Abstract
Training classification models in the medical domain is often difficult due to data heterogeneity (related to acquisition procedures) and due to the difficulty of getting sufficient amounts of annotations from specialized experts. It is particularly true in digital pathology, where models do not generalize easily. This paper presents a novel approach for the generalization of models in conditions where heterogeneity is high and annotations are few. The approach relies on a semi-supervised teacher/student paradigm to different datasets and annotations. The paradigm combines a small amount of strongly-annotated data, with a large amount of unlabeled data, for training two Convolutional Neural Networks (CNN): the teacher and the student model. The teacher model is trained with strong labels and used to generate pseudo-labeled samples from the unlabeled data. The student model is trained combining the pseudo-labeled samples and a small amount of strongly-annotated data. The paradigm is evaluated on the student model performance of Gleason pattern and Gleason score classification in prostate cancer images and compared with a fully-supervised learning approach for training the student model. In order to evaluate the capability of the approach to generalize, the datasets used are highly heterogeneous in visual characteristics and are collected from two different medical institutions. The models, trained with the teacher/student paradigm, show an improvement in performance above the fully-supervised training. The models generalize better on both the datasets, despite the inter-datasets heterogeneity, alleviating the overfitting. The classification performance shows an improvement both in the classification of Gleason pattern at patch level (\(\kappa \,=\,0.6129\, \pm \, 0.0127\) from \(\kappa \,=\,0.5608\pm 0.0308\)) and at in Gleason score classification, evaluated at WSI-level \(\kappa \,=\,0.4477\pm 0.0460\) from \(\kappa \,=\,0.2814\pm 0.1312\)).
Both authors contributed equally to this work. S. Otálora thanks to Colciencias for partially funding his Ph.D. studies through the call “756 - Doctorados en el exterior”.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
The lack of large datasets with local annotations and the highly-heterogeneous data represents a critical challenge for developing machine learning algorithms that generalize well in the digital pathology domain [8], despite the increasing amount of datasets available with repositories such as TCGA (The Cancer Genome Atlas).
Machine learning algorithms, particularly Convolutional Neural Networks (CNNs), are the state-of-the-art for analyzing digital pathology images [23, 35] (such as whole slide images, WSIs, or tissue-micro-arrays, TMAs). CNN models usually require large datasets with local annotations to train robust models [17] that generalize well to unseen data [6]. The annotation of digital pathology images is a time-consuming and expensive process that requires medical experts, such as the pathologists. Therefore, only a small amount among the publicly available datasets is locally annotated, e.g. the Camelyon dataset [22].
Despite the small amount of datasets that are locally annotated, an increasing number of histopathological images datasets is available, e.g. The Cancer Genome Atlas (TCGA)Footnote 1. Most of these datasets come without local annotations (strong annotations) of the region of interest for the diagnosis. Some of these datasets are released with medical reports and some are unlabeled. The reports include the final diagnosis, among other information, that can instead be used as weak annotations for digital pathology images.
The amount of strongly-annotated data is much smaller than the unlabeled and weakly-annotated data. This fact constitutes a challenge for training supervised CNN models in a fully-supervised fashion. Furthermore, histopathological images that come from different sources are highly-heterogeneous, as a consequence of the acquisition procedures applied to the samples. Hematoxylin and eosin (H&E) represent the golden standard for staining the samples within a WSI [10]. Although H&E is a standard, their preparation procedures are not fully standardized, often leading to inter-dataset heterogeneity [19, 37]. This heterogeneity leads to models that are more prone to overfit, compared with models trained in conditions where the inter-dataset heterogeneity is not present. Therefore, many CNN models, trained to analyze histopathological images, face a decrease in their performance when they are tested on data originated from a different source, as shown in previous works [32, 33].
Despite the lack of large datasets that are locally annotated and the highly-heterogeneous data, new methods were proposed recently for training the models with small datasets of local annotations, showing partial success, such as semi-supervised learning [3, 11, 14, 20, 21, 24, 26, 28, 31, 38], active learning [27, 29, 30, 39] and weakly supervised learning [1, 4, 6, 18, 25, 32, 36]. This paper represents a novelty in a domain where there is a lack of large datasets with local annotations and the data are highly heterogeneous. The semi-supervised teacher/student paradigm [13, 20, 31, 32] is applied to the digital pathology task of prostate cancer classification, using two datasets.
Prostate cancer (PCa) is the fourth most frequent cancer in the entire human populationFootnote 2. Prostate cancer is diagnosed using the Gleason grading system, which is based on two steps: first, the identification of Gleason patterns, second the computation of the Gleason Score. The identification of Gleason patterns is made to estimate the aggressiveness of cancer. The tissue structures in a sample are distinguished in different Gleason patterns, according to their cell abnormality and their gland deformation. The Gleason patterns range from 1 to 5. According to the guidelines described by the Union for International Cancer Control and the World Health Organization/International Society of Urological Pathology, the Gleason score is computed by evaluating the most diffused primary and secondary patterns. Typically, malignant prostate cancer has a Gleason score ranged from 6 to 10. The recent advancements in the digital pathology cancer prostate classification task are summarized in the Table 1.
In this paper, two highly-heterogeneous datasets are used for training the models: a small strongly-labeled dataset with pixel-wise annotations and a large unlabeled dataset of whole slide images. The strongly-annotated dataset is the Tissue Micro-Arrays Zurich dataset (TMAZ). The non locally annotated dataset is a cohort of The Cancer Genome Atlas PRostate ADenocarcinoma (TCGA-PRAD).
The approach proposed follows the teacher/student paradigm and consists of two models: a high-capacity model, called teacher model, and a smaller model, called the student model. The teacher model generates pseudo-labeled examples from the unlabeled data. The student model is trained combining the pseudo-labeled examples and the strongly-annotated data.
The teacher and the student models are implemented using large pre-trained models and following the paradigm constraints. The teacher model must be a high-capacity model, while the student model must be efficient at test time. The teacher model is a high-capacity ResNexT based model (22 million of parameters), pre-trained with a dataset of one billion natural images retrieved from Instagram [38]. The model is trained with the strongly-annotated data and it creates the pseudo-labeled examples annotating the unlabeled data. The student model is a DenseNet121, pre-trained with ImageNet weights. The student architecture is a small model, compared with the model used for implementing the teacher. The model is trained first with the pseudo-labeled data and then fine-tuned with the strongly-annotated data. The models’ performance is compared with the fully-supervised learning of the student model, considered as the baseline. The teacher/student paradigm, as shown in the experimental results, performs better than the fully-supervised CNN (trained only with strongly-annotated data), both at the Gleason pattern level and at the Gleason score level. The approach allows leveraging large unlabeled datasets as a source of supervision for training CNN models in digital pathology. This work is included in a bigger study on semi-supervised and semi-weakly supervised learning approaches, partly presented in Otálora et al. [26]. The difference between the approaches regards the steps included in order to train the teacher model: while the semi-weakly supervised learning approach previously described includes additional training components based on weak labels from the WSI, the semi-supervised approach described in this paper does not use any labels from the WSI dataset.
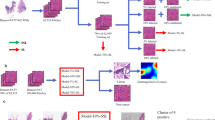
Overview of the teacher/student training model. In step one, the teacher is trained with strongly-annotated data. In step two, the teacher predicts the class probabilities for the unlabeled data. In step three, the samples with the highest probabilities are selected (pseudo-labeled data). In step four, the student model is trained using the pseudo-labeled data. In step five, the student model is trained using the strongly-annotated data.
2 Methods
2.1 Datasets
Two open-access datasets are adopted for the evaluation of the teacher/student paradigm. They are highly heterogeneous, which makes them similar to real clinical classification problems, and they are pre-processed with the same approach. In both datasets, the images are pre-processed dividing them into patches and removing the background regions. The images are divided into tiles of 750 \(\times \) 750 pixels, and then they are resized to 224 \(\times \) 224 pixels to fit as input to the chosen networks. Only the patches extracted from tissue regions are selected (background regions are non-informative). The HistoQC tool [16] is used for generating tissue masks of the images that come without local annotations so that only patches that include tissue are extracted. The two datasets are the tissue microarray dataset (TMAZ) released by Arvanity et al. [2] and a cohort of the TCGA-PRAD datasetFootnote 3. The TMAZ includes 886 prostate TMA core images with pixel-wise annotations, made by pathologists. Each TMA core has a size of \(3100^{2}\) pixels, scanned at 40x resolution (0.23 microns per pixel). The arrays are scanned at the same medical center, the University Hospital of Zurich (NanoZoomer-XR Digital slide scanner, Hamamatsu). The TMAZ includes four classes: benign, Gleason pattern 3, Gleason pattern 4, Gleason pattern 5. It is split into three partitions: the training partition is composed of 508 cores, the validation partition is composed of 133 cores, and the test partition of 245 cores. The partitions of the dataset are shown in the left part of Table 2. From each TMAZ core, 30 patches are randomly extracted. The number of patches to extract is chosen considering the trade-off between the patch size and the whole tissue covered within the TMA. The number of patches for each class is summarized in Table 3.
TCGA-PRADFootnote 4 is a data repository including up to 490 tissue slides of digitized prostactectomies (made up of \(100'000^{2}\) pixels), preserved with paraffin embeddings or frozen tissues, with no pixel-wise annotations. The cohort of the TCGA-PRAD dataset used in this work includes only 301 WSIs from the original dataset, preserved only with paraffin embeddings sections. The WSIs come without pixel-wise annotations and are paired with their primary and secondary Gleason pattern within the corresponding pathology report. The WSIs in the cohort are collected from 20 medical centers. This large number of medical centers leads to a highly heterogeneous WSIs. The dataset is split into three partitions (as shown in the right part of Table 2): the training set is composed of 171 WSIs, the validation set composed of 84 WSI, and the test set composed of 46 WSIs. In this paper, the TCGA-PRAD patches are annotated with pseudo-labels by the teacher model. It predicts a probability vector for each of the patches within the WSIs. The probability vectors are sorted in descending order by the class probabilities and the top-ranked K patches are selected. Different values of K are tested for the training partitions of pseudo-labeled data. They vary between 1000 and 10’000 patches per class and they are explored increasing the value of 1000 patches per class, between two consecutive K values. Therefore 1000 patches per class are included in the first subset and 2000 per class in the second one. The validation and test partition include both 8000 patches (2000 samples for each class).
2.2 Teacher/Student Paradigm
The presented semi-supervised learning approach is a pipeline based on teacher/student paradigm [13, 20]. Figure 1 shows an overview of the training schema. The paradigm includes two distinct CNNs, called respectively the teacher model and the student model. The teacher model is a high-capacity neural network, trained to annotate pseudo-labeled examples from the unlabeled data. The pseudo-labels are the labels predicted by a model, in this case, the teacher model [20]. They are assigned considering the prediction vector and selecting the class with the maximum predicted probability. The pseudo-labels do not come from experts, therefore some of them match with the correct class (relevant labels) and some of them do not (noisy labels) [14, 38]. Noisy labels can compromise the learning process [14]. The choice to use high-capacity models permits to better separate noisy labels from relevant labels [14]. Furthermore, high-capacity models can better leverage a large amount of data [38]. The teacher model annotates unlabeled data with pseudo-labels that are used for training the student model. The annotation process is made predicting the class probabilities of unlabeled data [20]. The relevant samples are labeled with the highest probabilities for separating them from noisy examples. The student model is a smaller (compared to the teacher) neural network, trained using a combination of pseudo-labeled and strongly-annotated data. The choice to use a smaller network is made so that the model can be highly efficient at test time, but guaranteeing performance comparable to the teacher [12].
The training schema is composed of a pipeline of operations that are summarized here:
-
1.
train the teacher with strongly-annotated data;
-
2.
predict pseudo-labeled data;
-
3.
select pseudo-labeled data;
-
4.
train the student with pseudo-labeled data;
-
5.
fine-tune the student with strongly-annotated data.
In the first step of the training schema, the teacher model is trained with strongly-annotated data. Thus, it learns how to select relevant examples from the unlabeled data. In the second step, the teacher annotates unseen data, generating a prediction vector of the class probabilities from a softmax layer. In the third step, the teacher selects the pseudo-labeled samples to present to the student model. The samples selected are the ones with the highest probability of belonging to a class. The vectors are sorted in descending order by the class probability. K samples per class are selected from the highest-ranked ones [38]. In this step, it is essential to minimize the number of noisy samples selected [14]. Therefore, the right K value must be selected. However, this value is not possible to be identified a priori. In the fourth step, the student model is trained using the pseudo-labeled data. In this step, it is possible to explore different K values. Therefore, the model is trained with different subsets of pseudo-labeled data, each one including a different number of pseudo-labels per class. Among these models, the one that shows the best performance is the one trained with the subset with fewer noisy labels. Indeed, this subset includes the smallest number of noisy labels, compared with the others. In the fifth step, the student model is fine-tuned using the strongly-annotated data. The learning paradigm is tested on the student model. The model is tested in two different steps of the pipeline and it is compared with fully-supervised learning approach. Firstly, it is tested after the training with only the pseudo-labeled data (Fig. 1, step 4). Secondly, it is tested after the training with the pseudo-labeled and the fine-tuning with the strongly annotated data (Fig. 1, step 5). In the fully-supervised learning approach, the student model is trained only with strongly-annotated data.
2.3 Implementation
The teacher model is Resnext50_32x4d, while the student model is DenseNet121 [15]. Both networks are implemented in PyTorch (version 1.1.0) and trained on the Cartesius cluster infrastructure, provided by the SURFsara HPC (High-Performance Computing) centreFootnote 5, using Tesla K40m GPUs. Both the architectures are trained with the same strategy to set the hyperparameters. In order to avoid overfitting, class-wise data augmentation is applied during the training, with a probabilistic rate. The strategy for training the models regards the hyperparameters of the network, the weights used for initializing the models and the replacement of the last layer. Both models are trained ten different times, in order to avoid the non-deterministic effects caused by the stochastic gradient descent and the data augmentation pipeline. The average and standard deviation of the models are reported. The teacher model used for annotating the unlabeled data is the one that shows the best performance in the TMAZ validation set among the ten repetitions. The student model, selected to be fine-tuned with strongly annotated data, is the one that shows the best performance on the TMAZ validation set among the ten repetitions. Each of these training repetitions is trained for 15 epochs with a batch size of 32 samples. The hyperparameters adopted are the same for both models: they are optimized using Adam optimizer with a learning rate of 0.001 and a decay rate of \(10^{-6}\). Both the models are initialized with pre-trained weights. The teacher model has the initialized weights pre-trained with the YFCC100M dataset [34], which includes almost 1 billion Instagram images [38]. The student model has the initialized weights pre-trained with ImageNet images [9]. In both models, the architecture is changed for adapting the problem to the number of classes. The last layer of the original network architecture (1000 nodes) is changed with a new dense layer of four nodes (the number of classes in this classification problem). A class-wise data augmentation (CWDA) solution is applied during the training phase of the CNNs. The class-wise data augmentation consists of three operations, applied in order to avoid overfitting. The operations of the pipeline are rotation, flipping and colour augmentation, implemented with the Albumentations open-source library [5]. They are applied to the training images with a probability of 0.5 on each batch. The unbalanced distribution of the classes, combined with the small amount of data, can lead to overfitting. Class-wise data augmentation (CWDA) is applied to reduce the effect of unbalanced classes on training. It is implemented by the GitHub open access repository of UfoynFootnote 6.
3 Results
The models trained with the teacher/student paradigm perform better than the one trained with the fully-supervised training. The performance is evaluated with the weighted Cohen \(\kappa \)-score. The models are trained to classify the Gleason patterns and the Gleason score of histopathological image patches. The Gleason patterns are evaluated on images with annotations manually made by a pathologist, while the Gleason score on the diagnosis included in the medical report. The performance is evaluated on the student model and compared with a fully-supervised learning approach.
The performance is measured by the weighted Cohen \(\kappa \)-score as a function of the amount of pseudo-labeled examples (per class) used for training the student model. The weighted Cohen \(\kappa \)-score is a metric for measuring agreement between raters. The quadratically weighted \(\kappa \) is adopted for penalizing stronger predictions far from their real class. The Gleason score classification is evaluated at the WSI level, while Gleason pattern classification is evaluated at the patch-level. The Gleason score is measured by the aggregation of Gleason patterns at the patch level, using a majority voting system and the rules of the American Urology AssociationFootnote 7. In this paper, the majority voting system is applied only on 1000 patches per WSI, selected with the Blue-ratio technique [7]. Blue-ratio permits to avoid the extraction of patches with a small number of nuclei, such as the ones that contain stroma or fat. The student models are tested also using the Wilcoxon Rank-Sum test, in order to determine if they have the same probabilistic distribution (null hypothesis) of the models trained with the fully-supervised approach. The null hypothesis is tested positive when the p-value > 0.05, while it is tested negative when the p-value < 0.05. Figures 2 and 3 show the performance of the training/student semi-supervised paradigm. In both figures, three curves are present. The blue curve represents the performance measured after training the student model with pseudo-labeled data. The green curve represents the performance measured after training the model with pseudo-labeled data and then fine-tuning it with strongly-annotated data. The dashed black line represents the performance of the fully-supervised training of the student model. The classification performance of Gleason patterns in the TMAZ dataset is presented in Fig. 2, while the classification performance of Gleason scores in TCGA-PRAD is presented in Fig. 3. In Fig. 2, the performance is measured on the TMAZ test set at the patch level. The baseline models (student model trained only with strongly–annotated data) reached a \(\kappa \) = 0.5608 ± 0.0308. Each curve has a peak value since the curves are not monotonically increasing. The performance of the student model trained only with pseudo-labeled data (blue curve) is below the baseline, for each one of the amounts of samples per class tested. The peak value is \(\kappa \) = 0.4434 ± 0.0547, reached with the pseudo-labeled training partition with 9000 patches pseudo-labeled per class. The performance of the student model trained with pseudo-labeled and fine-tuned with strongly-annotated data (green curve) exceeds the baseline, for each one of the amounts of pseudo-labeled data tested. The peak value is \(\kappa \) = 0.6129 ± 0.0127, reached with the pseudo-labeled training partition with 8000 patches pseudo-labeled per class. Therefore, the model trained with pseudo-labeled and fine-tuned with strongly-annotated data exceeds the baseline by 0.052 in \(\kappa \). The improvement obtained is statistically significant (p-value = 0.005 for the peak value). In Fig. 3, the performance is measured on the TCGA-PRAD test set at the WSI level. The baseline models (student model trained only with strongly–annotated data) reached a \(\kappa \) = 0.2814 ± 0.1312. Each curve has a peak value since the curves are not monotonically increasing. The performance of the student model trained only with pseudo-labeled data (blue curve) exceeds the baseline, for each one of the amounts of pseudo-labeled data tested. The peak value is \(\kappa \) = 0.4478 ± 0.0460, reached with the pseudo-labeled training partition with 6000 patches pseudo-labeled per class. The improvement obtained is statistically significant (p-value = 0.012 for the peak value). The lowest performance exceeds the baseline by 0.09 in \(\kappa \), where the model is trained with 5000 pseudo-labeled samples per class. The performance of the student model trained with pseudo-labeled and fine-tuned with strongly-annotated data (green curve) exceeds the baseline, only for a range (from 5000 to 8000) of pseudo-labeled samples per class tested. The peak value is \(\kappa \) = 0.3438 ± 0.0924, reached with the pseudo-labeled training partition with 5000 patches pseudo-labeled per class. The improvement obtained is not statistically significant (p-value = 0.200 for the peak value). Therefore, the baseline is exceeded by 0.062 in \(\kappa \) using the semi-supervised learning. The student model trained with the semi-supervised approach, in both the steps of the pipeline tested, exceed the baseline. The student model trained with pseudo-labeled data exceeds the baseline by 0.166 in \(\kappa \). The student model trained with pseudo-labeled and fine-tuned with strongly-annotated data exceeds the baseline by 0.062 in \(\kappa \). The results are summarized in Table 4.
4 Discussion
The teacher/student paradigm permits to leverage on a large amount of the unlabeled data for training a more robust CNN model and improving its performance in Gleason grading and Gleason scoring classification. The performance classification of the models trained with the paradigm is improved compared to a fully-supervised training schema. A trade-off is identified between the number of pseudo-labeled samples used for training and the model’s classification performance. The paradigm permits to face the heterogeneity between datasets, limiting the overfitting. As expected, in both the Gleason grading and the Gleason scoring, the models trained combining pseudo-labels and strongly-annotated data improve the performance, compared with the fully-supervised schema. This is explainable considering that the amount of data used (combining pseudo-labels and strongly-annotated) is increased. However, the metric curves are not monotonically increasing. A peak value in kappa is identified for each of the approaches tested. This peak value allows exploring the best P parameter for the paradigm. P represents the amount of pseudo-labeled samples per class in a subset. The subset that reaches the peak value has less noisy pseudo-labels, compared with the other subsets. The higher the peak value, the fewer noisy labels are included in pseudo-label samples. Therefore, the higher the peak value, the higher is the performance. The paradigm can alleviate overfitting caused by heterogeneity between datasets, although models tend to adapt their weights to the data with which they are trained (as it was expected). The results show that a model, trained on a dataset, does not generalize well for a different dataset. It is a consequence of the inter-dataset heterogeneity. This effect happens for both the datasets. The student model trained with the TMAZ patches reaches good results in its own set, but it fails to generalize in the TCGA-PRAD test partition, where it obtains some of the worst results (dashed line on Fig. 3). The student model, trained with the pseudo-labeled samples, reaches the best results in TCGA-PRAD test set, but it fails to generalize in the TMAZ test partition, where it reaches the worst results (blue curve in Fig. 2). The inter-dataset heterogeneity is the reason why the student model, trained only with pseudo-labeled data, performs better on TCGA-PRAD dataset, compared with the same model trained combining pseudo-labeled and strongly-annotated data. However, training the model combining the different data sources alleviates the overfitting. On the TMAZ dataset, the model trained with both the dataset obtains the best performance (\(\kappa \) = 0.6129 ± 0.0127), but it does not generalize well for the TCGA-PRAD dataset. The model’s performance is better than the fully-supervised training of the student. However, the same model, trained only with pseudo-labeled data, exceeds this performance by 0.096 in \(\kappa \).
5 Conclusion
In this paper, the classification of prostate cancer tissue is tackled with a novel approach, based on the semi-supervised teacher/student paradigm for training CNNs. It permits face data heterogeneity and alleviates the difficulty of obtaining a sufficient amount of locally annotated data for training the models. The approach is compared with a fully-supervised CNN learning approach. The teacher/student paradigm improves the performance of a CNN prostate cancer classification at the patch level and the WSI level. Therefore, it is possible to adopt it to leverage a large amount of unlabeled data and then improve the fully supervised classification performance of CNNs. Furthermore, the teacher/student paradigm permits to face the heterogeneity of the datasets used for training the models. It permits to generalize better in datasets that come from different medical sources, reducing the effects caused by the overfitting. In the future works, the teacher/student paradigm will be tested on different types of biopsy tissues, with larger values of K parameter and testing more training steps and within the pipeline. The code and all the pre-trained models are made publicly available on Github (https://github.com/ilmaro8/Semi_Supervised_Learning). The pseudo-labeled data are available from the corresponding author on request.
Notes
- 1.
https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga. Retrieved 9th of March, 2020.
- 2.
https://www.wcrf.org/dietandcancer/cancer-trends/worldwide-cancer-data. Retrieved 16th of March, 2020.
- 3.
https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga. Retrieved 9th of March, 2020.
- 4.
https://portal.gdc.cancer.gov/projects/TCGA-PRAD. Retrieved March 1, 2020.
- 5.
https://userinfo.surfsara.nl/systems/hpc-cloud. Retrieved 7th of February, 2020.
- 6.
https://github.com/ufoym/imbalanced-dataset-sampler. Retrieved 6th of February, 2020.
- 7.
References
Arvaniti, E., Claassen, M.: Coupling weak and strong supervision for classification of prostate cancer histopathology images. In: Medical Imaging Meets NIPS Workshop, NIPS 2018 (2018)
Arvaniti, E., et al.: Automated Gleason grading of prostate cancer tissue microarrays via deep learning. Sci. Rep. 8, 1–11 (2018)
Bagherzadeh, J., Asil, H.: A review of various semi-supervised learning models with a deep learning and memory approach. Iran J. Comput. Sci. 2(2), 65–80 (2018). https://doi.org/10.1007/s42044-018-00027-6
Bulten, W., et al.: Automated deep-learning system for Gleason grading of prostate cancer using biopsies: a diagnostic study. Lancet Oncol. 21(2), 233–241 (2020)
Buslaev, A., Parinov, A., Khvedchenya, E., Iglovikov, V.I., Kalinin, A.A.: Albumentations: fast and flexible image augmentations. ArXiv e-prints (2018)
Campanella, G., et al.: Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 25(8), 1301–1309 (2019)
Chang, H., Loss, L.A., Parvin, B.: Nuclear segmentation in H&E sections via multi-reference graph cut (MRGC). In: International Symposium Biomedical Imaging (2012)
Cheplygina, V., de Bruijne, M., Pluim, J.P.: Not-so-supervised: a survey of semi-supervised, multi-instance, and transfer learning in medical image analysis. Med. Image Anal. 54, 280–296 (2019)
Deng, J., Dong, W., Socher, R., Li, L.J., Li, K., Fei-Fei, L.: ImageNet: a large-scale hierarchical image database. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition, pp. 248–255. IEEE (2009)
Fischer, A.H., Jacobson, K.A., Rose, J., Zeller, R.: Hematoxylin and eosin staining of tissue and cell sections. Cold Spring Harb. Protoc. 2008(5), pdb-prot4986 (2008)
Foucart, A., Debeir, O., Decaestecker, C.: Snow: semi-supervised, noisy and/or weak data for deep learning in digital pathology. In: 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), pp. 1869–1872. IEEE (2019)
Guo, T., Xu, C., He, S., Shi, B., Xu, C., Tao, D.: Robust student network learning. IEEE Trans. Neural Netw. Learn. Syst. 31(7), 2455–2468 (2019)
Hady, M.F.A., Schwenker, F.: Semi-supervised learning. In: Bianchini, M., Maggini, M., Jain, L. (eds.) Handbook on Neural Information Processing. Intelligent Systems Reference Library, vol. 49, pp. 215–239. Springer, Heidelberg (2013). https://doi.org/10.1007/978-3-642-36657-4_7
Han, B., et al.: Co-teaching: robust training of deep neural networks with extremely noisy labels. In: Advances in Neural Information Processing Systems, pp. 8527–8537 (2018)
Huang, G., Liu, Z., Weinberger, K.Q.: Densely connected convolutional networks. CoRR abs/1608.06993 (2016). http://arxiv.org/abs/1608.06993
Janowczyk, A., Zuo, R., Gilmore, H., Feldman, M., Madabhushi, A.: HistoQC: an open-source quality control tool for digital pathology slides. JCO Clin. Cancer Inf. 3, 1–7 (2019)
Komura, D., Ishikawa, S.: Machine learning methods for histopathological image analysis. Comput. Struct. Biotechnol. J. 16, 34–42 (2018)
van der Laak, J., Ciompi, F., Litjens, G.: No pixel-level annotations needed. Nat. Biomed. Eng. 3, 1–2 (2019)
Larson, K., Ho, H.H., Anumolu, P.L., Chen, T.M.: Hematoxylin and eosin tissue stain in Mohs micrographic surgery: a review. Dermatol. Surg. 37(8), 1089–1099 (2011)
Lee, D.H.: Pseudo-label: the simple and efficient semi-supervised learning method for deep neural networks. In: Workshop on Challenges in Representation Learning, ICML, vol. 3, p. 2 (2013)
Li, J., et al.: An EM-based semi-supervised deep learning approach for semantic segmentation of histopathological images from radical prostatectomies. Comput. Med. Imaging Graph. 69, 125–133 (2018)
Litjens, G., et al.: 1399 H&E-stained sentinel lymph node sections of breast cancer patients: the CAMELYON dataset. GigaScience 7(6), giy065 (2018)
Litjens, G., et al.: A survey on deep learning in medical image analysis. Med. Image Anal. 42, 60–88 (2017)
Lu, M.Y., Chen, R.J., Wang, J., Dillon, D., Mahmood, F.: Semi-supervised histology classification using deep multiple instance learning and contrastive predictive coding. arXiv preprint arXiv:1910.10825 (2019)
Otálora, S., Atzori, M., Khan, A., Jimenez-del Toro, O., Andrearczyk, V., Müller, H.: A systematic comparison of deep learning strategies for weakly supervised Gleason grading. In: Medical Imaging 2020: Digital Pathology, vol. 11320, p. 113200L. International Society for Optics and Photonics (2020)
Otálora, S., Marini, N., Müller, H., Atzori, M.: Semi-weakly supervised learning for prostate cancer image classification with teacher-student deep convolutional networks. In: Cardoso, J., et al. (eds.) IMIMIC/MIL3ID/LABELS -2020. LNCS, vol. 12446, pp. 193–203. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-61166-8_21
Otálora, S., Perdomo, O., González, F., Müller, H.: Training deep convolutional neural networks with active learning for exudate classification in eye fundus images. In: Cardoso, M.J., et al. (eds.) LABELS/CVII/STENT-2017. LNCS, vol. 10552, pp. 146–154. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-67534-3_16
Peikari, M., Salama, S., Nofech-Mozes, S., Martel, A.L.: A cluster-then-label semi-supervised learning approach for pathology image classification. Sci. Rep. 8(1), 1–13 (2018)
Raczkowski, L., Mozejko, M., Zambonelli, J., Szczurek, E.: ARA: accurate, reliable and active histopathological image classification framework with Bayesian deep learning. Sci. Rep. 9(1), 1–12 (2019)
Shao, W., Sun, L., Zhang, D.: Deep active learning for nucleus classification in pathology images. In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), pp. 199–202. IEEE (2018)
Shaw, S., Pajak, M., Lisowska, A., Tsaftaris, S.A., O’Neil, A.Q.: Teacher-student chain for efficient semi-supervised histology image classification. arXiv preprint arXiv:2003.08797 (2020)
Ström, P., et al.: Pathologist-level grading of prostate biopsies with artificial intelligence. arXiv preprint arXiv:1907.01368 (2019)
Tellez, D., et al.: Quantifying the effects of data augmentation and stain color normalization in convolutional neural networks for computational pathology. Med. Image Anal. 58, 101544 (2019)
Thomee, B., et al.: The new data and new challenges in multimedia research. CoRR abs/1503.01817 (2015). http://arxiv.org/abs/1503.01817
Jimenez-del-Toro, O., Otálora, S., Atzori, M., Müller, H.: Deep multimodal case–based retrieval for large histopathology datasets. In: Wu, G., Munsell, B.C., Zhan, Y., Bai, W., Sanroma, G., Coupé, P. (eds.) Patch-MI 2017. LNCS, vol. 10530, pp. 149–157. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-67434-6_17
del Toro, O.J., et al.: Convolutional neural networks for an automatic classification of prostate tissue slides with high-grade Gleason score. In: Medical Imaging 2017: Digital Pathology, vol. 10140, p. 101400O. International Society for Optics and Photonics (2017)
Tsujikawa, T.: Robust cell detection and segmentation for image cytometry reveal Th17 cell heterogeneity. Cytom. Part A 95(4), 389–398 (2019)
Yalniz, I.Z., Jégou, H., Chen, K., Paluri, M., Mahajan, D.: Billion-scale semi-supervised learning for image classification. arXiv preprint arXiv:1905.00546 (2019)
Zhou, Z., Shin, J., Zhang, L., Gurudu, S., Gotway, M., Liang, J.: Fine-tuning convolutional neural networks for biomedical image analysis: actively and incrementally. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 7340–7351 (2017)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Marini, N., Otálora, S., Müller, H., Atzori, M. (2021). Semi-supervised Learning with a Teacher-Student Paradigm for Histopathology Classification: A Resource to Face Data Heterogeneity and Lack of Local Annotations. In: Del Bimbo, A., et al. Pattern Recognition. ICPR International Workshops and Challenges. ICPR 2021. Lecture Notes in Computer Science(), vol 12661. Springer, Cham. https://doi.org/10.1007/978-3-030-68763-2_9
Download citation
DOI: https://doi.org/10.1007/978-3-030-68763-2_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-68762-5
Online ISBN: 978-3-030-68763-2
eBook Packages: Computer ScienceComputer Science (R0)