Abstract
Total hip arthroplasty (THA) is a valid and reliable treatment for degenerative hip disease, and an elaborate preoperative planning is vital for such surgery. The key step of planning is to localize several anatomical landmarks in X-ray images for preoperative measurements. Conventionally, this work is almost conducted by surgeons manually that is labor-intensive and time-consuming. In this paper, we propose an automatic measurement method by detecting anatomical landmarks with the latest deep learning approaches. However, locating these landmarks automatically with high precision in X-ray images is challenging since image features of a certain landmark are subject to the variations of imaging postures and hip appearances. To this end, we impose the relative position constraints on each landmark by defining edges among landmarks according to the clinical significance. With multi-task learning, our method predicts the landmarks and edges simultaneously. Thus the correlations among these landmarks are exploited to correct the detection deviations implicitly in the network training. Extensive experiment results on two datasets have indicated the superiority of the anatomical constrained method and its potential for clinical applications.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Total hip arthroplasty is regarded as one of the most successful orthopedic operations, which improves quality of life in patients with hip pains. It is reported that more than 400,000 cases are implemented in China in 2018 [1] and there will be more cases in the future since the increasing number of aging population. An accurate and elaborate preoperative planning is vital for a successful surgery. Conventionally, surgeons need to take measurements such as neck-shaft angle, center-edge (CE) angle and femoral shaft axis on the anteroposterior and lateral X-ray images, which is time-consuming, labor-intensive and lacks of accuracy. A key step of these measurements is to localize the anatomical points on the femoral and the acetabulum, which could be achieved by identifying a set of landmarks on X-ray images. In this paper, we propose an automatic measurement method for THA preoperative planning by detecting accurate anatomical landmarks. Specifically, we train a deep convolutional neural network (CNN) to perform landmarks detection. By imposing anatomical constraints on the landmarks correlation, we achieve competitive results on both an in-house dataset and a public dataset as shown in experiments.
Anatomical landmarks detection in medical images is a prerequisite for many clinical applications, such as image registration, segmentation and computer-aided diagnosis (CAD). Due to the variations among patients and differences in image acquisition, it is difficult to detect the anatomical landmarks precisely. Traditional machine learning method mainly focused on predefined statistics priors, like classification of pixels [11, 12] and regression with forest [5, 6]. Recently, deep learning methods have demonstrated superior performances in medical image analysis to traditional methods. As for landmark detection, Zhang et al. [22] exploited two deep CNN to detect multiple landmarks on brain MR images, one learning associations between local image patches and target anatomical landmarks, and another predicting the coordinates of these landmarks. Yang et al. [21] utilized CNN to detect the geometric landmarks, and then segment femur surface with graph-cut optimization based on the identified landmarks. Bier et al. [2] detected the anatomical landmarks in pelvis X-ray images by a two-stage sequential CNN framework which is able to work in arbitrary viewing direction. Li et al. [8] presented a CNN model to learn spatial relationship between image patches and anatomical landmarks, and refining the positions by iterative optimization, which is effective for 3D ultrasound volumes. Noothout et al. [14] proposed a fully convolutional neural network (FCN) combined with regression and classification, in which they regressed the displacement vector from the identified image patch to the target landmark rather than its coordinates in cardiac CTA scans. Xu et al. [20] investigated the multi-task learning (MTL) method to perform view classification and landmark detection simultaneously by a single network on abdominal ultrasound images, and found that multi-task training outperforms the approaches that address each task individually. However, these methods hypothesized that the landmarks are independent, thus only using local information, without considering the relations between each other. To address such issue, Payer et al. [15] implicitly modeled the relationships by a spatial configurations block. Motivated by the intuition that anatomical landmarks generally lie on the boundaries of interested regions, Tuysuzoglu et al. [18] brought global context to landmarks detection by predicting their contours, and Zhang et al. [23] associated this task with bone segmentation. They have demonstrated the effectiveness of this idea, but the annotations of contours and segmentations are laborious especially for large datasets in practice.
Illustration of the defined 22 landmarks and 24 clinical relevant edges on a anteroposterior hip X-ray image. The notations of each landmarks and edges are listed on the right. 10 edges on each side are illustrated as yellow lines, and 4 interaction edges across two sides are connected by green lines. (Color figure online)
Different from previous method, we propose an accurate anatomical landmarks detection model for THA preoperative planning in this paper, which adds an extra relation loss to model the relationships among the anatomical landmarks. The contributions of this work are summarized as follows. First, an automatic and accurate framework is proposed for anatomical landmark detection with emphasis on THA preoperative planning. Second, the connections among landmarks are modeled to learn their relationships thus improving the detection precision. Third, extensive experiments on two datasets demonstrate that with our relation loss several state-of-the-art landmarks detection networks can be easily improved.
2 Method
To achieve the automatic measurement for THA preoperative planning, it is crucial to detect the anatomical landmark precisely. Rather than the conventional methods which only regress the coordinates or heatmaps of the landmarks, we further exploit the correlations of landmarks. As shown in Fig. 1, 11 landmarks (denoted as set V) on both left and right hip are selected on a anteroposterior X-ray image with the advice of orthopedics experts. Besides, the connection edges (denoted as set E) among these landmarks according to the clinical significance are introduced to model their relationships. Specifically, there are 10 edges on each side denoted as yellow lines, and 4 interaction edges across two sides are connected by green lines, totally 24 edges. These clinically relevant landmarks and edges are significant to the later measurements. Take the right hip as an example, the angle between the longitudinal axis of the femoral shaft (landmark 6 to landmark 9) and the central axis of the femoral neck (landmark 10 to landmark 11) is the neck-shaft angle, which is used to evaluate the force transmission of femur and normally lies in 125° to 140° [9]. The parallel lines of teardrops (landmark 1 to landmark 12) and lesser trochanter (landmark 4 to landmark 15) are utilized to assess the difference of two legs’ length [9]. Taking such anatomical constraint into consideration, i.e., the edges, the deviation of a landmark could be corrected by others, thus improving the location accuracy.
In order to predict the landmarks as well as the edges simultaneously, we adopt the MTL strategy. The framework is shown in Fig. 2, which outputs two branches: (1) landmarks detection branch, and (2) edges prediction branch. A 2D X-ray image with size of \(512\times 512\) is firstly input into the backbone network to extract high-level features for the following two branches. For landmarks detection, each landmark is converted into a heatmap with a 2D Gaussian distribution centered at its coordinates. The distribution is normalized to the range of 0 to 1 and the Standard Deviation (Std) \(\sigma \) decides the shape of distribution. Several convolutional layers are utilized to regress these heatmaps from the high-level features, and output 22 channel feature maps with size of \(128\times 128\) where each channel represents a heatmap of a corresponding landmark. For edges prediction, each edge connecting two landmarks is denoted as a vector, as follows
where \(v_i\) and \(v_j\) are the connected landmarks, \(x_{v_i}\) and \(y_{v_i}\) is the coordinates of \(v_i\) on x and y directions respectively. The high-level features are firstly fed into convolutional layers to reduce dimensions and extract more relevant features, and then a global average pooling layer is followed to predict the vectors. In addition, the edge vector is normalized during experiment for faster convergence.
The loss of landmarks detection denoted as \(L_{landmark}\) is formulated by Mean Square Error (MSE) that is consistent with previous literature [13, 17], while the edges prediction error \(L_{edge}\) is evaluated by smooth L1 loss which is inspired by object detection tasks. And the final objective function L is combined with \(L_{landmark}\) and \(L_{edge}\):
where \(\lambda \) is a balance factor. Added with the edges prediction loss that models the landmarks relations, the proposed method could correct the detection deviations implicitly during training.
3 Experiment and Results
Data. The proposed method is evaluated on an in-house dataset of 2D anteroposterior X-ray images. A total of 707 cases are collected from a hospital with an average size of \(2021\times 2021\) and a pixel spacing of 0.2 mm. All data are from different patients and with ethics approval. Each image is annotated by a clinical expert with more than 5 years of experience, and reviewed by a senior specialist. We adopt the simple cross-validation strategy in experiment and 80%, 10%, 20% of the data are randomly selected for training, validation, and testing. During preprocessing, all images are resized to \(512\times 512\) pixels with isotropic spacing of 0.79 mm. Besides, a supplementary experiment was conducted on a public dataset from the Grand Challenge [19] to further verify the effectiveness of our idea. The dataset contains 400 dental X-ray cephalometric images with 19 landmarks annotations from 2 experienced doctors, and the mean position of the two annotations is adopted as the ground truth. The resolution of an image is 1935 by 2400 pixels, and each pixel is about 0.1 mm. The data is divided into 3 subsets, 150 images for training data, 150 images for Test 1 data and 100 images for Test 2 data. As shown in Fig. 3, we also define 18 edges among the 19 landmarks according to the 8 measurement methods for detecting anomalies in the challenge. Likewise, all the images are firstly cropped to squares (\(1935\times 1935\) pixels) and then resampled into the size of \(512\times 512\) with isotropic resolution of 0.38 mm during training.
Backbone Networks. The backbone network is utilized to extract high-level features from images for the following two branches of specific tasks. To eliminate the influence from backbone, we test 3 prevalent backbone networks in our experiment. The first is U-Net [16], a symmetrical downsample-upsample FCN framework with features concatenation, which is firstly applied to cell segmentation. The second is Hourglass [13], named for its hourglass like shape, in which features are processed across all scales to capture the various spatial relationships of human pose. Specifically, we used the stacked Hourglass network consisting of two Hourglass blocks. And the last is newly HRNet [17], which connects high-to-low resolution sub-networks in parallel and outputs reliable high-resolution representations with repeated multi-scale fusions. The small network HRNet-W32 from [17] is evaluated in our experiments.
Experiment Setup. The proposed model is implemented using Pytorch framework and running on a machine with 4 Nvidia P100 GPUs. Due to the limited data, the parameters of the network are initialized with the pre-trained model from the large public dataset ImageNet [4]. In addition, training datasets are augmented by flipping (with possibility of 0.5), random rotation (up to 45°) and random scaling (up to 50% difference). The network is optimized by Adam [7] with an initial learning rate (LR) of 1e-3, and is decreased to 1e-4 and 1e-5 at the 120th and 170th epochs. Optimization is carried out for 200 epochs with a batch size N of 32. The hyperparameter \(\sigma \) of Gaussian heatmaps is chosen to 2, and the balance factor \(\lambda \) is set to 1e-3 empirically.
Results. We compared landmarks detection performance of three backbone networks with the proposed edge constraints against their original ones without edges prediction branch on testing dataset. The quantitative comparison is measured by mean radial error (MRE), defined as
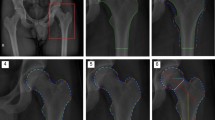
where n denotes the number of detected landmarks and \(R_{i}\) is the Euclidean distance between the predicted landmarks coordinates obtained by extracting the maxima on heatmaps and the ground-truth. The comparison results are reported in Table 1, and four randomly selected examples of predictions are given by each method as illustrated in Fig. 4. In Table 1, MREs of each individual landmark and the mean results are listed. It is obvious that the three backbone networks with the proposed edges prediction branch outperform their original ones, which validates our hypothesis that the relationships among anatomical landmarks are helpful for improving the locating precision. With the constraints of the landmarks connections, the mean MRE improvement percentiles of each backbone are: 2.3% (U-Net), 11.7% (Hourglass) and 5.1% (HRNet), which overpass the original state-of-the-art methods by a considerable margin. Among the three backbone networks, the HRNet model and Hourglass model work better than U-Net model since they have more sophisticated network structures. Furthermore, by using the intermediate supervision, Hourglass model with edge predictions get the best performance with mean MRE of 2.10 mm, which meets the clinical acceptable error 3 mm suggested by our orthopedics experts.
Qualitative results of four randomly selected examples by each method: U-Net (top), Hourglass (middle) and HRNet (bottom). The predictions of original methods are shown in yellow circles, while the results of the methods with edge branches are shown in green circles. And the dots in red are ground truth.
Having the accurate method for anatomical landmarks detection, we develop a simple demo for automatic THA preoperative measurement. Given an anteroposterior X-ray image, the demo can output the angles and lengths in 0.3 seconds, which is much faster than specialists. As shown in Fig. 5, the neck-shaft angle, the lateral CE angle and abduction angle are 131.76°, 36.83° and 55.93.93 on the right side, and 127.79°, 39.64° and 44.64.64 on the left side, respectively. The femoral offsets (FO) on each side are 39.94 and 42.80 mm, and the leg length difference is 9.12 mm.
To verify our idea, we further applied our method to the supplementary cephalometric dateset as described before. The results are show in Table 2 where successful detection rate (SDR) are adopted as the evaluation metric. The SDR shows the percentage of landmarks successfully localized within the radius of 2.0 mm, 2.5 mm, 3.0 mm, 4 mm. We use the Hourglass as the backbone network in this testing, and maximum iterations is reduced to 100 epochs to avoid overfitting due to the limited training data. Besides, three method are used for comparison. The first method from Lindner et al. [10] won the first place of the challenge, which explored the Random Forest regression-voting to detect cephalometric landmarks, and the other two methods are based on deep learning approach. The former [24] regressed the heatmaps of landmarks from coarse to fine by a two-stages U-Net embeded with attention mechanism, and applied expansive exploration strategy to improve robustness during inferring. The later [3] achieved higher accuracy than existing deep learning-based methods with the help of a attentive feature pyramid fusion module. In terms of results, our method has achieved comparable performance with the others, especially in the case of higher location requirement, i.e., 2 mm and 2.5 mm. Meanwhile, it also indicates that our idea could generalized to wider clinical applications related to anatomical landmarks detection.
4 Discussion
In this paper, we propose an accurate anatomical landmarks detection method for THA preoperative planning, in which the connections among the landmarks are defined as the edges according to the clinical significance to model their relationships. By imposing such anatomical constraints, the deviation position of a landmark could be corrected by others implicitly, thus improving the detection accuracy. The extensive experiment results on two datasets have shown the superiority of the proposed method as well as the potential for clinical applications. In the future, we will extend this idea to more general landmark detection tasks.
References
How many total hip arthroplasty surgeries have been performed in china in 2018? https://www.sohu.com/a/299015396_100281680
Bier, B., et al.: X-ray-transform invariant anatomical landmark detection for pelvic trauma surgery. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11073, pp. 55–63. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00937-3_7
Chen, R., Ma, Y., Chen, N., Lee, D., Wang, W.: Cephalometric landmark detection by attentive feature pyramid fusion and regression-voting. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11766, pp. 873–881. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32248-9_97
Deng, J., Dong, W., Socher, R., Li, L.J., Li, K., Fei-Fei, L.: Imagenet: A large-scale hierarchical image database. In: CVPR 2009, pp. 248–255. IEEE (2009)
Gao, Y., Shen, D.: Collaborative regression-based anatomical landmark detection. Phys. Med. Biol. 60(24), 9377 (2015)
Han, D., Gao, Y., Wu, G., Yap, P.-T., Shen, D.: Robust anatomical landmark detection for MR brain image registration. In: Golland, P., Hata, N., Barillot, C., Hornegger, J., Howe, R. (eds.) MICCAI 2014. LNCS, vol. 8673, pp. 186–193. Springer, Cham (2014). https://doi.org/10.1007/978-3-319-10404-1_24
Kingma, D.P., Ba, J.: Adam: A method for stochastic optimization (2014). arXiv preprint arXiv:1412.6980
Li, Y., et al.: Fast multiple landmark localisation using a patch-based iterative network. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11070, pp. 563–571. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00928-1_64
Lim, S.J., Park, Y.S.: Plain radiography of the hip: a review of radiographic techniques and image features. Hip Pelvis 27(3), 125–134 (2015)
Lindner, C., Cootes, T.F.: Fully automatic cephalometric evaluation using random forest regression-voting. In: ISBI 2015. Citeseer (2015)
Lu, X., Jolly, M.-P.: Discriminative context modeling using auxiliary markers for LV landmark detection from a single MR image. In: Camara, O., Mansi, T., Pop, M., Rhode, K., Sermesant, M., Young, A. (eds.) STACOM 2012. LNCS, vol. 7746, pp. 105–114. Springer, Heidelberg (2013). https://doi.org/10.1007/978-3-642-36961-2_13
Mahapatra, D.: Landmark detection in cardiac MRI using learned local image statistics. In: Camara, O., Mansi, T., Pop, M., Rhode, K., Sermesant, M., Young, A. (eds.) STACOM 2012. LNCS, vol. 7746, pp. 115–124. Springer, Heidelberg (2013). https://doi.org/10.1007/978-3-642-36961-2_14
Newell, A., Yang, K., Deng, J.: Stacked hourglass networks for human pose estimation. In: Leibe, B., Matas, J., Sebe, N., Welling, M. (eds.) ECCV 2016. LNCS, vol. 9912, pp. 483–499. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46484-8_29
Noothout, J.M., de Vos, B.D., Wolterink, J.M., Leiner, T., Išgum, I.: CNN-based landmark detection in cardiac CTA scans (2018). arXiv preprint arXiv:1804.04963
Payer, C., Štern, D., Bischof, H., Urschler, M.: Regressing heatmaps for multiple landmark localization using CNNs. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 230–238. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_27
Ronneberger, O., Fischer, P., Brox, T.: U-Net: Convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Sun, K., Xiao, B., Liu, D., Wang, J.: Deep high-resolution representation learning for human pose estimation. CVPR 2019, 5693–5703 (2019)
Tuysuzoglu, A., Tan, J., Eissa, K., Kiraly, A.P., Diallo, M., Kamen, A.: Deep adversarial context-aware landmark detection for ultrasound imaging. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11073, pp. 151–158. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00937-3_18
Wang, C.W., et al.: Evaluation and comparison of anatomical landmark detection methods for cephalometric x-ray images: a grand challenge. IEEE Trans. Med. Imaging 34(9), 1890–1900 (2015)
Xu, Z., et al.: Less is More: Simultaneous view classification and landmark detection for abdominal ultrasound images. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11071, pp. 711–719. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00934-2_79
Yang, D., Zhang, S., Yan, Z., Tan, C., Li, K., Metaxas, D.: Automated anatomical landmark detection ondistal femur surface using convolutional neural network. In: ISBI 2015, pp. 17–21. IEEE (2015)
Zhang, J., Liu, M., Shen, D.: Detecting anatomical landmarks from limited medical imaging data using two-stage task-oriented deep neural networks. IEEE Trans. Image Process. 26(10), 4753–4764 (2017)
Zhang, J., et al.: Context-guided fully convolutional networks for joint craniomaxillofacial bone segmentation and landmark digitization. Med. Image Anal. 60, 101621 (2020)
Zhong, Z., Li, J., Zhang, Z., Jiao, Z., Gao, X.: An attention-guided deep regression model for landmark detection in Cephalograms. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11769, pp. 540–548. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32226-7_60
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Liu, W., Wang, Y., Jiang, T., Chi, Y., Zhang, L., Hua, XS. (2020). Landmarks Detection with Anatomical Constraints for Total Hip Arthroplasty Preoperative Measurements. In: Martel, A.L., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2020. MICCAI 2020. Lecture Notes in Computer Science(), vol 12264. Springer, Cham. https://doi.org/10.1007/978-3-030-59719-1_65
Download citation
DOI: https://doi.org/10.1007/978-3-030-59719-1_65
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-59718-4
Online ISBN: 978-3-030-59719-1
eBook Packages: Computer ScienceComputer Science (R0)