achieves superior results to existing HJC prediction approaches.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- Hip joint center
- Fast deep stacked network
- Dynamic registration graph
- Anatomical landmarks
- Multiverse optimizer
1 Introduction
The hip joint center (HJC) is an important reference point for decisions regarding the treatment of hip dysplasia. Unlike prominent bony landmarks, the HJC cannot be palpated, and thus, its determination must be calculated [1]. In the clinic, the HJC location is manually estimated by expert orthopedists, which is time consuming and labor intensive. It has become increasingly important to automatically and efficiently determine the HJC coordinates.
Current state-of-the-art methods for HJC determination can be categorized into three classes: Mose circle methods, functional methods and regression methods. (1) The Mose circle method is a quantitative approach used to estimate the HJC by using a template of concentric circles [2], and some modified Mose circle techniques have been proven to provide a more accurate approximation of the HJC than manual prediction [3]. However, when the ossification of the femoral skull epiphysis is insufficient, the HJC position determined by this method may be eccentric. (2) Functional methods calculate the trajectories of markers attached to the thigh and then locate the HJC by estimating the relative motion of the thigh and pelvis [4]. Many optimization approaches have been proposed to improve the accuracy and calculation time [5], such as the quadratic best sphere fitting [6], bias-compensated quartic best fitted sphere [7], and Reuleaux [8] methods. However, the estimation of the HJC relies heavily on the quality and range of movement during calibration trials. Due to soft tissue artifacts and limited circumduction movement, it is difficult to establish a reliable local coordinate system (LCS) of the femur or pelvis. A functional method would result in HJC location errors of 26 mm when performing substantially limited hip circumduction movement [4]. Functional calibration is challenging for those with impaired movement control or reduced range of motion, and recent studies suggest that it may be less accurate in clinical populations [9,10,11]. (3) Regression methods use empirical regression equations between palpable bone landmarks and the HJC as an anthropometric quantity function, and regression parameters are obtained by pelvis samples isolated in vivo and via medical imaging. However, these equations are not applicable universally since HJC-related measurements are quite variable between races or even communities [12]. The mean error is estimated in the range of 15–30 mm when using available regression equations [5, 10, 11, 13, 14]. Thus, more accurate and adaptive prediction approaches are desired.
However, within the context of HJC determination, the challenges stem from three aspects. (1) The first aspect is large-scale cartilage. Due to insufficient ossification of the femoral head, the acetabular curvature displayed on the orthographic image of the hip joint is far smaller than the actual curvature. This property brings a new challenge to general regression methods. (2) The second aspect is intraclass variations. There are obvious differences between study subjects, such as the width and height of the pelvis, the ossification degree of the acetabulum and femur, inconsistent abduction/adduction performance, and an unexpectedly tilted or rotated pelvis when examinations are performed. These differences often fail to yield effective results. (3) The third aspect is adaptive HJC prediction. Different empirical regression equations have been proposed in the past to identify the HJC, but these methods are suitable only for certain groups of people [15]. How to adaptively predict HJC using intelligent learning models remains unsolved. Note that the deep learning approaches have been widely used in the field of medical images [16,17,18], but there are few studies to explore the deep learning approaches for the HJC prediction.
To accurately identify the HJC, this paper advocates the idea that static to dynamic prominent bony landmarks contribute to object localization. This idea is motivated by the following intuitive concepts: pelvis bone landmarks remain unchanged regardless of what movement the legs perform, and bone landmarks from the hip joint are dynamic when the legs perform large abduction/adduction and flexion/extension movements. Thus, it is desirable to investigate static to dynamic landmarks to achieve better predictions.
To this end, we propose an innovative hybrid framework for HJC identification. It is equipped with a fast deep stacked network (FDSN)-based fast prediction module and a dynamic registration graph-based accurate positioning module with the following four aspects.
-
(1)
Comprehensive prominent bony landmarks are extracted. Our method is based on X-ray images, and the extraction of the most comprehensive bone anatomical landmarks is desired. The coordinate system is built on multipose X-ray images (including abduction images and adduction images), and eight anatomical landmarks on each hip are annotated by expert orthopedists.
-
(2)
We introduce a new attribute optimization module, guiding the network to focus on useful anatomical landmarks. By ranking the correlation coefficients between the prominent bony landmarks and the HJC using the grey relational analysis (GRA) method, we select the high-importance attributes to train our model since some landmarks may be damaged.
-
(3)
The proposed FDSN method considers adaptive HJC prediction to tackle the problem of automated model optimization. Specifically, a regularized extreme learning machine (RELM) is employed to train the modules, and random feature mapping is shared among the modules to speed up the FDSN training. We initialized the model with optimal parameters obtained by the multiverse optimizer (MVO) method [19]. This MVO can be easily added to model architectures, increasing prediction accuracy.
-
(4)
The proposed dynamic registration graph-based positioning method is based on multipose X-ray images. A two-step registration approach is used to align two pelvis images, and we fit the circle center in a dynamic abduction/adduction manner for accurate HJC positioning. The proposed innovative hybrid framework features better accuracy and reliability than previously developed approaches.
2 The Proposed Method
2.1 Method Architecture
Our goal is to automatically and efficiently determine the position coordinates of the HJC. This goal requires that both static and dynamic anatomical landmarks be investigated for the best prediction of the HJC. To this end, we propose an innovative hybrid prediction framework, as shown in Fig. 1. It involves one landmark extraction module for the comprehensive prominent bony landmarks, one GRA optimization module for the importance measure of landmarks, one FDSN module that achieves fast HJC prediction in static cases and one dynamic registration strategy to fit the HJC in dynamic cases. Next, we will detail these modules.
2.2 Landmark Extraction
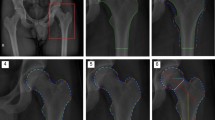
Comprehensive prominent bony landmarks should be extracted from multipose X-ray images of hip joints. Considering that there were obvious differences between the study subjects, such as the width and height of the pelvis, the ossification degree of the acetabulum and femur, and inconsistent abduction/adduction performance, we built a coordinate system on each image according to the posterior inferior iliac spine on both sides, as shown in Fig. 2(a). Specifically, the subjects were asked to perform dynamic abduction/adduction movements. Sixteen prominent bony landmarks were annotated by orthopedists, as displayed in Fig. 2(b) and Table 1, and the HJC was determined by Mose circles with a template of concentric circles, as displayed in Fig. 2(c) and (d).
2.3 Grey Relational Analysis
GRA is a quantitative analysis and development trend comparison method, and it is used to analyze the relevance between input attributes and output predictions. The basic idea of the GRA method is to judge the closeness degree based on the geometric similarity between the reference sequence and the multiple comparison sequences [20]. In our study, the input attributes are the coordinates of prominent landmarks (also called the comparison sequence), and the output predictions are the coordinates of the HJC determined by Mose circles (also called the reference sequence). Note that our model takes the most prominent bone anatomical landmarks as the predictors, and the GRA optimization module can exclude some incorrect landmarks, such as damaged or broken landmarks.
The core ideas of GRA are as follows: the reference sequence is described as \({{\varvec{x}}}_{0}=\left\{{x}_{0}\left(1\right),{x}_{0}\left(2\right),\dots {x}_{0}(n)\right\}\), and the comparison sequence is represented by \({{\varvec{x}}}_{i}=\left\{{x}_{i}\left(1\right), {x}_{i}\left(2\right),\dots {x}_{i}(n)\right\}\), where n represents the number of the samples and i represents the i-th input attribute. We normalize the original sequence and can obtain the initialized reference sequence and the comparison sequences, \({{\varvec{y}}}_{0}=\left\{{y}_{0}\left(1\right),{y}_{0}\left(2\right), \dots y(n)\right\}\) and \({{\varvec{y}}}_{i}=\left\{{y}_{i}\left(1\right),{y}_{i}\left(2\right),\dots {y}_{i}(n)\right\}\), respectively. After the nondimensional transformation, the correlation coefficient between y0(t) and yi(t) at time k can be calculated:
where Δi\(\left(min\right)={min}_{k}(\left|{y}_{0}\left(k\right)-{y}_{i}(k)\right|)\), Δi\(\left(max\right)={max}_{k}(\left|{y}_{0}\left(k\right)-{y}_{i}(k)\right|)\), and β represent the distinguishing coefficients. The grey correlation grade γi between the reference sequence and the multiple comparison sequences is the average value of all the correlation coefficients.
If the coordinates of a prominent landmark obtain the highest grey correlation grade with the coordinates of the HJC, this landmark is most relevant to the HJC. Therefore, we introduce a new attribute selection module, guiding the network to focus on meaningful landmarks at input variables. The correlation coefficient is estimated to determine whether prominent landmarks should be added to train the models. By ranking the correlation coefficients between the prominent landmark and the HJC using the GRA method, we select the high-importance landmarks to train the model.
2.4 Fast HJC Prediction Module
The proposed FDSN method is shown in Fig. 3, which takes the prominent landmark coordinates as model inputs and achieves fast HJC prediction. Specifically, a regularized extreme learning machine (RELM) is employed to train the modules of the FDSN. This idea is motivated by the following intuitive concepts: (1) As an efficient approach to train neural networks, ELMs have low computational cost and good generalization performance [21]. (2) ELM-based random feature mapping is shared among the modules in the FDSN to speed up the training process. In the FDSN, the first module is trained using the RELM-based approach with additive nodes: random weights W1 and biases v1. Matrix H1 is calculated as shown in Eq. (2).
where G is an operator, X1 is the input, and N1 is the v1 vector in each row. As the weight matrix connecting the output and hidden layers, B1 is calculated using H1 and the chosen RELM-based algorithm. The first module output is described by \(\overline{{\varvec{T}}}_{{1}} = {\varvec{H}}_{{1}} {\varvec{B}}_{{1}}\).
Note that the modules of the FDSN are stacked on top of each other, and the input and the output of a module make up the input of the following module [22]. The FDSN can suffer from a badly chosen initial weight since some weights and calculations are reused when accelerating the training of the DSN and reducing memory usage. The FDSN algorithm has three extremely important parameters (including the number of hidden neurons (N), regularization parameter (r), and maximal number of modules (s)), and the manual tuning of these parameters can be a time-consuming and tedious process. To automatically and efficiently determine the optimal FDSN model parameters, we added an MVO global optimization module to the framework. The main inspirations of this MVO algorithm are based on three concepts in cosmology: white holes, black holes, and wormholes. A universe with a high expansion rate is considered to have white holes, and a universe with a low expansion rate is considered to have black holes. Each universe has wormholes to transport its objects through space randomly, and the objects are transferred from the black hole in the source universe to the black hole in the target universe. Mathematical models of these three concepts are developed to perform exploration, exploitation, and a local search.
In the MVO algorithm, a set of random universes is created to start the optimization process. During each iteration, objects in the universes with high inflation rates tend to move to the universes with low inflation rates via white/black holes. Note that every single universe performs random teleportation in its objects through wormholes towards the best universe [19]. The computational complexity of the MVO algorithms mainly depends on four factors (including the number of iterations, number of universes, roulette wheel mechanism, and universe sorting mechanism). Note that universes are sorted in every iteration, and the Quicksort algorithm is employed in this study, which has the complexity of O(nlog n) and O(n2) in the best and worst cases, respectively. We conduct roulette wheel selection in every universe over the iterations and can obtain the complexity O(n) or O(logn) based on implementation. Finally, the overall computational complexity can be obtained:
where n represents the number of universes, l represents the maximum number of iterations, and d represents the number of objects. This MVO module can be easily added to model architectures for the optimal model parameters and increasing prediction accuracy.
2.5 Accurate HJC Positioning Module
Calculating the trajectories of annotated bone landmarks under dynamic behaviors is desired. However, this calculation also introduces two issues. One issue is that bone landmarks are manually annotated by orthopedists. The manual tuning of landmark coordinates can be a time-consuming and tedious process. Moreover, it is difficult to obtain optimal fixed points for accurate registration. The other issue is that two pairs of dynamic points (dynamic landmarks in hip joints in abduction images and adduction images, as shown in Table 1) are too weak to identify the HJC, leading to an amplified distance error after fitting circles. Thus, it is necessary to further tune the control point locations (static landmarks in the pelvis in Table 1) and select enough dynamic landmarks (dynamic landmarks in hip joints in Table 1) as informative spatial locations for HJC localization.
To align two pelvis images (abduction images and adduction images), a two-step registration approach is used in this study. First, we tune the control point locations (static landmarks in the pelvis in Table 1) using the cross-correlation method, and the adjusted coordinates are accurate up to one-tenth of a pixel, which more closely matches the positions of the fixed landmarks than previous methods can achieve [23]. Note that the cross-correlation method is used to obtain subpixel accuracy from the image content and coarse control point selection. Second, we fit a geometric transformation to the adjusted control point pairs (adjusted static landmarks in the pelvis in abduction images and adduction images). Finally, every two pairs of dynamic landmarks (landmarks 5, 6 and 8 in abduction images and adduction images) are fitted to a circle center, and the average value of all the centers is taken as the final HJC, as shown in Fig. 4.
3 Experimental Results
3.1 Data Processing and Analysis
To show the effectiveness of our proposed HJC prediction method, we validated it on a dataset from the Wuhan Women and Children’s Health Care Center in China. In 100 subjects’ X-ray images and a total of 200 hips, a coordinate system was established on each X-ray image according to the posterior inferior iliac spine on both sides. Sixteen prominent bony landmarks were annotated by orthopedists. Taking the left hip joint as an example, the correlation between the coordinates of each landmark and the HJC was analyzed using the GRA method, as shown in Fig. 5. A relational grade value closer to 1 indicates a better corresponding landmark. Those landmarks with strong correlation (comprehensive relational grade value > 0.7) were retained as the input variables of the model.
3.2 Experimental Settings
Our static prediction module is based on the implementation of the FDSN. By considering the number of landmarks and the HJC, the end-to-end parameters of our model include the number of input neurons (16) and the number of output neurons (2). Note that 70% of the samples are used to train the model, and the remaining 30% of the samples are used for testing. Our optimization objective function is the average root mean square error (RMSE) from ten 10-fold validation processes performed on the training data using the FDSN method. Figure 6 shows the convergence curve of the global optimization obtained with the MVO. The optimal parameters obtained by the MVO include the number of hidden neurons (443), regularization parameter (1) and maximum number of modules (10). Our dynamic positioning module takes the average value of three fitted circle centers as the final HJC. All experiments are performed on the same computer with an Intel Core i7-8700 3.20 GHz CPU and 16 GB memory.
3.3 Comparison with State-of-the-Art Methods
The experimental results of our proposed FDSN method are shown in Fig. 7. We compared the proposed FDSN model with the Elman [24], radial basis function (RBF) [25], ELM [21], RELM [26], stacked ELM (SELM) [27], SELM-based autoencoder (AE-SELM) [27], and DSNELM [28] models. All experiments are performed with optimal parameters obtained by the MVO algorithm. We used the RMSE, mean absolute percentage error (MAPE), and R-squared (R2) value as evaluation metrics. We used the mean and standard deviation (SD) to analyze the results. We ran the experiment 1000 times (independently), and the average results for the testing sets are shown in Table 2. Obviously, (1) our FDSN method outperformed the seven state-of-the-art methods. Our method achieved the smallest RMSE, the smallest MAPE, and the largest R2 value. In addition, our algorithm achieved the smallest standard deviation among the three indicators. These results imply the effectiveness and robustness of our algorithm. (2) The ELM model achieved inferior performances. The reason for this is that the ELM is a single-layer feedforward network (SLFN) and is prone to underfitting or overfitting.
In addition, we compared the proposed dynamic registration approach with traditional functional and regression methods. The proposed dynamic registration graph method does not participate in the training process and achieves the following results: RMSE = 0.4139 mm, MAPE = 1.4660, and R2 = 0.9962. The traditional functional method results in unsatisfactory location errors of up to 26 mm [4], the mean error of the traditional regression method is in the range of 15–30 mm [5, 10, 11, 13, 14], and the modified Ranawat method and pelvic height ratio method have estimation errors <5 mm [29]. We can clearly observe that the proposed static FDSN model and dynamic registration approach have better accuracy than traditional functional methods and traditional regression methods; in particular, the proposed dynamic registration graph features superior accuracy (RMSE = 0.4139 mm, MAPE = 1.4660, and R2 = 0.9962). These findings show that our framework obtains coordinates much closer to the real HJC than the previously developed approaches.
3.4 Ablation Analysis
To better demonstrate the effectiveness of the proposed FDSN method, we performed ablation experiments on our model, as shown in Table 3. The results indicate that (1) the model performance is evidently improved by combining the static landmarks in the pelvis and dynamic landmarks in the hip joint. Clearly, both the static landmarks and dynamic landmarks can help identify the HJC, and our fusion strategy can provide richer feature representations than single static/dynamic landmarks. (2) Applying an MVO global optimization module further optimizes the prediction of the proposed FDSN method (complexity analysis: 9.2598e−04 s with the MVO module and 2.8210e−04 s without the MVO module). Obviously, our proposed FDSN method greatly enhances the effectiveness and robustness in HJC identification.
4 Conclusion
This paper proposes a novel framework to address the problem of HJC determination. First, comprehensive prominent landmarks from multipose X-ray images are extracted, and grey relational analysis (GRA) is used to guide the network to focus on useful variables. Then, a fast deep stacked network (FDSN) with a multiverse optimizer (MVO) is designed to achieve fast HJC prediction, and it considers adaptive HJC prediction to tackle the problem of automated model optimization. Finally, using a dynamic registration graph, the HJC localization ability of the whole framework is enhanced. Validation on a dataset from the China Wuhan Women and Children’s Health Care Center demonstrates the effectiveness of our hip joint center (HJC) determination method on X-ray images.
References
Harrington, M.E., Zavatsky, A.B., Lawson, S.E.M., Yuan, Z., Theologis, T.N.: Prediction of the hip joint centre in adults, children, and patients with cerebral palsy based on magnetic resonance imaging. J. Biomech. 40(3), 595–602 (2007)
Mose, K.: Methods of measuring in Legg-Calvé-Perthes disease with special regard to the prognosis. Clin. Orthop. Relat. Res. 150, 103–109 (1980)
Cuomo, A.V., Moseley, C.F., Fedorak, G.T.: A practical approach to determining the center of the femoral head in subluxated and dislocated hips. J. Pediatr. Orthop. 35(6), 556–560 (2015)
Piazza, S.J., Erdemir, A., Okita, N., Cavanagh, P.R.: Assessment of the functional method of hip joint center location subject to reduced range of hip motion. J. Biomech. 37(3), 349–356 (2004)
Camomilla, V., Cereatti, A., Vannozzi, G., Cappozzo, A.: An optimized protocol for hip joint centre determination using the functional method. J. Biomech. 39(6), 1096–1106 (2006)
Silaghi, M.-C., Plänkers, R., Boulic, R., Fua, P., Thalmann, D.: Local and global skeleton fitting techniques for optical motion capture. In: Magnenat-Thalmann, N., Thalmann, D. (eds.) CAPTECH 1998. LNCS (LNAI), vol. 1537, pp. 26–40. Springer, Heidelberg (1998). https://doi.org/10.1007/3-540-49384-0_3
Gamage, S.S.H.U., Lasenby, J.: New least squares solutions for estimating the average centre of rotation and the axis of rotation. J. Biomech. 35(1), 87–93 (2002)
Halvorsen, K., Lesser, M., Lundberg, A.: A new method for estimating the axis of rotation and the center of rotation. J. Biomech. 32(11), 1221–1227 (1999)
Assi, A., et al.: Validation of hip joint center localization methods during gait analysis using 3D EOS imaging in typically developing and cerebral palsy children. Gait Posture 48, 30–35 (2016)
Sangeux, M., Pillet, H., Skalli, W.: Which method of hip joint centre localisation should be used in gait analysis? Gait Posture 40(1), 20–25 (2014)
Peters, A., Baker, R., Morris, M.E., Sangeux, M.: A comparison of hip joint centre localisation techniques with 3-DUS for clinical gait analysis in children with cerebral palsy. Gait Posture 36(2), 282–286 (2012)
Miller, E.J., Kaufman, K.R.: Verification of an improved hip joint center prediction method. Gait Posture 59, 174–176 (2018)
Sangeux, M.: On the implementation of predictive methods to locate the hip joint centres. Gait Posture 42(3), 402–405 (2015)
Sangeux, M., Peters, A., Baker, R.: Hip joint centre localization: evaluation on normal subjects in the context of gait analysis. Gait Posture 34(3), 324–328 (2011)
Bombaci, H., Simsek, B., Soyarslan, M., Murat Yildirim, M.: Determination of the hip rotation centre from landmarks in pelvic radiograph. Acta Orthop. Traumatol. Turc. 51(6), 470–473 (2017)
Wang, X., et al.: Obstructive sleep apnea detection using ecg-sensor with convolutional neural networks. Multimedia Tools Appl. 79(23), 15813–15827 (2020)
Shi, W., Liu, S., Jiang, F., Zhao, D., Tian, Z.: Anchored neighborhood deep network for single-image super-resolution. EURASIP J. Image Video Process. 2018(1), 34 (2018)
Jiang, F., et al.: Medical image semantic segmentation based on deep learning. Neural Comput. Appl. 29(5), 1257–1265 (2018)
Mirjalili, S., Mirjalili, S.M., Hatamlou, A.: Multi-verse optimizer: a nature-inspired algorithm for global optimization. Neural Comput. Appl. 27(2), 495–513 (2015). https://doi.org/10.1007/s00521-015-1870-7
Sun, G., Xin, G., Xiao, Y., Zheng, Z.: Grey relational analysis between hesitant fuzzy sets with applications to pattern recognition. Exp. Syst. Appl. 92(9), 521–532 (2018)
Huang, G., Zhou, H., Ding, X., Zhang, R.: Extreme learning machine for regression and multiclass classification. IEEE Trans. Syst. Man Cybernet. Part B (Cybernet.) 42(2), 513–529 (2012)
da Silva, B.L.S., Inaba, F.K., Salles, E.O.T., Ciarelli, P.M.: Fast deep stacked networks based on extreme learning machine applied to regression problems. Neural Netw. 131, 14–28 (2020)
Goshtasby, A.: Image registration by local approximation methods. Image Vis. Comput. 6(4), 255–261 (1988)
Yuan-Chu, C., Wei-Min, Q., Wei-You, C.: Dynamic properties of Elman and modified Elman neural network. In: Proceedings of the International Conference on Machine Learning and Cybernetics, pp. 637–640 (2002)
Park, J., Sandberg, I.W.: Universal approximation using radial-basis-function networks. Neural Comput. 3(2), 246–257 (1991)
Deng, W., Zheng, Q., Chen, L.: Regularized extreme learning machine. In: 2009 IEEE Symposium on Computational Intelligence and Data Mining, pp. 389–395 (2009)
Zhou, H., Huang, G., Lin, Z., Wang, H., Soh, Y.C.: Stacked extreme learning machines. IEEE Trans. Cybernet. 45(9), 2013–2025 (2015)
Li, D.: A tutorial survey of architectures, algorithms, and applications for deep learning. Apsipa Trans. Signal Inf. Process. 3, e2 (2014)
Fujii, M., Nakamura, T., Hara, T., Nakashima, Y.: Is Ranawat triangle method accurate in estimating hip joint center in Japanese population? J. Orthop. Sci. 26(2), 219–224 (2021)
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No.61772556), National Key R&D Program of China (No.2018YFB1107100, No.2016 YFC1100600), Postgraduate Research and Innovation Project of Hunan (No.CX20200321) and Fundamental Research Funds for the Central Universities of Central South University (2020zzts140).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Han, F., Liao, S., Wu, R., Liu, S., Zhao, Y., Shen, X. (2021). Radiological Identification of Hip Joint Centers from X-ray Images Using Fast Deep Stacked Network and Dynamic Registration Graph. In: Farkaš, I., Masulli, P., Otte, S., Wermter, S. (eds) Artificial Neural Networks and Machine Learning – ICANN 2021. ICANN 2021. Lecture Notes in Computer Science(), vol 12893. Springer, Cham. https://doi.org/10.1007/978-3-030-86365-4_52
Download citation
DOI: https://doi.org/10.1007/978-3-030-86365-4_52
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-86364-7
Online ISBN: 978-3-030-86365-4
eBook Packages: Computer ScienceComputer Science (R0)