Abstract
In this chapter, we review and compare existing theoretical models of the relationship between genetic and phenotypic variation or genotype-phenotype map (GPM). By doing that, we introduce the reader to concepts and assumptions of evolutionary genetics and contrast them with concepts and models coming from developmental biology. Although these two approaches can be regarded as complementary to study the same underlying problem, phenotypic variation and evolution, they contradict each other in a number of ways.
The evolutionary genetics models on the GPM consider genetic interactions but not epigenetic interactions. This simplicity has been used to argue that they are the most general (Wagner, Trends Ecol Evol 26: 577–584, 2011). We argue, in contrast, that epigenetic factors are crucial to understand the GPM. We understand epigenetic factors as nongenetic factors that are instrumental in building the phenotype during development (Waddington, Beyond reductionism, 1968). We argue that models including epigenetic factors exhibit features found in real GPMs that are not found in purely genetic models. Since these features are widely found in real GPMs, no model can be considered general without those and, then, models including epigenetic factors are more general than purely genetic models, even when the details of the former are specific of certain types of phenotypes.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- G matrix
- Mendelian genetics
- Michaelis-Menten
- RNA secondary structure
- Alleles
- Animal development
- Artificial selection experiments
- Assumption of linearity
- Asymmetric spatial distribution
- Bio-mechanics
- Biological functions
- Causal role
- Cell adhesion
- CELL behaviors
- Cell collectives
- CELL contraction
- Cell division
- Cell mechanical interactions
- Cell-cell signalling
- Complex morphologies
- Conservative natural selection
- Constructional constraints
- Cusps
- Developmental patterns
- Developmental resources
- Developmental stages
- Developmental time
- Dimensionality
- Egg shape
- Embryo morphology
- Embryonic development
- Environmental robustness
- Environmental variation
- Epigenesis
- Epigenetic factors
- Epigenetic interactions
- Epigenetic mechanisms
- Epigenotype
- Epistatic coefficients
- Epithelial buckling
- Evolutionary genetics
- Experimental knowledge
- Explanatory power
- Explicit spatial context
- Extracellular diffusible signals
- Extracellular matrix
- Fixation
- Fly segmentation
- Gene expression levels
- Gene network
- Gene network dynamics
- Gene network topologies
- Gene networks
- Gene product interactions
- Genetic factors
- Genetic interactions
- Genetic models
- Genetic pathways
- Genetic variation
- Genetic-epigenetic models
- Genome
- Genotype-phenotype map
- Infinitesimal model
- Initial condition
- Initial developmental pattern
- Intrinsic phenotypic effects
- Lattice model
- Lattice pattern formation model
- Limb morphogenesis
- Linearity
- Locus
- Mammal blastulation
- Mammalian teeth
- Material properties
- Mathematical models
- Mechanical properties
- Modern evolutionary synthesis
- Morphogenetic fields
- Morphological phenotypes
- Morphological variation
- Morphology
- Multilinear model
- Multilocus
- Natural populations
- Network dynamics
- Neurophysiology
- Non-general
- Non-genetic factors
- Non-linear genotype-phenotype map
- Non-motile cells
- Novelty
- Organ physiology
- Pattern formation
- Phenogenesis
- Phenome
- Phenotypic changes
- Phenotypic effect
- Phenotypic trait
- Phenotypic variation
- Polygenic
- Population genetics
- Protein conformation
- Quantitative changes
- Quantitative genetics
- Quantitative traits
- Quantitative variation
- Realism
- Secondary RNA structure models
- Segmentation mechanisms
- Shape
- Short-range signaling
- Sigmoidal function
- Signal secretion
- Skin coat patterns
- Small adaptive mutations
- Soft-matter properties
- Spatial distribution
- Spatial epigenetic factors
- Spatio-temporal regulation
- Theoretical models
- Tooth morphogenesis
- Traits
1 Introduction
The twentieth century can be seen as the century of genetics. We have learned that phenotypes and most of their variation have, ultimately, a specific genetic basis. We know what is at the bottom, the genome, and what is at the top, the phenome, but we do not understand well enough the processes in between to explain which changes at the genetic level lead to which specific changes at the phenotypic level (and why to those changes and not to others). It is our perception, and that of many others (e.g., Houle et al., 2010), that early twenty-first century biology would be, largely, about understanding the genotype-phenotype map.
The genotype-phenotype map, or GPM, is simply the pattern of association between each specific phenotypic variant and its underlying genetic variant. Its domain of application is not fixed and, thus, one can talk of the GPM of an organism, a part of an organism or even of a gene network affecting some aspect of the phenotype.
A central tenet of developmental evolutionary biology is that the GPM is, at least for the case of morphology, determined by embryonic development (Alberch, 1991). By morphology, we mean the distribution of cells and extracellular matrix, ECM, in space. During the process of embryonic development, a single zygotic cell gives rise to a functional organism characterized by a complex distribution of cell types in space. If one defines morphology as the distribution of cells and extracellular matrix in space, it follows that cells have to do things to change their position from the early embryo to the adult. These things are the cell behaviors (cell division, cell adhesion, cell contraction, etc.) and interactions, either mechanical or chemical through cell–cell signaling. In other words, specific morphologies arise in development because individual cells do specific things (e.g., divide, die, secrete ECM) in specific places and times along development. Genes and gene networks have an effect on morphology because they affect the spatiotemporal regulation of these cell behaviors and interactions (Forgacs & Newman, 2005).
There is a complex interdependence between gene networks, cell behaviors, and cell interactions (Salazar-Ciudad et al., 2003). On one hand, some of the gene products in networks are extracellular diffusible signals. These can alter the behavior and network dynamics of the neighboring cells that receive them. On the other hand, changes in cell behaviors and cell mechanical properties lead to changes in mechanical interactions between cells. These lead to changes in the shape of the space in which these signals are diffusing (i.e., the embryo morphology). By affecting this shape, biomechanics indirectly affect the spatial distribution of signals and then which cells are receiving which extracellular signals. This, in turn, affects which genes are expressed and where. In that sense, gene networks, signaling, and biomechanics reciprocally affect each other.
As a result of these interdependent interactions between gene networks and cellular processes, gene expression, cell location, cell shape and ECM change over time and space to produce the complex morphology of the adult.
Since morphology arises through complex networks of gene, cell, and tissue-level interactions, it follows that single genes do not have intrinsic morphological effects. In other words, no individual gene codes, or has the information, for a specific phenotype or phenotypic variant. On the contrary, the morphological effect of a gene, and by extension of genetic variation in it, is totally dependent on the gene networks in which it is embedded and on the effect of these networks on the cell behaviors and interactions by which morphology is built (Oster & Alberch, 1981; Alberch, 1991). We call a gene network regulating cell behaviors and mechanical properties a developmental mechanism (Salazar-Ciudad et al., 2003). The cell behaviors and properties are also part of the developmental mechanism.
Natural selection is a crucial factor determining the direction of evolutionary change. Natural selection, however, can only act by eliminating phenotypic variation in each generation. Within an individual, the ultimate cause of phenotypic variation is genetic and environmental variation (Griffiths, 2002). Which phenotypic variation will arise from such genetic and environmental variation, however, is determined by development since, as we come to discuss, development has a crucial role in determining the GPM. It follows then that development, in addition to natural selection, is crucial to understand the direction of phenotypic evolution (de Beer, 1930; Goldschmidt, 1940; Waddington, 1957; Alberch, 1982; Arthur, 2001; Salazar-Ciudad, 2006a), at least for the case of morphological phenotypes.
In this chapter, we review existing models of the GPM. We aim to show that many prevailing genetic models, although claiming to be general, fail to recreate some fundamental properties of real GPMs. We will argue that this is largely because they do not consider epigenetic factors (e.g., cell behaviors). Instead, phenotypes are often conceptualized as arising purely from gene product interactions or even from genes themselves. We will argue that it is only because of epigenetic factors that complex multicellular phenotypes, or even just phenotypes, are possible at all. Based on these premises, we will also argue that models including epigenetic factors are not only more realistic, they also uncover general characteristics of real GPMs that are simply invisible to purely genetic models.
In this article, we will focus on the GPMs for morphological phenotypes but we will also discuss similarities between these GPMs and those studied at other phenotypic levels. The article is arranged in sections: a section describing what we mean by epigenetic factors, one section per each kind of GPM model and sections describing each main difference between the models that include epigenetic factors and the models that do not.
2 Epigenesis and Epigenetic Factors
The concept of epigenetic factors has two meanings. An epigenetic factor can be anything that is not in the DNA sequence (such as the patterns of methylation on the DNA) but that is heritable and has a causal role in development, physiology or any other biological process. Epigenetic is also the adjective of epigenesis (Haig, 2004) and, thus, epigenetic factor would be any “factor” related to epigenesis. Epigenesis is one of the two alternative views Aristotle proposed about embryonic development. Epigenesis is the view that during development new organization arises from previously existing organization that was not equal or trivially similar to it (Jablonka & Lamb, 2005; Müller, 2007). The other view is preformationism: the view that nothing really new arises in development and that most body parts and organization are already present, at a smaller scale, within the gametes.
At the most general conceptual level, development can be described as the process by which specific arrangements of cell types, what we call developmental patterns (Salazar-Ciudad et al., 2003), transform into other developmental patterns. Over developmental time, these pattern transformations occur constantly and lead from a simple developmental pattern, the zygote, to a quite complex one, the adult organism. In that sense, embryonic development is in concordance with Aristotle’s epigenesis, except that there is something that remains constant during development and that is faithfully copied between generations: the genotype.
An example of an epigenetic factor, in the epigenesis sense, is the asymmetric spatial distribution of many proteins and RNAs in the oocytes of many species (Newman, 2011a). These distributions are relatively simple but most animals have an asymmetry along the animal-vegetal pole and many of them have also asymmetries along other axes (Gilbert & Raunio, 1997; Gilbert & Barresi, 2016). These asymmetries are absolutely required for development, if they are experimentally disturbed, embryos become spherically symmetric and their development gets arrested very early (Kandler-Singer & Kalthoff, 1976; Gilbert & Barresi, 2016). These spatial asymmetries arise from spatial asymmetries present in the gonads of the parents, typically their cell-level apical-basal asymmetries (Bastock & St Johnston, 2008), or are transferred to the oocyte from an apical-basally polarized epithelium through short-range signaling (Neuman-Silberberg & Schupbach, 1993; Roth & Lynch, 2009).
One may argue that the asymmetries in the mother’s gonads are due to gene product interactions in the earlier development of the mother. This is indeed the case, but these asymmetries in the mother’s gonad required also that the oocyte that gave rise to the mother had the same spatial asymmetries, otherwise, the mother’s development would have arrested early on. The spatial asymmetries in the oocyte are, thus, not reducible to gene product interactions. This interdependence between genetic and spatial epigenetic factors is not exclusive of multicellular organisms, but applies to all organisms (Jablonka & Lamb, 2005).
Another example of epigenetic factors is the developmental patterns themselves (see Figs. 1 and 2). Gene product interactions are crucial in determining which developmental patterns arise from which previous developmental patterns, but so are these previous developmental patterns themselves. The same developmental mechanism (i.e., gene network plus cell behaviors and mechanical properties) can lead to different final developmental patterns depending on which developmental pattern it acts on (Salazar-Ciudad et al., 2000, 2003). In each developmental stage, existing developmental patterns depend on previous gene product interactions acting on previous developmental patterns. Thus, these patterns, starting from the asymmetries in the oocyte, are both a consequence and a cause of developmental dynamics. In this process, genes and epigenetic factors are intricately interdependent, but not reducible to each other.
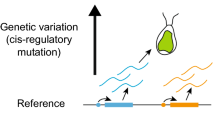
Schema of the interplay between genetic and epigenetic factors along successive stages of organismal development. The top row shows different developmental patterns, different distributions of cell types in space, from the zygote to the adult of one organism and its parent. In the case of the zygote, the developmental pattern comes from the asymmetric distribution of gene products mediated by the mother during gametogenesis. The plot below shows the amount of genetic and epigenetic accumulated change within an individual and between generations in respect to the zygote. The genotype does not change per se during development, it is the interplay between it and the epigenetic factors (solid and dashed arrows) that builds the developing organism. This interaction (the arrows) should be understood, as discussed in the text, as different epigenetic factors affecting where and when different genes get expressed, while at the same time, which epigenetic factors are encountered in each time and place in the embryo depends on previous genetic and epigenetic interactions (as the arrows abstractly depict)
Example of a genetic-epigenetic model. The figure shows, in the left, an initial developmental pattern the model takes as input and the developmental mechanism it implements, top. In the right, the resulting developmental pattern is shown (the output of the model). In the left, each purple sphere represents the apical side of an epithelial cell and each sphere in blue the basal side. In the developmental mechanism, positive gene regulation is shown by green arrows and negative gene regulation by red arrows. Each sphere is a different gene product, gene product in red can diffuse between cells. In the final developmental pattern, the colors show the expression of gene 2 in the developmental mechanism (yellow for the maximal expression and blue for the minimal expression). The model is implemented in EmbryoMaker (Marin-Riera et al., 2016)
Other epigenetic factors relevant for embryonic development include the mechanical properties of cell collectives (Newman & Comper, 1990; Newman & Müller, 2000) and basic cell behaviors such as cell division, cell adhesion, apoptosis, extracellular signal secretion, etc. (Salazar-Ciudad et al., 2003). All these factors are often regulated by gene products, but their existence is not due to, or merely reducible to, genes or genetic interactions. In fact, many cells and tissue mechanical properties that are relevant to understand morphogenesis are also found, in some rudimentary form, in other systems such as in liposomes devoid of proteins (Newman & Comper, 1990). Gene interactions can then be seen as just a way to more precisely regulate properties and behaviors that are intrinsic to cell clusters (Newman & Müller, 2000). Understanding those cell properties and behaviors is fundamental to understand development, as has been widely discussed (Newman & Comper, 1990; Beloussov, 1998; Oyama, 2000; Newman, 2011b; Guillot & Lecuit, 2013) under different names and slightly different concepts: epigenetic mechanisms (Newman & Müller, 2000), soft-matter properties (Newman & Comper, 1990), developmental resources (Oyama, 2000), phenogenesis (Weiss & Fullerton, 2000), and epigenotype (Waddington, 1942).
Epigenetic factors are heritable but their variation is usually not (Jablonka & Lamb, 2005; Salazar-Ciudad, 2008). One could then argue that the epigenetic factors are not necessary to understand the GPM. After all, the GPM is defined as the association between genetic variation and phenotypic variation. However, as we come to explain, genetic and epigenetic factors need to interact for complex phenotypes and their variation to be possible at all. In other words, genetic variation does not have phenotypic effects unless it affects some epigenetic factors. Thus, to understand the phenotypic consequences of a genetic change, and then the GPM, the interaction between genetic and epigenetic factors needs to be considered too (Waddington, 1968; Oster & Alberch, 1981).
3 GPM Models
Models about the GPM coming from an evolutionary genetics tradition do not consider epigenetic factors (Wagner, 1994; Hansen & Wagner, 2001). In fact, it has been claimed that these purely genetic models (Wagner, 2011) are the most general because, allegedly, they make fewer assumptions and do not include epigenetic factors that are always specific of certain phenotypic levels. In this chapter, we will argue that purely genetic models are not more general, but more particular because they do not exhibit some general features of real phenotypic variation and GPMs. In addition, they make a number of simple but strong assumptions. When these assumptions are considered explicitly, as we will do in this article, it becomes clear that the purely genetic models are not more general than the models that consider epigenetic factors.
4 Mendelian and Quantitative Genetics
The study of the GPM has not been central to evolutionary genetics and, by extension, to the modern evolutionary synthesis, also called neo-Darwinism (Mayr & Provine, 1980). Although it was very early recognized (Wright, 1932) that such a map was likely to be complex, most models in evolutionary genetics assume or require a simple GPM. Such an assumption may be justified depending on the questions being addressed. Thus, for example, if the aim is to show that natural selection can lead to the fixation of some alleles (i.e., some variants of a gene), then one may ignore the GPM and consider only the average phenotypic effect of an allele on the phenotype (Fisher, 1930). For more advanced questions, as we will discuss, this assumption is not tenable.
Evolutionary genetics is based in population genetics. The way the relationship between genetic and phenotypic variation is understood in population genetics, is based on Mendelian genetics (Griffiths, 2002). Specific discrete phenotypic variation was conceived to be associated with some particles in the nucleus, the genes. Later on, these particles were found to be made of DNA (Griffiths, 2002). Finding this association is a major achievement of twentieth century biology. However, it is rarely the case that a discrete phenotypic character can be simply associated with a gene as in Mendelian genetics. Even in the best cases, there is substantial penetrance and expressivity: different proportions of the individuals bearing a gene exhibit the corresponding phenotype and when they do it, they do it to different degrees.
Most characters do not depend on one but several genes, i.e., they are polygenic (Griffiths, 2002). In that case, Mendelian genetics considers that each different combination of alleles could be associated with a different state in that character, a different variant phenotype. Mendelian genetics, however, has no way to tell which these character states would be. In other words, Mendelian genetics can tell us about the frequency of certain phenotypic states (e.g., a pea being green) given that those states are statistically associated with specific combinations of alleles, but it cannot tell us why some alleles are associated with some specific character states, and it cannot tell us which character states, or phenotypic variation, are possible (Alberch, 1982). This relevant information has to be found by other means, i.e., observation or other theories. In essence, genes in Mendelian genetics are defined based on their statistical association with phenotypic variants. However, in some of the applications of Mendelian genetics that we will discuss, genes become understood as actual bearers of the information necessary to build specific phenotypes and phenotypic variants.
Quantitative genetics is a theory that aims at providing such information. Quantitative genetics is concerned with the inheritance of quantitative characters or traits. These are characters that can be described by continuous, rather than discrete, values (e.g., weight, limb length). Quantitative genetics was originally conceived as an extension of multilocus (i.e., polygenic) population genetics following the so-called infinitesimal model (Rice, 2004), that in its turn, was based in Mendelian genetics. According to this model, each phenotypic trait is determined by the sum of the phenotypic effect of a large number of Mendelian loci (i.e., genes) of small quantitative effect on the phenotype. In addition, loci’s phenotypic effects are supposed to be additive. This means that the phenotypic effect of a locus is independent from the effect of other loci. Thus, each different allele in a locus (i.e., each variant of a gene) adds a value to the phenotype independently from the other alleles in the other loci in the genome. Contrary to what we proposed in the introduction, then, genes are conceived to have intrinsic phenotypic effects. Note also that the assumption that the effects of alleles are small and independent from each other is also a mechanistic assumption on how genes interact in order to construct the phenotype.
The infinitesimal model leads to a clear view on which phenotypic variation should be possible by genetic variation. This model is, thus, implicitly a model about the GPM. Mutations would affect genes and lead to new alleles. These would have slightly larger or smaller intrinsic phenotypic effects than their non-mutated version. It is then assumed that the values of phenotypic traits are able to increase or decrease by the replacement of old alleles by specific new alleles over time. This simple model implies that, in principle, all quantitative traits should be able to vary gradually and increase or decrease forever as long as there is selection acting on them. Since the phenotypic effect of alleles is small and additive (independent) one is assuming a linear GPM: small genetic changes lead to small phenotypic changes.
In practice, quantitative genetics uses information about the genetic relatedness between individuals to estimate which proportion of the phenotypic variation between these individuals is due to shared genetics. This information is based on genealogies or on direct information of the genotypes of individuals (such as in GWAS studies) (Uricchio, 2020). The GPM is conceptualized through the G matrix, the matrix of covariances between traits due to shared genetics (Lande & Arnold, 1983). Describing the GPM through covariances is convenient if the GPM is assumed to be linear. In such an approach, all traits can change indefinitely, but not all of them are necessarily equally variable. Since some genes may be affecting several traits at the same time these traits may not be able to vary independently from each other.
It is important to note that although the quantitative genetics approach is eminently statistical and claimed to be applicable to any phenotype (Roff, 2007), the assumption of linearity implies that it should only be accurate when the GPM is linear. When the GPM is nonlinear, quantitative genetics will still provide predictions but these would often be inaccurate (Pigliucci, 2006; Milocco & Salazar-Ciudad, 2020).
There is the general perception that the predictions of quantitative genetics are experimentally accurate (Roff, 2007). Artificial selection experiments are usually seen as supporting this perception (Roff, 2007). This evidence, however, exists only for selection on single traits. Some authors claim that most studies only show that the quantitative genetics approach works better than nothing since no alternative approaches are used (Pigliucci, 2006). In addition, more general experiments selecting for a trait and looking at the response in others or experiments selecting for multiple traits at the same time are rare. The few studies that attempt that are sometimes compatible with the expectations of multivariate quantitative genetics, but some other times they are not (Roff, 2007).
5 Evolutionary Genetics Models on Epistasis
There are several theoretical models that are conceptually related to quantitative genetics but that explicitly consider that the phenotypic effects of loci depend on each other, although linearly (Barton & Turelli, 1987; Jones et al., 2004). As an example, we will discuss one such model in some more detail: Hansen’s and Wagner’s multilinear model (Hansen & Wagner, 2001). The multilinear model was developed with the explicit intention of (quoting this original work; Hansen & Wagner, 2001, p. 76):
The current body of theory in quantitative genetics lacks an operational theory of gene interaction […] The multilinear theory is […] the only current suggestion that allows for a systematic non-statistical way of incorporating gene interactions in quantitative genetic theory.
The multilinear model bears an assumption about the GPM: The interactions between gene effects are linear; changing the genetic background for a locus causes a linear transformation of the effects of all substitutions at this locus. Thus, the value of each trait is determined by a fixed phenotypic (i y) effect from each locus i, defined as the phenotypic change of a substitution in that locus in respect to an arbitrary reference genotype, and a term multiplying all combinations of phenotypic effects by an epistatic coefficient ε. This coefficient can be different for each gene interaction (ij ε for two loci interactions, ijk ε for three loci interactions, etc.).
For several traits we have
Where a, b, c … are the successive traits conforming the phenotype. There are a number of models that make similar linear assumptions (Zhivotovsky & Feldman, 1992; Gavrilets & Dejong, 1993; Turelli & Barton, 1994; Nowak et al., 1997; Wagner et al., 1997). Other than linearity, this model does not propose much about how the GPM is (i.e., nothing is said about the distribution of the epistatic coefficients or about whether epistasis happens between two, three or more loci at the same time). However, the actual applications of the model do take more detailed assumptions about the GPM: they assume that (Hansen et al., 2006; Fierst & Hansen, 2010) all epistatic coefficients are equally likely to change by mutation. In other words, since the coefficients in Eqs. (1) and (2) can acquire any arbitrary value through mutations, the GPM is totally plastic to change in any direction over generations. This is a common feature of the models based on quantitative genetics (Barton & Turelli, 1987; Jones et al., 2004). These models consider that loci’s phenotypic effects depend on each other but they assume that these dependencies are free to change, over time, by mutation in any way. This is a sort of second-order neo-Darwinism: Traits are not assumed to all be equally likely to change as in the neo-Darwinian approach. There are genetic covariances between traits but these covariances are free to change in any way by the accumulation of small adaptive mutations. This is like applying the infinitesimal model to the evolution of the GPM rather than directly to the evolution of the phenotype (Roff, 2000; Wagner et al., 2007; Cheverud, 2007; Crow, 2010).
6 Wagner’s Model
Wagner’s purely genetic model (Wagner, 1994) has also been used to study the evolution of the GPM and its effect on phenotypic evolution (Le Cunff & Pakdaman, 2012; Fierst, 2011; Pinho et al., 2012; Draghi & Whitlock, 2012). In contrast to Hansen’s model, this is a nonlinear model in which gene products regulate each other expression. The phenotype is the level of expression of the genes in the network.
The level of expression of a gene i in a given time t + 1, S i(t + 1), is a nonlinear function, σ, of the sum of the regulatory inputs from each other gene j, w ij, each multiplied by the expression of those genes S j(t).
The σ function is a sigmoidal function that it effectively acts as a threshold: it gives −1 if its argument is smaller than 0 and 1 if its argument is larger than 0 (and 0 if its argument is zero). The matrix of all w ij values effectively determines gene network topology and the interaction strength between genes.
In Wagner’s model, the number of traits is the same as the number of genes. For that model to be generally applicable to the GPM, it is required that the phenotype can be explained, at least in its main features, solely on the bases of gene expression levels. To explain the phenotype, thus, Wagner’s model still takes the idea that genes have intrinsic phenotypic effects. In this case, simply, it is not the genes but their expression that have intrinsic phenotypic effects. Thus, there are no epigenetic factors nor space.
The main improvement of this model in respect to previous models is that there are gene networks and that the gene interactions in this network are nonlinear. Although this approach is an improvement over other purely genetic approaches, it still fails to capture some important features of GPMs, as we later discuss.
7 Genetic-Epigenetic Models
In this section, we include those models in which the phenotype is understood to arise from genetic factors, epigenetic factors and their interactions. The models we will discuss in most detail are those of embryonic development. Most models of embryonic development (e.g., Odell et al., 1981; Meinhardt, 1982; Jaeger et al., 2004; Honda et al., 2008; Salazar-Ciudad & Jernvall, 2010; Zhu et al., 2010; Osterfield et al., 2013) have some elements in common: gene networks, cells, and some epigenetic factors in an explicit spatial context. These models always include some initial developmental pattern (i.e., an initial condition), and often, some cell behaviors (e.g., cell division, cell contraction, cell adhesion, etc.). There is often the diffusion of extracellular signals in space. Similar models can be applied to other phenotypic levels such as cell biology (Vik et al., 2011; Karr et al., 2012), organ physiology (Noble, 2002; Gjuvsland et al., 2013) and neurophysiology (Skinner, 2012; Goldberg & Bergman, 2011), RNA secondary structure (Schuster et al., 1994; Schuster et al., 1994; Cowperthwaite & Meyers, 2007) and protein conformation (Silva et al., 2009; Ferrada & Wagner, 2012). Models at each phenotypic level include epigenetic factors that are level-specific (e.g., cell signaling and other cell behaviors, the rules of nucleotide base pairing in RNA models, or the stereochemistry of amino acids in proteins).
In the case of development, most models focus on a specific organ and incorporate experimental information on the genetic, cellular, and other epigenetic factors known to be important for that organ development, just to cite a few: epithelial buckling (Odell et al., 1981), skin coat patterns (Meinhardt, 1982), early fly segmentation (Jaeger et al., 2004), mammal blastulation (Honda et al., 2008), tooth morphogenesis (Salazar-Ciudad & Jernvall, 2010), limb morphogenesis (Zhu et al., 2010), and egg shape (Osterfield et al., 2013).
There are genetic-epigenetic models that are not specific to any organ but that intend to apply to the whole of animal development (Hogeweg, 2000; Hagolani et al., 2019). They implement basic equations for gene network dynamics, cell behaviors, extracellular signal diffusion, and cell mechanical interactions. One can then explore the different GPMs arising from the theoretically possible developmental mechanisms (Hogeweg, 2000; Marin-Riera et al., 2016; Hagolani et al., 2019). With these models, it has been recently suggested that the development of complex morphologies can be achieved if most cells in an embryo activate cell behaviors (mostly cell division and contraction). For these complex morphologies to be also stable (i.e., that they develop in the same way in spite of noise), it is required that the embryo is partitioned in relatively small areas of gene expression where cell behaviors are regulated differently (Hagolani et al., 2019). These models, however, have not yet been applied to better understand the GPM directly.
One limitation of the genetic-epigenetic models is that there is no genotype as such. There are parameter’s that specify the strength of some genetic interactions in the model. Changes in the values of these parameters are usually used as proxies for genetic variation but, in fact, the relationship between the values of these parameters and genetic variation is likely to be quite complex since it may depend in process such as protein folding, RNA folding, enzymatic catalysis, etc.
8 The Lattice Pattern Formation Model
The simplest genetic-epigenetic models, are those that include gene networks and two epigenetic factors: the cell behavior of cell signaling, and the physical process of signal diffusion in the extracellular space. These models are framed in a spatial cellular context, e.g., a lattice of cells, and the phenotype of interest is the spatial distribution of gene expression. There are several of those models (Mjolsness et al., 1991; Jaeger et al., 2004; Cotterell & Sharpe, 2010; Jiménez et al., 2015; Rothschild et al., 2016) but only one is being discussed here, the Salazar-Ciudad’s lattice model (from here on just the lattice model), because it intends to be general (i.e., applying to all animal development not to a specific organ) and it explicitly focuses in the GPM and in its evolution (Salazar-Ciudad et al., 2000, 2001a). In essence, this and other models can be seen as adding epigenetic factors to Wagner’s model.
In this model, a number of nonmotile cells, each one including an identical gene network, occupy different positions in a 2D regular lattice. Some of the gene products are extracellular diffusible signals that affect gene expression in neighboring cells. In the lattice model the (continuous) change in concentration of a given gene product “i” in a given cell “j,” that is g ij, does not only depend on the gene network dynamics of that cell, but also on the contribution of diffusible gene product coming from other cells:
As in Wagner’s model, w jk values determine the gene network topology and the interaction strength between genes. The heaviside function ф prevents negative concentrations in the gene products (ф(x) is such that if x > 0 then ф(x) = x and if x ≤ 0 then ф(x) = 0). The first term is a Michaelis–Menten saturating function with a K m coefficient. The degradation rate of the gene products is specified by the μ parameter and D l is the specific diffusion rate of the gene product l.
The model starts from a very simple initial developmental pattern in which only one gene is expressed and only in a cell in the middle of the lattice (Salazar-Ciudad et al., 2000). As a result of genetic interactions in the network, cell signaling and diffusion, a new developmental pattern arises (i.e., new distributions of gene expression in space). This developmental pattern is the phenotype in this model. There is, thus, some basic morphology in the sense of the distribution of cell types in space where cell types are described by their gene expression. Despite its relative simplicity, lattice models have been used to study the segmentation mechanisms in the early embryos of Drosophila and other insects (Salazar-Ciudad et al., 2001b; Jaeger et al., 2004).
The main result of the analysis of the Salazar-Ciudad lattice model is that there are a limited number of gene network topologies that can lead to pattern formation (Salazar-Ciudad et al., 2000). In other words, most gene networks one could build (for example by wiring genes at random as in Salazar-Ciudad et al., 2000) simply do not lead to any changes in gene expression over space, no pattern, even if they promote cell–cell signaling. Somehow there are mathematical constraints or rules on which gene networks can lead to pattern formation. These rules exist, however, because genes affect some epigenetic factors (e.g., the physical process of extracellular signal diffusion). These epigenetic factors allow for phenotypes (i.e., patterns) to arise rather than just being assumed as in the purely genetic models. These constraints simplify the study of the GPM, since one only needs to worry about the GPM in this subset of topologies.
9 Models Including Morphogenesis: The Tooth Model
The Salazar-Ciudad lattice and similar models including cell signaling are still rather unrealistic models of the GPM for morphological phenotypes. First, because morphological variation does not involve just differences in the location of gene expression but also differences in the location of cells themselves and the former cannot be defined without the latter unless one leaves morphology as such as unexplained. Second, during development, cells not only signal to each other, they also activate cell behaviors. In fact, cells only change their position, and thus morphology, as a consequence of these other cell behaviors. There are a number of specific models doing that (Odell et al., 1981; Honda et al., 2008; Salazar-Ciudad & Jernvall, 2010; Osterfield et al., 2013) but most of them are only concerned with wild-type phenotypes or with mutations of large phenotypic effect that are unlikely to be found in natural populations due to their likely highly deleterious effects.
One exception is the tooth model (Salazar-Ciudad & Jernvall, 2010; Salazar-Ciudad & Marín-Riera, 2013). This model integrates experimental knowledge on how genes and cells interact in a specific system, the mammalian teeth, to reproduce their development, their adult morphologies, and their variation. In addition, the model is used to reproduce the multivariate and three-dimensional morphological variation in natural populations. This allows to suggest which changes in development, and possibly in which genetic pathways, may be responsible for the observed patterns of micro-evolutionary phenotypic variation. This model also allows to obtain some mechanistic understanding on why some morphological variation arises and some other does not arise from mutations affecting development.
General models of development including many, or even most, cell behaviors do exist (Hogeweg, 2000; Marin-Riera et al., 2016; Delile et al., 2017) but so far their potential to address general questions about the GPM itself has not been fully explored (except for two attempts, Hogeweg, 2000; Hagolani et al., 2019).
10 The Differences Between Purely Genetic Models and Genetic-Epigenetic Models
Overall, genetic-epigenetic models provide a view on the GPM and the phenotypic variation that differs from that of the purely genetic models in a number of fundamental ways:
10.1 Genetic-Epigenetic Models Reproduce Individual Phenotypes and the GPM
The first difference is that in genetic-epigenetic models the phenotypes arise from the modeling of specific epigenetic factors or even physical interactions (e.g., between gene products, between cells, between nucleotides; see Fig. 1 for an example) while in the purely genetic models the phenotypes arise from the assumed intrinsic phenotypic effect of genes and the distribution of genes among individuals in a population. Wagner’s model is somehow in between because it simulates gene product interactions but in order to be general it assumes that gene expression has an intrinsic effect on the phenotype (i.e., it does not depend on epigenetic factors) and that these can arise without considering the interactions between cells (i.e., Wagner’s model is a unicellular model).
In the genetic-epigenetic models, the GPM is studied by exploring which phenotypes arise from which changes in its parameters. These models make assumptions at the level of which genetic interactions and epigenetic factors are relevant to explain the phenotype and its variation. Given these assumptions, the GPM arises from the model, i.e., no assumptions are made directly on the nature of the phenotype or the GPM. In the purely genetic models, in contrast, there are features of the GPM that do not arise from the model, they are simply assumed, most notably linearity and that genes have intrinsic phenotypic effects. This inevitably makes the genetic-epigenetic models more general since they make fewer assumptions on the nature of the GPM.
Genetic-epigenetic models often have enough realism as to be quantitatively comparable with a real organ’s morphology (Odell et al., 1981; Meinhardt, 1982; Jaeger et al., 2004; Harris et al., 2005; Honda et al., 2008; Newman et al., 2008; Salazar-Ciudad & Jernvall, 2010; Osterfield et al., 2013; Moustakas-Verho et al., 2014; Ray et al., 2015; Onimaru et al., 2016; Brun-Usan et al., 2017; Marin-Riera et al., 2018).
It is relevant to note that genetic-epigenetic models reproducing population-level phenotypic variation can also reproduce the statistical properties of the population that purely genetic models aim at. All models have parameters, e.g., the diffusivity of an extracellular signal in the lattice models that can be varied. This variation leads, through the model, to phenotypic variation. Statistics can then be applied to this variation, as it is done in population and quantitative genetics, and these can be compared to the ones observed in natural populations (as for example in Salazar-Ciudad & Jernvall, 2010; Milocco & Salazar-Ciudad, 2020).
10.2 Epigenetic Factors Inform About the Space of Possible Networks
Epigenetic factors provide informative rules on which networks are biologically plausible. In the lattice model, for example, we have found that cell signaling and signal diffusion allows for pattern formation but only for specific types, or families, of gene network topologies (Salazar-Ciudad et al., 2000). In other words, by including the epigenetic factor of signaling, we learn that not all gene network topologies can be found in nature to perform a given function (e.g., lead to pattern formation in lattices of cells). This is not the case in purely genetic models, not even in Wagner’s model. Since these include no epigenetic factors, the space of gene network topologies that can lead to plausible phenotypes is much larger. This is relevant because different conclusions would be reached when studying one space of gene networks or the other. For example, some studies (Wagner, 1996; Draghi & Whitlock, 2012) claim that phenotypes become more robust to environmental changes as a result of conservative natural selection (i.e., selection for a phenotype to remain unchanged over generations). For that to occur there has to be many gene networks that can lead to the same specific phenotype and they have to differ in their environmental robustness. This is the case for Wagner’s model. This is not necessarily the case in genetic-epigenetic models. In the genetic-epigenetic models where this has been studied (Salazar-Ciudad & Jernvall, 2005; Hagolani et al., 2019), many phenotypes are only possible for specific genetic-epigenetic networks and for very specific values in their parameters. Conservative natural selection is, thus, unlikely to increase environmental robustness by choosing among the networks that can produce the same morphology. This is because there is not many of them and they are not connected to each other by a small number of mutations. Thus, the result that conservative selection increases robustness depends on a specific unconstrained space of gene networks that does not seem to be realistic.
10.3 In Genetic-Epigenetic Models Not All Aspects of the Phenotype Can Change
The third main difference between genetic-epigenetic models and purely genetic models is that, in the former, not all aspects of the phenotype are equally likely to change by mutation, and in fact, some aspects may not be changeable at all, at least in the short term. This is a general property of genetic-epigenetic models (Oster & Alberch, 1981), although it has been explicitly studied only in a subset of these models (Hogeweg, 2000; Salazar-Ciudad & Marín-Riera, 2013; Crombach et al., 2016; Verd et al., 2019). This coincides with the evo-devo view that, because of how genes and epigenetic factors interact during development, some aspects of morphology are more variable than others (Alberch, 1982; Horder, 1989). Similar results are obtained in the GPM models of RNA and protein folding (Schuster et al., 1994). In this case, there are also many studies measuring how different phenotypes are more or less likely to arise by mutations and how the same phenotype can arise from different genotypes (Ahnert, 2017; Ferrada & Wagner, 2012).
As explained in the previous sections, the purely genetic models, except for Wagner’s model, either assume that all aspects of the phenotype are equally likely to change or have no way to explain why some aspects of the phenotype are more variable than others, other than natural selection-based arguments. In addition, they assume that all traits can change unlimitedly by mutation.
This difference implies that the genetic-epigenetic models provide a more general depiction of the GPM and of the variability of the phenotype. In other words, the purely genetic models only consider the possibility that all traits are equally likely to vary while the genetic-epigenetic models can explain, even within a single model (Salazar-Ciudad & Jernvall, 2005; Cotterell & Sharpe, 2010), many other possibilities. Again, the purely genetic models do in fact make an assumption, that all traits are free to vary indefinitely, that the genetic-epigenetic models do not usually make.
10.4 Genetic-Epigenetic Models Can Explain Changes in Phenotypic Dimensionality and Novelty
The fourth main difference between genetic-epigenetic models and purely genetic models is related to the dimensionality of the phenotypic changes they can consider. Quantitative and evolutionary genetics conceptualize the phenotype as a set of quantitative traits. Although these approaches consider that the value of each trait can vary without limit, they do not consider that the nature and number of these traits can itself evolve (Müller & Wagner, 1991). In Hansen’s model, for example, one has to specify the number of traits from the beginning and this number will not change over time. In Wagner’s model, since each phenotypic trait is the expression of a gene, the number of genes defines the dimensionality of the phenotype.
Evolution, however, cannot be reduced to quantitative changes in previously existing traits. As an extreme example, it is clear that one cannot derive a human from gradually changing the traits one could measure in a distant bacteria-like ancestor: the nature and number of traits has dramatically changed in evolution. This kind of changes are sometimes called, or related to the concept of, novelty (Müller & Wagner, 1991).
In many genetic-epigenetic models, some novelty can arise. This is specially evident in models that include cell division or growth since there are then new traits being created over the time in the model. In the tooth model, for example, mutations from one individual can lead to individuals with novel cusps. These are due to simple changes on how strongly genes interact with each other or on how strongly they affect cell division (Salazar-Ciudad & Jernvall, 2004, 2010). These new cusps cannot be defined as arising from other cusps or as arising from some traits measurable in its ancestors: they arise where there was a feature-less flat area on the tooth and the show no relationship to previously existing cusps.
Again the genetic-epigenetic models turn out to be more general since they do not necessarily assume that variation occurs only as quantitative variation in existing traits but can consider a much larger class of phenotypic changes.
10.5 Genetic-Epigenetic Models Can Explain How the GPM Evolves
Ideally, any model on the GPM should give some hints about how the GPM itself evolves, otherwise its utility is restricted to the short time scale in which the GPM itself is not expected to evolve much. Here again, the differences between the two approaches are fundamental. The purely genetic models have no way to address this question other that assuming, as in the second-order neo-Darwinian view described above, that all aspects of the GPM are equally likely to change by mutation and that then, the GPM smoothly changes in the direction imposed by natural selection. From this perspective past natural selection would be the only factor determining the evolution of the GPM.
In the case of the genetic-epigenetic models, as explained, the nature of the GPM arises from the genetic and epigenetic interactions included in the model. The evolution of the GPM can then be studied by making changes in the genetic interactions, i.e., the gene network. This approach has been taken in a number of articles simulating evolution and development (Hogeweg, 2000; Salazar-Ciudad, 2001a; Salazar-Ciudad & Marín-Riera, 2013; Crombach et al., 2016; Hagolani et al., 2019). Some of these articles show, for example, that the most parsimonious way to evolve a complex phenotypes leads to a highly complex, non-linear GPM where parts of the phenotype cannot vary independently (Salazar-Ciudad et al., 2001a, b).
Many other rules or trends of change in development and the GPM have been hypothesized over the years, either based on models or not: constructional constraints on how developmental stages can be put onto each other over developmental time (Alberch & Blanco, 1996), rules arising from the intrinsic material properties of cells and tissues (Newman & Müller, 2000), from the structure of morphogenetic fields (Webster & Goodwin, 1996) from the logic of gene networks (Kauffman, 1993) or from the limited number of ways in which genes and cells can be wired to lead to pattern formation and morphogenesis (Salazar-Ciudad et al., 2000; Von Dassow et al., 2000).
11 Conclusions
From the previous discussion, it should become apparent that the genetic-epigenetic models describe some important and general properties of GPMs that cannot be described by models that do not include epigenetic factors. First, they explain phenotypes and the GPM based on genetic interactions and epigenetic factors without making assumptions on the nature of the GPM itself. Second, they restrict the space of possible gene network topologies to the ones capable of performing biological functions. Third, they can help to explain which directions of phenotypic variation are more likely by genetic mutation and why. Fourth, they can explain some novelty, and, thus, changes in the dimension of phenotypic variation. Fifth, they can explain how the GPM itself evolves and, thus, they have a stronger explanatory power in evolution.
We think that the generality of models should be measured based on the number of features of reality they can reproduce. Purely genetic models cannot reproduce many of the features of GPMs and, thus, should be considered non-general. Genetic-epigenetic models are specific of a specific phenotypic level. In spite of that, however, many of them reproduce more features of real GPMs than purely genetic models. Thus, even if the details may differ between genetic-epigenetic models, they should be regarded, overall, as a more general description of the GPM than purely genetic models. This view has already been put in practice in some research. Models of specific phenotypes, the tooth model, have been used as a general models of evolution under realistically complex GPMs (Salazar-Ciudad & Marín-Riera, 2013; Milocco & Salazar-Ciudad, 2020). Other authors have gone even further and suggested that secondary RNA structure models can be used to model evolution at other phenotypic levels such as morphology because they capture crucial features of the GPM that are not captured by purely genetic models (Fontana, 2002).
References
Ahnert, S. E. (2017). Structural properties of genotype-phenotype maps. Journal of the Royal Society Interface, 14(132), 132–141.
Alberch, P. (1982). Developmental constraints in evolutionary processes. In J. T. Bonner (Ed.), Evolution and development. Dahlem Konferenzen (pp. 313–332). Springer.
Alberch, P. (1991). From genes to phenotype: Dynamical systems and evolvability. Genetica, 84, 5–11.
Alberch, P., & Blanco, M. J. (1996). Evolutionary patterns in ontogenetic transformation: From laws to regularities. The International Journal of Developmental Biology, 40(4), 845–858.
Arthur, W. (2001). Developmental drive: An important determinant of the direction of phenotypic evolution. Evolution & Development, 3, 271–278.
Barton, N. H., & Turelli, M. (1987). Adaptive landscapes, genetic distance and the evolution of quantitative characters. Genetical Research, 49(2), 157–173.
Bastock, R., & St Johnston, D. (2008). Drosophila oogenesis. Current Biology, 18, R1082–R1087.
Beloussov, L. V. (1998). The dynamic architecture of a developing organism: An interdisciplinary approach to the development of organisms. Kluwer Academic Publishers.
Brun-Usan, M., Marin-Riera, M., Grande, C., Truchado-Garcia, M., & Salazar-Ciudad, I. (2017). A set of simple cell processes is sufficient to model spiral cleavage. Development, 144, 54–56.
Cheverud, J. M. (2007). The relationship between development and evolution through heritable variation. In G. Bock & J. Goode (Eds.), Tinkering: The microevolution of development. John Wiley & Sons, Ltd.
Crombach, A., Wotton, K. R., Jiménez-Guri, E., & Jaeger, J. (2016). Gap gene regulatory dynamics evolve along a genotype network. Molecular Biology and Evolution, 33, 1293–1307.
Cotterell, J., & Sharpe, J. (2010). An atlas of gene regulatory networks reveals multiple three-gene mechanisms for interpreting morphogen gradients. Molecular Systems Biology, 6, 425.
Cowperthwaite, M. C., & Meyers, L. A. (2007). How mutational networks shape evolution: Lessons from RNA models. Annual Review of Ecology, Evolution, and Systematics, 38, 203–230.
Crow, J. F. (2010). On epistasis: Why it is unimportant in polygenic directional selection. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 365, 1241–1244.
de Beer, G. R. (1930). Embryology and evolution. Claredon Press.
Delile, J., Herrmann, M., Peyriéras, N., & Doursat, R. (2017). A cell-based computational model of early embryogenesis coupling mechanical behaviour and gene regulation. Nature Communications, 8, 13929.
Draghi, J. A., & Whitlock, M. C. (2012). Phenotypic plasticity facilitates mutational variance, genetic variance, and evolvability along the major axis of environmental variation. Evolution, 66(9), 2891–2902.
Ferrada, E., & Wagner, A. (2012). A comparison of genotype-phenotype maps for RNA and proteins. Biophysical Journal, 102, 1916–1925.
Fierst, J. L. (2011). A history of phenotypic plasticity accelerates adaptation to a new environment. Journal of Evolutionary Biology, 24, 1992–2001.
Fierst, J. L., & Hansen, T. F. (2010). Genetic architecture and postzygotic reproductive isolation: Evolution of Bateson-Dobzhansky-Muller incompatibilities in a polygenic model. Evolution, 64, 675–693.
Fisher, R. A. (1930). The genetical theory of natural selection. Oxford University Press.
Fontana, W. (2002). Modelling ‘evo-devo’ with RNA. BioEssays, 24(12), 1164–1177.
Forgacs, G., & Newman, S. A. (2005). Biological physics of the developing embryo. Cambridge University Press.
Gavrilets, S., & Dejong, G. (1993). Pleiotropic models of polygenic variation, stabilizing selection, and epistasis. Genetics, 134, 609–625.
Gilbert, S. F., & Barresi, M. J. F. (2016). Developmental biology. Oxford University Press.
Gilbert, S. F., & Raunio, A. M. (1997). Embryology: Constructing the organism. Sinauer Associates.
Gjuvsland, A. B., Vik, J. O., Beard, D. A., Hunter, P. J., & Omholt, S. W. (2013). Bridging the genotype-phenotype gap: What does it take? The Journal of Physiology, 591, 2055–2066.
Goldberg, J. A., & Bergman, H. (2011). Computational physiology of the neural networks of the primate globus pallidus: Function and dysfunction. Neuroscience, 198, 171–192.
Goldschmidt, R. (1940). The material basis of evolution. Yale University Press.
Griffiths, A. J. (2002). Modern genetic analysis: Integrating genes and genomes (Vol. 2). Macmillan.
Guillot, C., & Lecuit, T. (2013). Mechanics of epithelial tissue homeostasis and morphogenesis. Science, 340, 1185–1189.
Hagolani, P. F., Zimm, R., Marin-Riera, M., & Salazar-Ciudad, I. (2019). Cell signaling stabilizes morphogenesis against noise. Development, 146(20), 18.
Haig, D. (2004). The (dual) origin of epigenetics. Cold Spring Harbor Symposia on Quantitative Biology, 69, 67–70.
Hansen, T. F., Alvarez-Castro, J. M., Carter, A. J., Hermisson, J., & Wagner, G. P. (2006). Evolution of genetic architecture under directional selection. Evolution, 60, 1523–1536.
Hansen, T. F., & Wagner, G. P. (2001). Modeling genetic architecture: A multilinear theory of gene interaction. Theoretical Population Biology, 59, 61–86.
Harris, M. P., Williamson, S., Fallon, J. F., Meinhardt, H., & Prum, R. O. (2005). Molecular evidence for an activator-inhibitor mechanism in development of embryonic feather branching. Proceedings of the National Academic Science U S A, 102, 11734–11739.
Hogeweg, P. (2000). Evolving mechanisms of morphogenesis: On the interplay between differential adhesion and cell differentiation. Journal of Theoretical Biology, 203, 317–333.
Honda, H., Motosugi, N., Nagai, T., Tanemura, M., & Hiiragi, T. (2008). Computer simulation of emerging asymmetry in the mouse blastocyst. Development, 135, 1407–1414.
Horder, T. J. (1989). Syllabus for an embryological synthesis. In D. B. In Wake & G. Roth (Eds.), Complex organismal functions: Integration and evolution (pp. 315–348). John Wiley.
Houle, D., Govindaraju, D. R., & Omholt, S. (2010). Phenomics: The next challenge. Nature Reviews. Genetics, 11, 855–866.
Jablonka, E., & Lamb, M. J. (2005). Evolution in four dimensions: Genetic, epigenetic, behavioral, and symbolic variation in the history of life. MIT Press.
Jaeger, J., Blagov, M., Kosman, D., Kozlov, K. N., Myasnikova, E., Surkova, S., Vanario-Alonso, C. E., Samsonova, M., Sharp, D. H., & Reinitz, J. (2004). Dynamical analysis of regulatory interactions in the gap gene system of Drosophila melanogaster. Genetics, 167, 1721–1737.
Jiménez, A., Cotterell, J., Munteanu, A., & Sharpe, J. (2015). Dynamics of gene circuits shapes evolvability. Proceedings of the National Academy of Sciences of the United States of America, 112(7), 2103–2108.
Jones, A. G., Arnold, S. J., & Bürger, R. (2004). Evolution and stability of the G-matrix on a landscape with a moving optimum. Evolution, 58(8), 1639–1654.
Kandler-Singer, I., & Kalthoff, K. (1976). RNase sensitivity of an anterior morphogenetic determinant in an insect egg (Smittia sp, Chironomidae, Diptera). Proceedings of the National Academic Science U S A, 73, 3739–3743.
Karr, J. R., Sanghvi, J. C., Macklin, D. N., Gutschow, M. V., Jacobs, J. M., Bolival, B., Jr., Assad-Garcia, N., Glass, J. I., & Covert, M. W. (2012). A whole-cell computational model predicts phenotype from genotype. Cell, 150, 389–401.
Kauffman, S. A. (1993). The origins of order. Oxford University Press.
Lande, R., & Arnold, S. J. (1983). The measurement of selection on correlated characters. Evolution, 37, 1210–1226.
Le Cunff, Y., & Pakdaman, K. (2012). Phenotype-genotype relation in Wagner’s canalization model. Journal of Theoretical Biology, 314, 69–83.
Marin-Riera, M., Brun-Usan, M., Zimm, R., Välikangas, T., & Salazar-Ciudad, I. (2016). Computational modeling of development by epithelia, mesenchyme and their interactions: A unified model. Bioinformatics, 32(2), 219–225.
Marin-Riera, M., Moustakas-Verho, J., Savriama, Y., Jernvall, J., & Salazar-Ciudad, I. (2018). Differential tissue growth and cell adhesion alone drive early tooth morphogenesis: An ex vivo and in silico study. PLoS Computational Biology, 14(2), e1005981.
Mayr, E., & Provine, W. (Eds.). (1980). The evolutionary synthesis: Perspectives on the unification of biology. Harvard University Press.
Meinhardt, H. (1982). Models of biological pattern formation. Academic Press.
Milocco, L., & Salazar-Ciudad, I. (2020). Is evolution predictable? Quantitative genetics under complex genotype-phenotype maps. Evolution, 74(2), 230–244.
Mjolsness, E., Sharp, D. H., & Reinitz, J. (1991). A connectionist model of development. Journal of Theoretical Biology, 152, 429–453.
Moustakas-Verho, J. E., Zimm, R., Cebra-Thomas, J., Lempiäinen, N. K., Kallonen, A., Mitchell, K. L., Hämäläinen, K., Jernvall, J., & Gilbert, S. F. (2014). The origin and loss of periodic patterning in the turtle shell. Development, 141(15), 3033–3039.
Müller, G. B. (2007). Evo-devo: Extending the evolutionary synthesis. Nature Reviews. Genetics, 8, 943–949.
Müller, G. B., & Wagner, G. P. (1991). Novelty in evolution: Restructuring the concept. Annual Review of Ecology and Systematics, 22, 229–256.
Neuman-Silberberg, F. S., & Schupbach, T. (1993). The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGF-alfa like protein. Cell, 75, 165–174.
Newman, S. A. (2011a). Animal egg as evolutionary innovation: A solution to the “embryonic hourglass” puzzle. Journal of Experimental Zoology. Part B, Molecular and Developmental Evolution, 316, 467–483.
Newman, S. A. (2011b). The developmental specificity of physical mechanisms. Ludus Vitalis, 36, 343–351.
Newman, S. A., Christley, S., Glimm, T., Hentschel, H. G., Kazmierczak, B., Zhang, Y. T., Zhu, J., & Alber, M. (2008). Multiscale models for vertebrate limb development. Current Topics in Developmental Biology, 81, 311–340.
Newman, S. A., & Comper, W. D. (1990). ‘Generic’ physical mechanisms of morphogenesis and pattern formation. Development, 110, 1–18.
Newman, S. A., & Müller, G. B. (2000). Epigenetic mechanisms of character origination. The Journal of Experimental Zoology, 288, 304–317.
Noble, D. (2002). Modeling the heart – From genes to cells to the whole organ. Science, 295, 1678–1682.
Nowak, M. A., Boerlijst, M. C., Cooke, J., & Smith, J. M. (1997). Evolution of genetic redundancy. Nature, 388, 167–171.
Odell, G. M., Oster, G., Alberch, P., & Burnside, B. (1981). The mechanical basis of morphogenesis: I Epithelial folding and invagination. Developmental Biology, 85, 446–462.
Onimaru, K., Marcon, L., Musy, M., Tanaka, M., & Sharpe, J. (2016). The fin-to-limb transition as the re-organization of a Turing pattern. Nature Communications, 7, 11582.
Oster, G. F., & Alberch, P. (1981). Evolution and bifurcation of developmental programs. Evolution, 36, 444–459.
Osterfield, M., Du, X., Schüpbach, T., Wieschaus, E., & Shvartsman, S. Y. (2013). Three-dimensional epithelial morphogenesis in the developing Drosophila egg. Developmental Cell, 24, 400–410.
Oyama, S. (2000). The ontogeny of information: Developmental systems and evolution (2nd ed.). Duke University Press.
Pigliucci, M. (2006). Genetic variance–covariance matrices: A critique of the evolutionary quantitative genetics research program. Biology and Philosophy, 21(1), 1–23.
Pinho, R., Borenstein, E., & Feldman, M. W. (2012). Most networks in Wagner’s model are cycling. PLoS One, 7, e34285.
Ray, R. P., Matamoro-Vidal, A., Ribeiro, P. S., Tapon, N., Houle, D., Salazar-Ciudad, I., & Thompson, B. J. (2015). Patterned anchorage to the apical extracellular matrix defines tissue shape in the developing appendages of Drosophila. Developmental Cell, 34(3), 310–322.
Rice, S. H. (2004). Developmental associations between traits: Covariance and beyond. Genetics, 166(1), 513–526.
Roff, D. A. (2000). The evolution of G-Matrix: Selection or drift? Heredity, 84, 135–142.
Roff, D. A. (2007). A centennial celebration for quantitative genetics. Evolution, 61, 1017–1032.
Roth, S., & Lynch, J. A. (2009). Symmetry breaking during Drosophila oogenesis. Cold Spring Harbor Perspectives in Biology, 1, a001891.
Rothschild, J. B., Tsimiklis, P., Siggia, E. D., & François, P. (2016). Predicting ancestral segmentation phenotypes from Drosophila to Anopheles using in silico evolution. PLoS Genetics, 12(5), e1006052.
Salazar-Ciudad, I. (2006a). Developmental constraints vs variational properties: How pattern formation can help to understand evolution and development. Journal of Experimental Zoology. Part B, Molecular and Developmental Evolution, 306, 107–125.
Salazar-Ciudad, I. (2008). Evolution in biological and nonbiological systems under different mechanisms of generation and inheritance. Theory in Biosciences, 127(4), 343–358.
Salazar-Ciudad, I., Garcia-Fernandez, J., & Solé, R. V. (2000). Gene networks capable of pattern formation: From induction to reaction-diffusion. Journal of Theoretical Biology, 205, 587–603.
Salazar-Ciudad, I., & Jernvall, J. (2004). How different types of pattern formation mechanisms affect the evolution of form and development. Evolution & Development, 6, 6–16.
Salazar-Ciudad, I., & Jernvall, J. (2005). Graduality and innovation in the evolution of complex phenotypes: Insights from development. Journal of Experimental Zoology. Part B, Molecular and Developmental Evolution, 304, 619–631.
Salazar-Ciudad, I., & Jernvall, J. (2010). A computational model of teeth and the developmental origins of morphological variation. Nature, 464, 583–586.
Salazar-Ciudad, I., Jernvall, J., & Newman, S. A. (2003). Mechanisms of pattern formation in development and evolution. Development, 130, 2027–2037.
Salazar-Ciudad, I., & Marín-Riera, M. (2013). Adaptive dynamics under development-based genotype-phenotype maps. Nature, 497, 361–364.
Salazar-Ciudad, I., Newman, S. A., & Solé, R. V. (2001a). Phenotypic and dynamical transitions in model genetic networks. I Emergence of patterns and genotype-phenotype relationships. Evolution & Development, 3, 84–94.
Salazar-Ciudad, I., Solé, R. V., & Newman, S. A. (2001b). Phenotypic and dynamical transitions in model genetic networks. II Application to the evolution of segmentation mechanisms. Evolution & Development, 3(2), 95–103.
Schuster, P., Fontana, W., Stadler, P. F., & Hofacker, I. L. (1994). From sequences to shapes and back: A case study in RNA secondary structures. Proceedings of the Royal Society of London B, 255, 279–284.
Silva, J. R., Pan, H., Wu, D., Nekouzadeh, A., Decker, K. F., Cui, J., Baker, N. A., Sept, D., & Rudy, Y. (2009). A multiscale model linking ion-channel molecular dynamics and electrostatics to the cardiac action potential. Proceedings of the National Academic Science U S A, 106, 11102–11106.
Turelli, M., & Barton, N. H. (1994). Genetic and statistical analyses of strong selection on polygenic traits: What, me normal? Genetics, 138, 913–941.
Uricchio, L. H. (2020). Evolutionary perspectives on polygenic selection, missing heritability, and GWAS. Human Genetics, 139(1), 5–21.
Verd, B., Monk, N. A. M., & Jaeger, J. (2019). Modularity, criticality, and evolvability of a developmental gene regulatory network. eLife, 8, e42832.
Vik, J. O., Gjuvsland, A. B., Li, L., Tøndel, K., Niederer, S., Smith, N. P., Hunter, P. J., & Omholt, S. W. (2011). Genotype-phenotype map characteristics of an in silico heart cell. Frontiers in Physiology, 2, 106.
Von Dassow, G., Meir, E., Munro, E. M., & Odell, G. M. (2000). The segment polarity network is a robust developmental module. Nature, 406, 188–192.
Waddington, C. H. (1942). The epigenotype. Endeavour, 1, 18–20.
Waddington, C. H. (1957). The strategy of the genes (Vol. 63, pp. 375–384). Allen and Unwin.
Waddington, C. H. (1968). The theory of evolution today. In A. Koestler & J. R. Smythies (Eds.), Beyond reductionism. Columbia Univ. Press.
Wagner, A. (1994). Evolution of gene networks by gene duplications: A mathematical model and its implications on genome organization. Proceedings of the National Academic Science U S A, 91, 4387–4391.
Wagner, A. (1996). Does evolutionary plasticity evolve? Evolution, 50, 1008–1023.
Wagner, A. (2011). Genotype networks shed light on evolutionary constraints. Trends in Ecology & Evolution, 26, 577–584.
Wagner, G. P., Booth, G., & Bagheri, H. C. (1997). A population genetic theory of canalization. Evolution, 51, 329–347.
Wagner, G. P., Pavlicev, M., & Cheverud, J. M. (2007). The road to modularity. Nature Reviews. Genetics, 8, 921–931.
Webster, G., & Goodwin, B. (1996). Form and transformation. In Generative and relational principles in biology. Cambridge University Press.
Weiss, K. M., & Fullerton, S. M. (2000). Phenogenetic drift and the evolution of genotype-phenotype relationships. Theoretical Population Biology, 57, 187–195.
Wright, S. (1932). The roles of mutation, inbreeding, crossbreeding and selection in evolution. In D. F. Jones (Ed.), Proceedings of the Sixth International Congress of Genetics (Vol. 1, pp. 356–366).
Zhivotovsky, L. A., & Feldman, M. W. (1992). On models of quantitative genetic variability: A stabilizing selection-balance models. Genetics, 130, 947–955.
Zhu, J., Zhang, Y. T., Alber, M. S., & Newman, S. A. (2010). Bare bones pattern formation: A core regulatory network in varying geometries reproduces major features of vertebrate limb development and evolution. PLoS One, 5, e10892.
Acknowledgments
We thank J. Jernvall, I. Salvador-Martinez, P. Hagolani, T. Välikangas, K. Ruiz-Mirazo, A. Moreno, S. A. Newman and R. Zimm for comments. This research was funded by the Finnish Academy (WBS 1250271) and the Spanish Ministry of Science and Innovation (BFU2010-17044) to I.S.C., the Spanish Ministry of Science and Innovation (BES2011-046641) to M.B.U., and the Generalitat de Catalunya (2013FI_B00439) to M.M.R.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Salazar-Ciudad, I., Marín-Riera, M., Brun-Usan, M. (2021). Understanding the Genotype-Phenotype Map: Contrasting Mathematical Models. In: Crombach, A. (eds) Evolutionary Systems Biology. Springer, Cham. https://doi.org/10.1007/978-3-030-71737-7_10
Download citation
DOI: https://doi.org/10.1007/978-3-030-71737-7_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-71736-0
Online ISBN: 978-3-030-71737-7
eBook Packages: Computer ScienceComputer Science (R0)